Laccase Gene Family in Cerrena sp. HYB07: Sequences, Heterologous Expression and Transcriptional Analysis
Abstract
:1. Introduction
2. Results
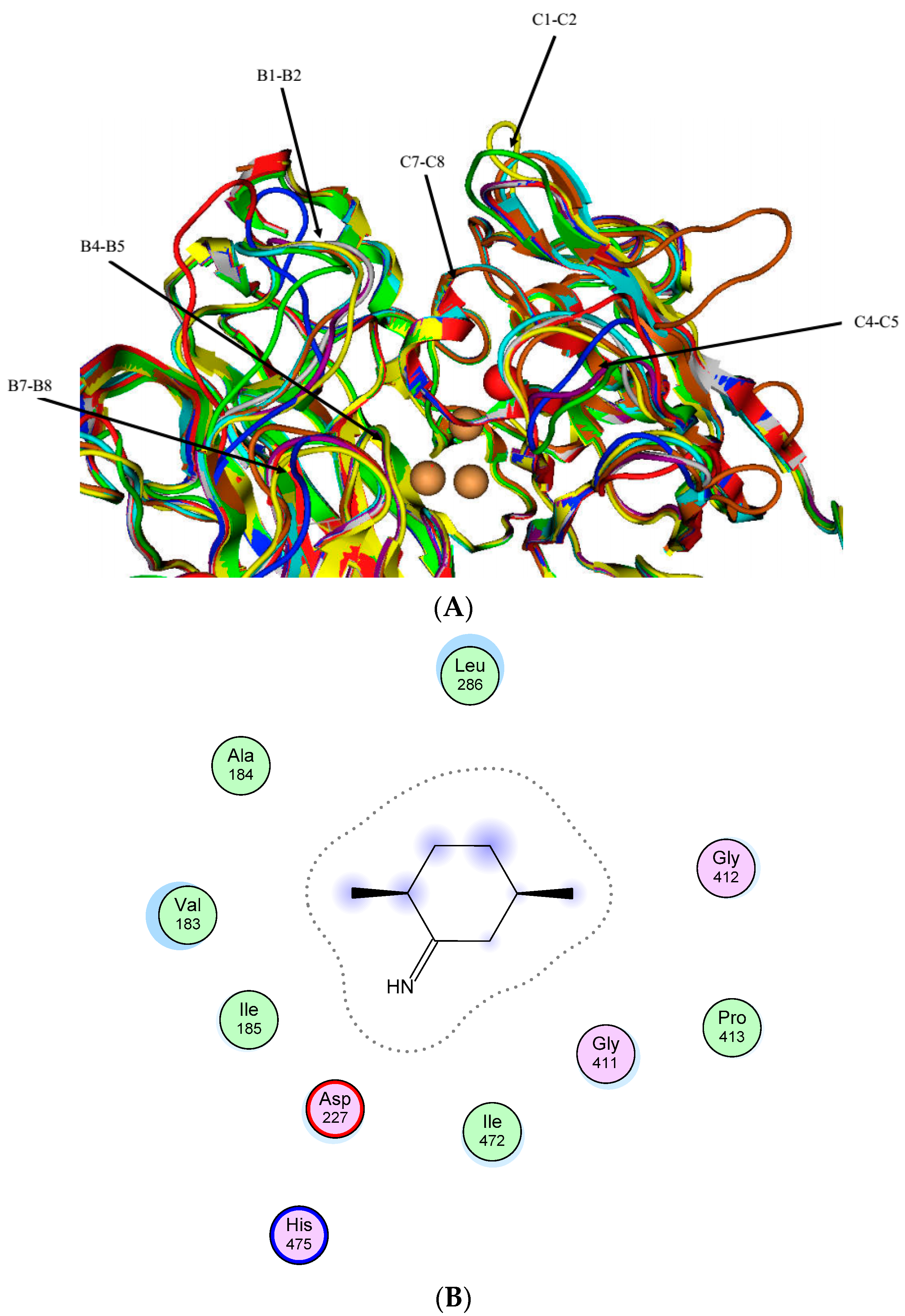
2.1. The Laccase Family of Cerrena sp. HYB07
2.2. Heterologous Expression of HYB07 Laccase Genes in P. pastoris
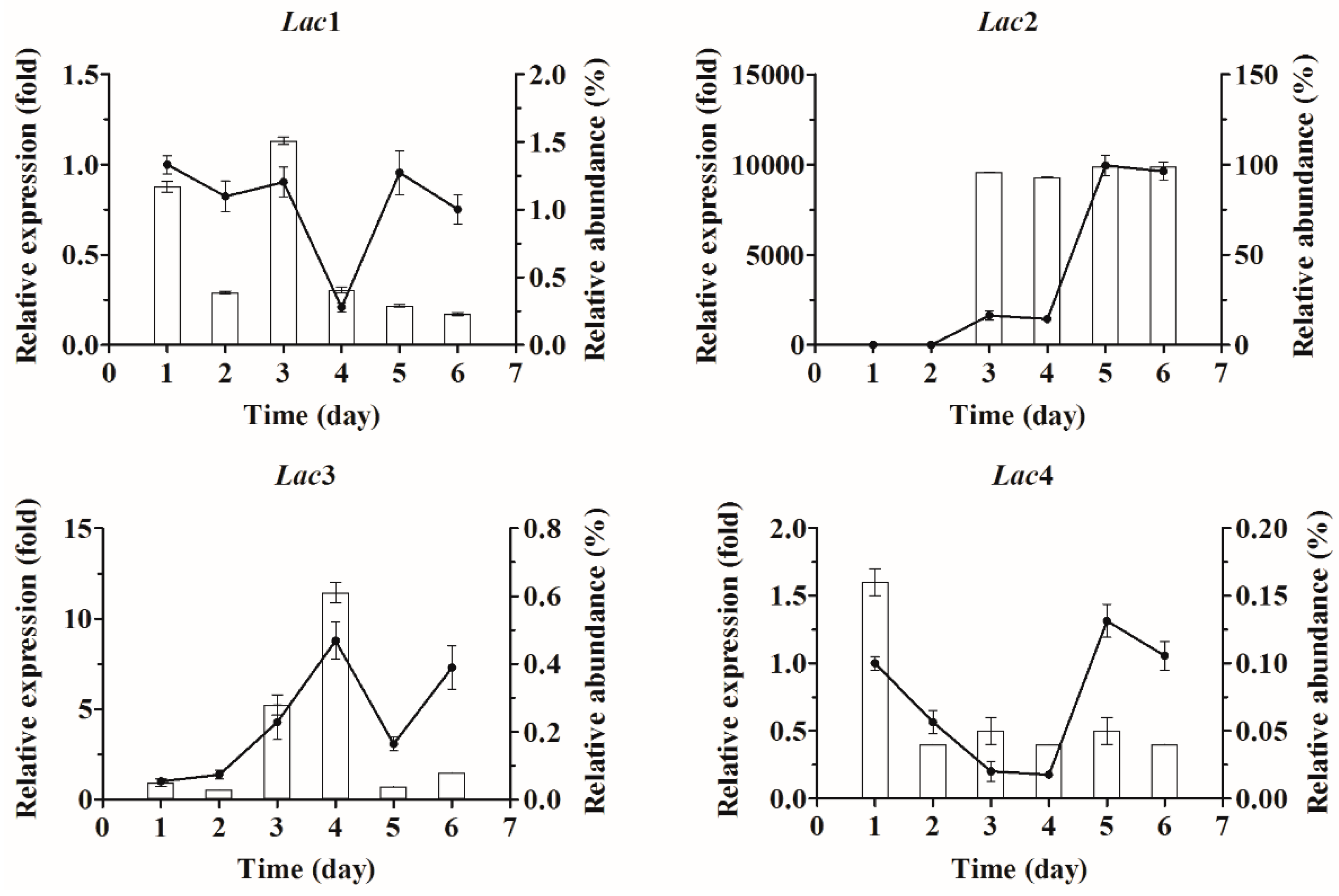
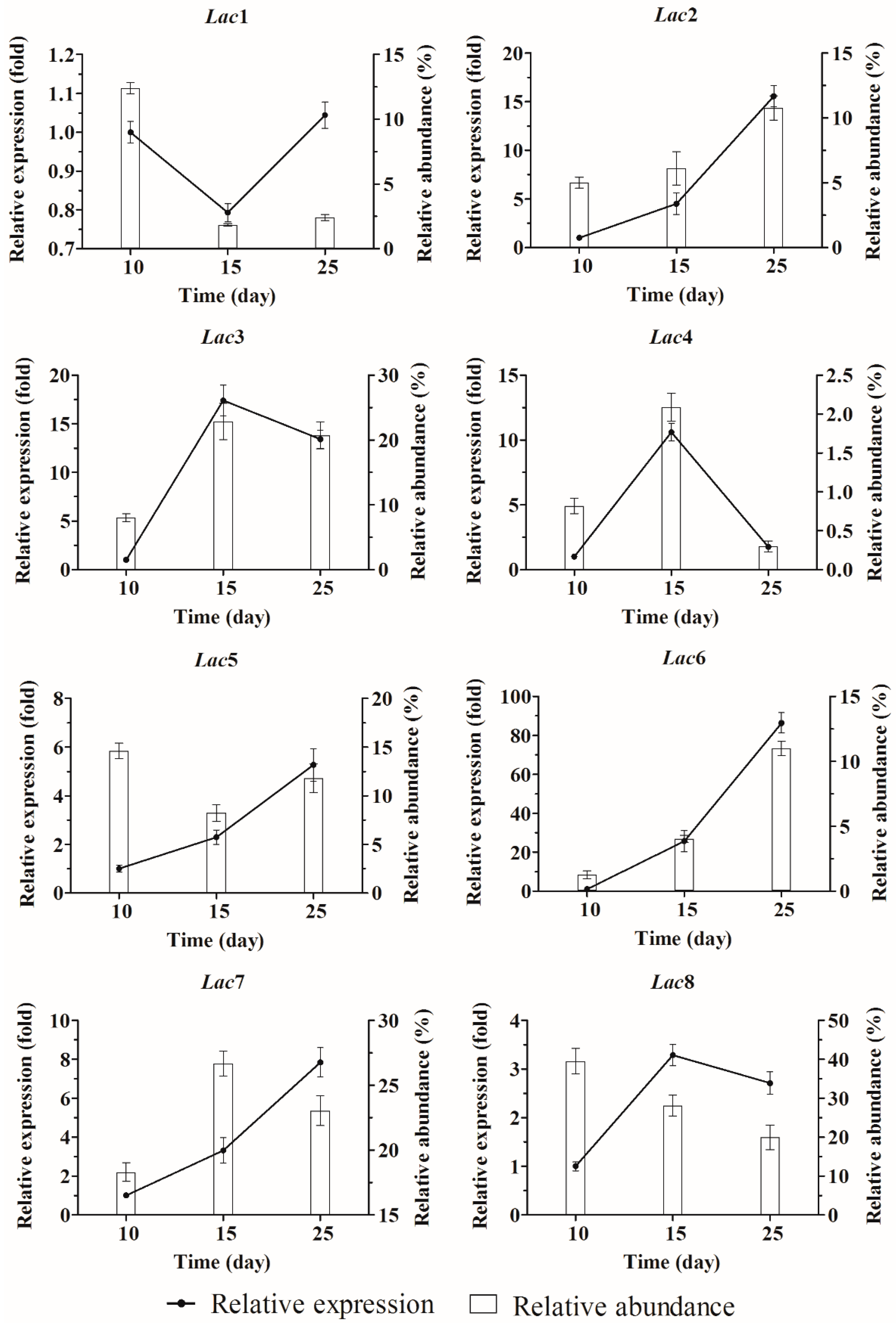
2.3. Quantification of Laccase Transcripts with qPCR
3. Discussion
4. Materials and Methods
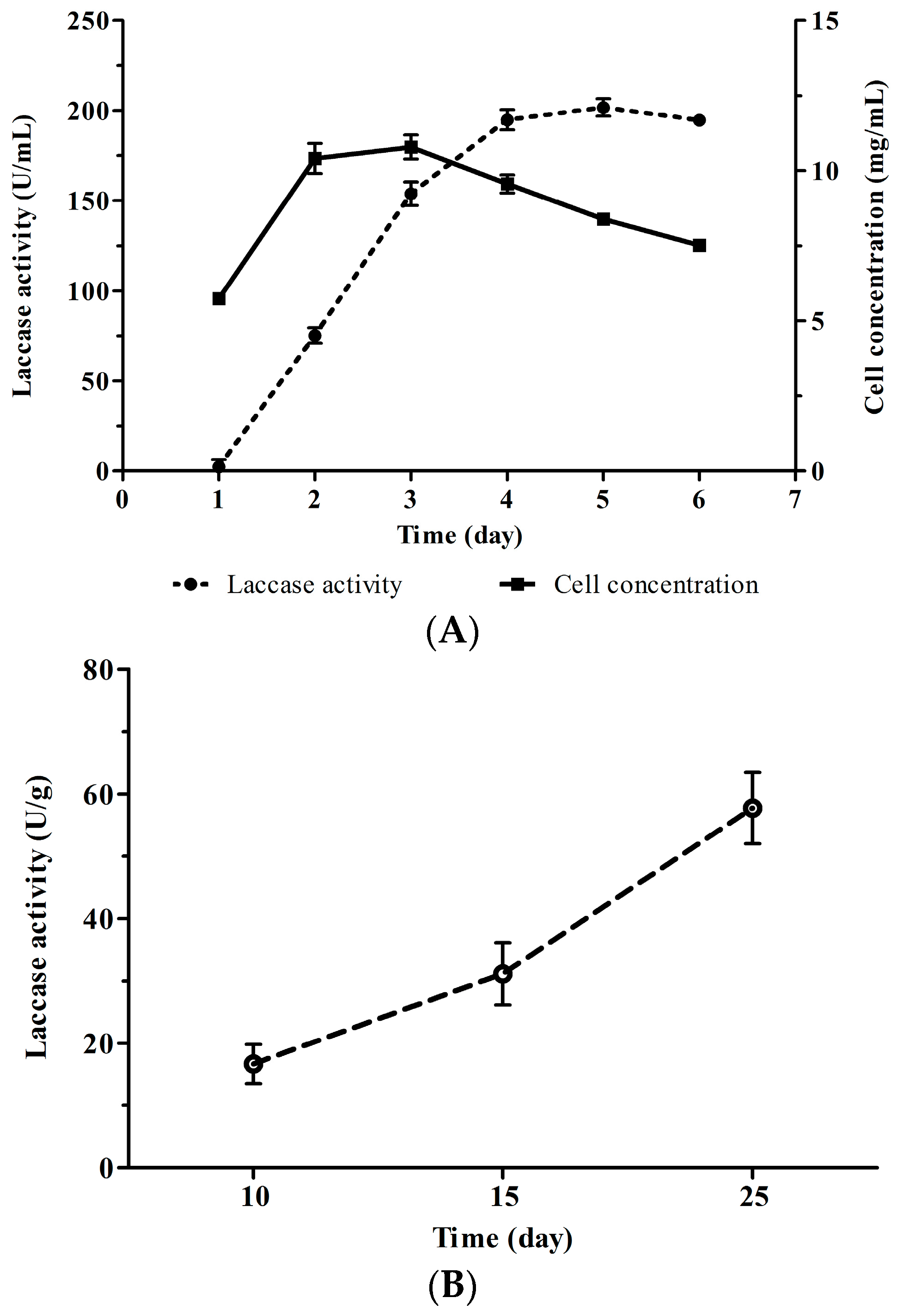
4.1. Fermentation of Cerrena sp. HYB07
4.2. Enzyme Activity Assay
4.3. Nucleic Acid Extraction
4.4. Cloning of Laccase Genes
4.5. Bioinformatics Analysis
4.6. Heterologous Expression of Laccase Genes in Pichia pastoris
4.7. Enzymatic Properties of Recombinant Laccases
4.8. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
4.9. Reference Gene Validation
4.10. LC-MS/MS Analysis of Secreted Laccases
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Arora, D.S.; Sharma, R.K. Ligninolytic fungal laccases and their biotechnological applications. Appl. Biochem. Biotechnol. 2010, 160, 1760–1788. [Google Scholar] [CrossRef]
- Couto, S.R.; Herrera, J.L.T. Industrial and biotechnological applications of laccases: A review. Biotechnol. Adv. 2006, 24, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2010, 67, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Kües, U.; Rühl, M. Multiple multi-copper oxidase gene families in basidiomycetes—What for? Curr. Genomics 2011, 12, 72–94. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhou, Y.; Xiao, Y.; Xu, Z.; Bian, Y. Cloning, expression and phylogenetic analysis of a divergent laccase multigene family in Auricularia auricula-judae. Microbiol. Res. 2014, 169, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.-R.; Wang, S.-S.; Zhu, M.-J.; Ning, Y.-J.; Wang, S.-N.; Li, B.; Yang, A.-Z.; Zhang, G.-Q.; Zhao, X.-M. An extracellular laccase with potent dye decolorizing ability from white rot fungus Trametes sp. LAC-01. Int. J. Biol. Macromol. 2015, 81, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhu, M.; Chen, X.; Wang, H.; Zhang, G. A novel laccase from fresh fruiting bodies of the wild medicinal mushroom Tricholoma matsutake. Acta Biochim. Pol. 2015, 62, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Zheng, F.; Long, L.; Wang, J.; Ding, S. Engineering the expression and characterization of two novel laccase isoenzymes from Coprinus comatus in Pichia pastoris by fusing an additional ten amino acids tag at N-terminus. PLoS ONE 2014, 9, e93912. [Google Scholar] [CrossRef] [PubMed]
- Castanera, R.; Péreza, G.; Omarini, A.; Alfaro, M.; Pisabarro, A.G.; Faraco, V.; Amore, A.; Ramíreza, L. Transcriptional and enzymatic profiling of Pleurotus ostreatus laccase genes in submerged and solid-state fermentation cultures. Appl. Environ. Microbiol. 2012, 78, 4037–4045. [Google Scholar] [CrossRef] [PubMed]
- Pezzella, C.; Lettera, V.; Piscitelli, A.; Giardina, P.; Sannia, G. Transcriptional analysis of Pleurotus ostreatus laccase genes. Appl. Microbiol. Biotechnol. 2012, 97, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Pezzella, C.; Autore, F.; Giardina, P.; Piscitelli, A.; Sannia, G.; Faraco, V. The Pleurotus ostreatus laccase multi-gene family: Isolation and heterologous expression of new family members. Curr. Genet. 2009, 55, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Vasina, D.V.; Mustafaev, O.N.; Moiseenko, K.V.; Sadovskaya, N.S.; Glazunova, O.A.; Tyurin, А.А.; Fedorova, T.V.; Pavlov, A.R.; Tyazhelova, T.V.; Goldenkova-Pavlova, I.V.; et al. The Trametes hirsuta 072 laccase multigene family: Genes identification and transcriptional analysis under copper ions induction. Biochimie 2015, 116, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Kilaru, S.; Hoegger, P.; Kües, U. The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr. Genet. 2006, 50, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, F.; Jiang, Y.; Wu, G.; Guo, L.; Chen, R.; Chen, B.; Lu, Y.; Dai, Y.; Xie, B. The multigene family of fungal laccases and their expression in the white rot basidiomycete Flammulina velutipes. Gene 2015, 563, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Courty, P.E.; Hoegger, P.J.; Kilaru, S.; Kohler, A.; Buée, M.; Garbaye, J.; Martin, F.; Kües, U. Phylogenetic analysis, genomic organization, and expression analysis of multi-copper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor. New. Phytol. 2009, 182, 736–750. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, Y.; Nakade, K.; Yoshida, K.; Natsume, S.; Miyazaki, K.; Sato, S.; van Peer, A.F.; Konno, N. Grouping of multicopper oxidases in Lentinula edodes by sequence similarities and expression patterns. AMB Express 2015, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bao, D.; Gong, M.; Zheng, H.; Chen, M.; Zhang, L.; Wang, H.; Jiang, J.; Wu, L.; Zhu, Y.; Zhu, G.; et al. Sequencing and comparative analysis of the straw mushroom (Volvariella volvacea) genome. PLoS ONE 2013, 8, e58294. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhu, G.; Chen, M.; Wang, H.; Bao, D. The bioinformatic analyses and the gene expression induced by Cu2+ of 11 laccase homologous genes from Volvariella volvacea. Mycosystema 2013, 33, 323–333. [Google Scholar]
- Kucharzyk, K.H.; Janusz, G.; Karczmarczyk, I.; Rogalski, J. Chemical modifications of laccase from white-rot basidiomycete Cerrena unicolor. Appl. Biochem. Biotechnol. 2012, 168, 1989–2003. [Google Scholar] [CrossRef] [PubMed]
- Lisova, Z.A.; Lisov, A.V.; Leontievsky, A.A. Two laccase isoforms of the basidiomycete Cerrena unicolor VKMF-Induction, isolation and properties. J. Basic Microbiol. 2010, 50, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Michniewicz, A.; Ullrich, R.; Ledakowicz, S.; Hofrichter, M. The white-rot fungus Cerrena unicolor strain 137 produces two laccase isoforms with different physico-chemical and catalytic properties. Appl. Microbiol. Biotechnol. 2006, 69, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Songulashvili, G.; Jimenéz-Tobón, G.A.; Jaspers, C.; Penninckx, M.J. Immobilized laccase of Cerrena unicolor for elimination of endocrine disruptor micropollutants. Fungal Biol. 2012, 116, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Elisashvili, V.; Kachlishvili, E.; Khardziani, T.; Agathos, S.N. Effect of aromatic compounds on the production of laccase and manganese peroxidase by white-rot basidiomycetes. J. Ind. Microbiol. Biotechnol. 2010, 37, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- D'Souza-Ticlo, D.; Sharma, D.; Raghukumar, C. A thermostable metal-tolerant laccase with bioremediation potential from a marine-derived fungus. Mar. Biotechnol. 2009, 11, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-C.; Wu, P.-H.; Su, Y.-C.; Wen, T.-N.; Wei, Y.-S.; Wang, N.-C.; Hsu, C.-A.; Wang, A.H.-J.; Shyur, L.-F. Biochemical characterization of a novel laccase from the basidiomycete fungus Cerrena sp. WR1. Protein Eng. Des. Sel. 2012, 25, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lin, Q.; Ng, T.B.; Ye, X.; Lin, J. Purification and characterization of a novel laccase from Cerrena sp. HYB07 with dye decolorizing ability. PLoS ONE 2014, 9, e110834. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ng, T.B.; Lin, J.; Ye, X. A novel laccase from basidiomycete Cerrena sp.: Cloning, heterologous expression, and characterization. Int. J. Biol. Macromol. 2015, 77, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Hakulinen, N.; Kiiskinen, L.-L.; Kruus, K.; Saloheimo, M.; Paananen, A.; Koivula, A.; Rouvinen, J. Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear copper site. Nat. Struct. Biol. 2002, 9, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Eggert, C.; LaFayette, P.R.; Temp, U.; Eriksson, K.-E.L.; Dean, J.F.D. Molecular analysis of a laccase gene from the white rot fungus Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 1998, 64, 1766–1772. [Google Scholar] [PubMed]
- Xu, F.; Berka, R.M.; Wahleithner, J.A.; Nelson, B.A.; Shuster, J.R.; Brown, S.H.; Palmer, A.E.; Solomon, E.I. Site-directed mutations in fungal laccase: Effect on redox potential, activity and pH profile. Biochem. J. 1998, 334, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Piontek, K.; Antorini, M.; Choinowski, T. Crystal structure of a laccase from the fungus Trametes versicolor at 1.90-Å resolution containing a full complement of coppers. J. Biol. Chem. 2002, 277, 37663–37669. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, B.; Martínez-Morales, F.; Tinoco-Valencia, R.; Rojas, S.; Acosta-Urdapilleta, L.; Trejo-Hernández, M.R. Biochemical and molecular characterization of laccase isoforms produced by the white-rot fungus Trametes versicolor under submerged culture conditions. J. Mol. Catal. B Enzym. 2015, 122, 339–347. [Google Scholar] [CrossRef]
- Bertrand, T.; Jolivalt, C.; Briozzo, P.; Caminade, E.; Joly, N.; Madzak, C.; Mougin, C. Crystal structure of a four-copper laccase complexed with an arylamine: Insights into substrate recognition and correlation with kinetics. Biochemistry (Mosc) 2002, 41, 7325–7333. [Google Scholar] [CrossRef]
- Piscitelli, A.; Pezzella, C.; Giardina, P.; Faraco, V.; Sannia, G. Heterologous laccase production and its role in industrial applications. Bioeng. Bugs 2010, 1, 252. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, R.; Ma, L.; Fan, F.; Gong, Y.; Wan, X.; Jiang, M.; Zhang, X.; Yang, Y. Decolorization of different dyes by a newly isolated white-rot fungi strain Ganoderma sp. En3 and cloning and functional analysis of its laccase gene. J. Hazard. Mater. 2011, 192, 855–873. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.-Z.; Zhou, H.-M.; Tu, X.-M.; Li, J.-F.; Xiao, Y.-Z. Cloning of a laccase gene from a novel basidiomycete Trametes sp. 420 and its heterologous expression in Pichia pastoris. Curr. Microbiol. 2007, 54, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Si, J.; Cui, B.-K. Study of the physiological characteristics of the medicinal mushroom Trametes pubescens (higher basidiomycetes) during the laccase-producing process. Int. J. Med. Mushrooms 2013, 15, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Rühl, M.; Majcherczyk, A.; Kües, U. Lcc1 and Lcc5 are the main laccases secreted in liquid cultures of Coprinopsis cinerea strains. Antonie van Leeuwenhoek 2013, 103, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Bourbonnais, R.; Paice, M.G. Demethylation and delignification of kraft pulp by Trametes versicolor laccase in the presence of 2,2′-azinobis-(3-ethylbenzthiazoline-6-sulphonate). Appl. Microbiol. Biotechnol. 1992, 36, 823–827. [Google Scholar] [CrossRef]
- D’Souza, T.M.; Boominathan, K.; Reddy, C.A. Isolation of laccase gene-specific sequences from white rot and brown rot fungi by PCR. Appl. Environ. Microbiol. 1996, 62, 3739–3744. [Google Scholar] [PubMed]
- Liu, Z.; Sun, X.; Qu, Y. Cloning Cellobiohydrolase I from Penicillium decumbens 114–2 with TAIL-PCR and comparing with its derepressed mutant JU-A10. Acta Microbiol. Sin. 2008, 48, 667–671. [Google Scholar]
- Liu, Y.-G.; Whittier, R.F. Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 1995, 25, 674–681. [Google Scholar] [CrossRef]
- BLAST. Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 3 August 2016).
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved prediction of signal peptides: SignalP 3. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- SOPMA Secondary Structure Prediction Method. Available online: http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html (accessed on 3 August 2016).
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.D.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1–research0034.11. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Q.; Dai, J.; Wu, Y.; Tang, H.; Zeng, R. BuildSummary: Using a group-based approach to improve the sensitivity of peptide/protein identification in shotgun proteomics. J. Proteome Res. 2012, 11, 1494–1502. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are not available from the authors.

| Laccase Gene | GenBank # | DNA Length (bp) a | Intron # | Mature Protein (aa) | Signal Peptide (aa) | MW (kD) | pI | Glycosylation Sites |
|---|---|---|---|---|---|---|---|---|
| Lac1 | KC540913 | 2491 | 11 | 497 | 20 | 54.1 | 5.5 | 1 |
| Lac2 | KF317944 | 2212 | 11 | 505 | 21 | 54.7 | 5.2 | 1 |
| Lac3 | KF317945 | 1892 | 6 | 497 | 21 | 53.1 | 4.4 | 5 |
| Lac4 | KF317946 | 2170 | 11 | 496 | 20 | 53.6 | 5.4 | 0 |
| Lac5 | KF317947 | 2199 | 11 | 504 | 21 | 55.4 | 5.3 | 1 |
| Lac6 | KF317943 | 2179 | 10 | 524 | 18 | 57.9 | 5.5 | 6 |
| Lac7 | KF317949 | 2168 | 11 | 495 | 21 | 52.9 | 5.6 | 1 |
| Lac8 | KF317948 | 2181 | 11 | 498 | 21 | 53.7 | 5.0 | 4 |
| Laccase | Lac1 | Lac2 | Lac3 | Lac4 | Lac5 | Lac6 | Lac7 | Lac8 | 193382 | 310741 | 357631 | 364416 | 383410 | 390832 | 390880 | 395909 | 408157 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lac1 | 69.5 | 66.0 | 69.3 | 65.4 | 60.3 | 76.3 | 71.6 | 71.1 | 72.4 | 63.8 | 65.8 | 69.4 | 67.8 | 52.7 | 85.1 | 65.7 | |
| Lac2 | 63.8 | 75.3 | 72.5 | 61.5 | 73.0 | 67.3 | 68.4 | 69.8 | 62.0 | 88.4 | 64.8 | 74.4 | 62.5 | 68.2 | 73.8 | ||
| Lac3 | 63.7 | 58.5 | 56.6 | 68.5 | 66.7 | 66.5 | 63.3 | 88.8 | 61.5 | 61.7 | 63.0 | 46.9 | 64.9 | 58.4 | |||
| Lac4 | 68.9 | 60.9 | 72.2 | 67.8 | 69.2 | 71.4 | 62.8 | 72.3 | 67.6 | 92.1 | 57.0 | 69.7 | 68.1 | ||||
| Lac5 | 59.8 | 68.6 | 64.6 | 64.1 | 65.1 | 57.8 | 71.2 | 61.9 | 67.3 | 59.0 | 63.8 | 92.8 | |||||
| Lac6 | 60.4 | 58.9 | 55.8 | 56.5 | 53.7 | 60.0 | 54.5 | 56.7 | 47.0 | 55.1 | 57.9 | ||||||
| Lac7 | 76.7 | 82.5 | 78.1 | 66.5 | 69.6 | 75.2 | 72.2 | 54.0 | 73.8 | 68.1 | |||||||
| Lac8 | 73.2 | 71.5 | 64.5 | 64.5 | 70.6 | 67.2 | 50.9 | 68.8 | 65.0 | ||||||||
| 193382 | 75.8 | 64.5 | 68.2 | 71.7 | 69.9 | 50.6 | 71.7 | 65.3 | |||||||||
| 310741 | 62.3 | 68.3 | 73.9 | 71.2 | 54.0 | 72.8 | 66.3 | ||||||||||
| 357631 | 60.4 | 61.7 | 63.9 | 46.5 | 63.6 | 58.6 | |||||||||||
| 364416 | 63.8 | 74.1 | 61.9 | 66.5 | 72.5 | ||||||||||||
| 383410 | 66.0 | 51.5 | 69.0 | 63.3 | |||||||||||||
| 390832 | 57.8 | 69.9 | 67.9 | ||||||||||||||
| 390880 | 53.3 | 60.6 | |||||||||||||||
| 395909 | 65.2 | ||||||||||||||||
| 408157 |
| Laccase | Laccase Yield (U/mL) | Medium | Cu2+ Concentration | Production Cycle (days) | Optimum pH | Optimum Temp (°C) | Reference |
|---|---|---|---|---|---|---|---|
| rLac1 | 6.3 | YP | 0.4 | 9 | 3.5 | 55 | [27] |
| rLac3 | 0.2 | YP | 0.5 | 6 | NA | NA | This study |
| rLac6 | 2.9 | YP | 1.0 | 16 | 3.0 | 55 | This study |
| rLac7 | 0.5 | BMM | 0.4 | 10 | NA | NA | This study |
| rLac8 | 0.7 | YP | 0.5 | 7 | NA | NA | This study |
| Laccase | Score | Coverage | Unique Peptides | Total Peptides | PSMs |
|---|---|---|---|---|---|
| Lac2 | 66.67 | 27.72 | 9 | 9 | 22 |
| Lac6 | 2.55 | 2.86 | 1 | 1 | 1 |
| Lac7 | 676.22 | 41.82 | 13 | 13 | 205 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Xu, X.; Ng, T.B.; Lin, J.; Ye, X. Laccase Gene Family in Cerrena sp. HYB07: Sequences, Heterologous Expression and Transcriptional Analysis. Molecules 2016, 21, 1017. https://doi.org/10.3390/molecules21081017
Yang J, Xu X, Ng TB, Lin J, Ye X. Laccase Gene Family in Cerrena sp. HYB07: Sequences, Heterologous Expression and Transcriptional Analysis. Molecules. 2016; 21(8):1017. https://doi.org/10.3390/molecules21081017
Chicago/Turabian StyleYang, Jie, Xinqi Xu, Tzi Bun Ng, Juan Lin, and Xiuyun Ye. 2016. "Laccase Gene Family in Cerrena sp. HYB07: Sequences, Heterologous Expression and Transcriptional Analysis" Molecules 21, no. 8: 1017. https://doi.org/10.3390/molecules21081017
APA StyleYang, J., Xu, X., Ng, T. B., Lin, J., & Ye, X. (2016). Laccase Gene Family in Cerrena sp. HYB07: Sequences, Heterologous Expression and Transcriptional Analysis. Molecules, 21(8), 1017. https://doi.org/10.3390/molecules21081017






