A Novel Biomolecule-Mediated Reduction of Graphene Oxide: A Multifunctional Anti-Cancer Agent
Abstract
:1. Introduction
2. Results and Discussion
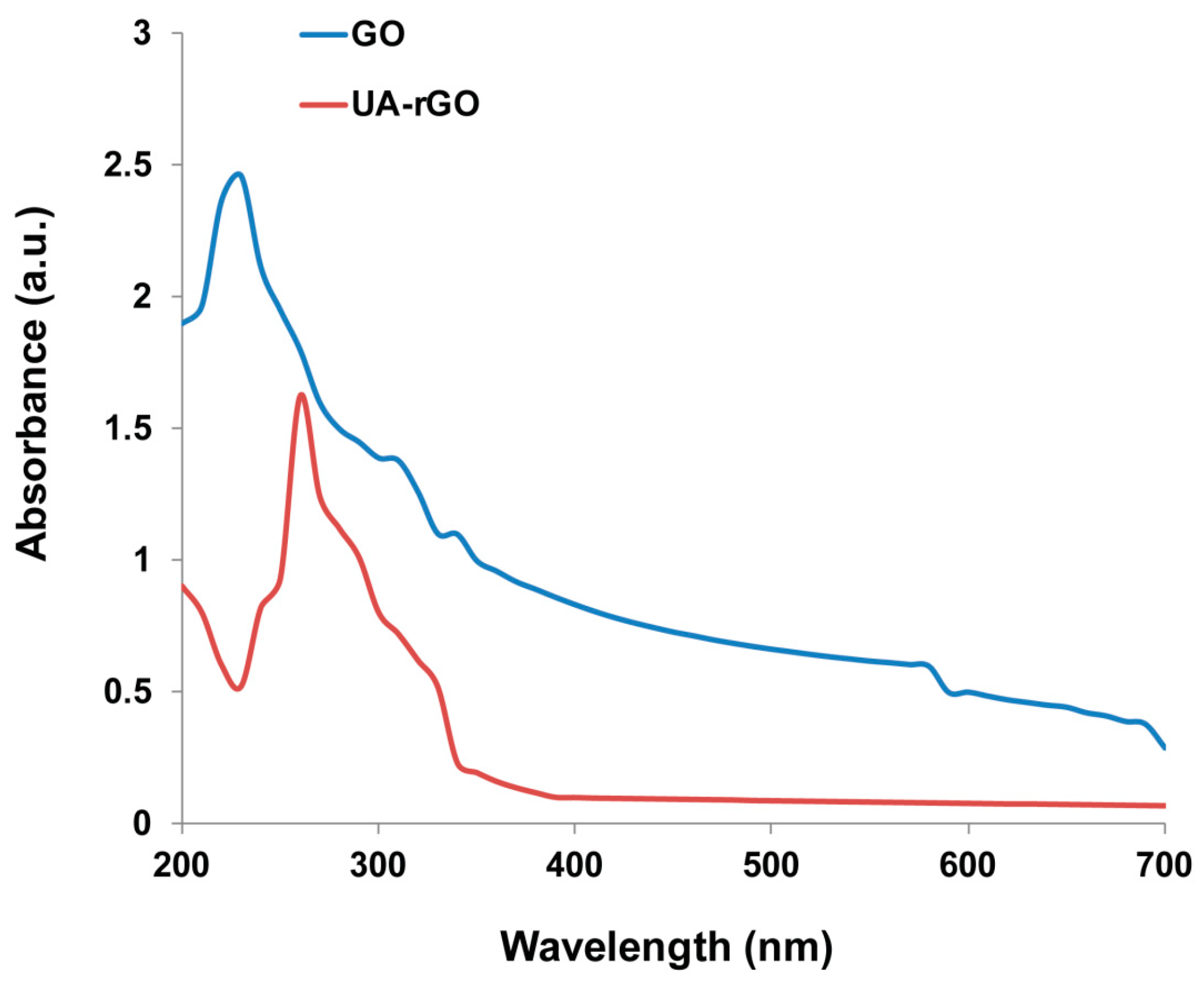
2.1. Synthesis and Characterization of UA-rGO by UV-Vis Spectroscopy
2.2. X-ray Diffraction (XRD) Analysis of GO and UA-rGO
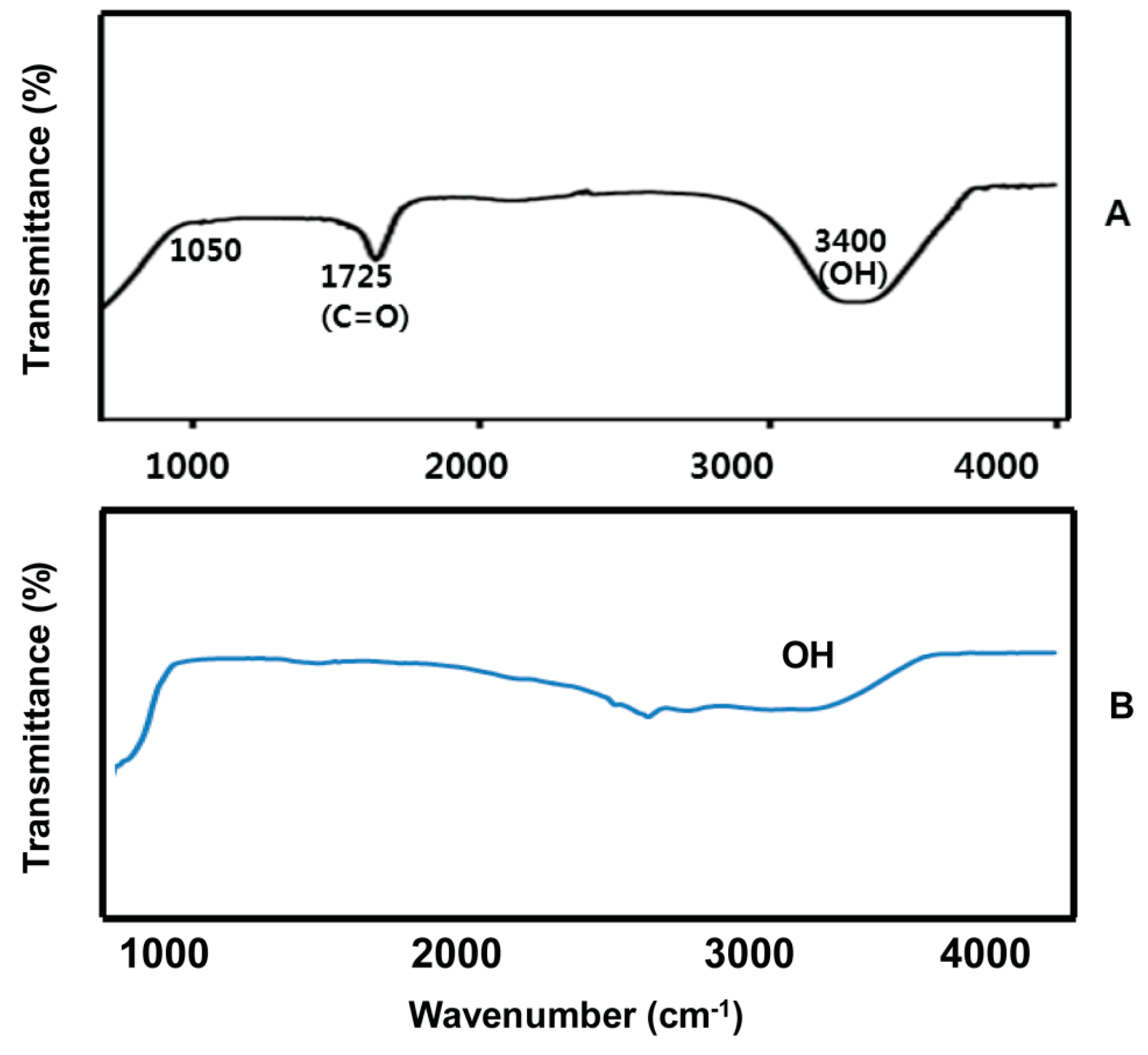
2.3. FTIR Spectra of GO and UA-rGO
2.4. Size Analysis of GO and UA-rGO by Dynamic Light Scattering (DLS)
2.5. Surface Analysis of GO and UA-rGO
2.6. Raman Spectroscopy Analysis of GO and UA-rGO
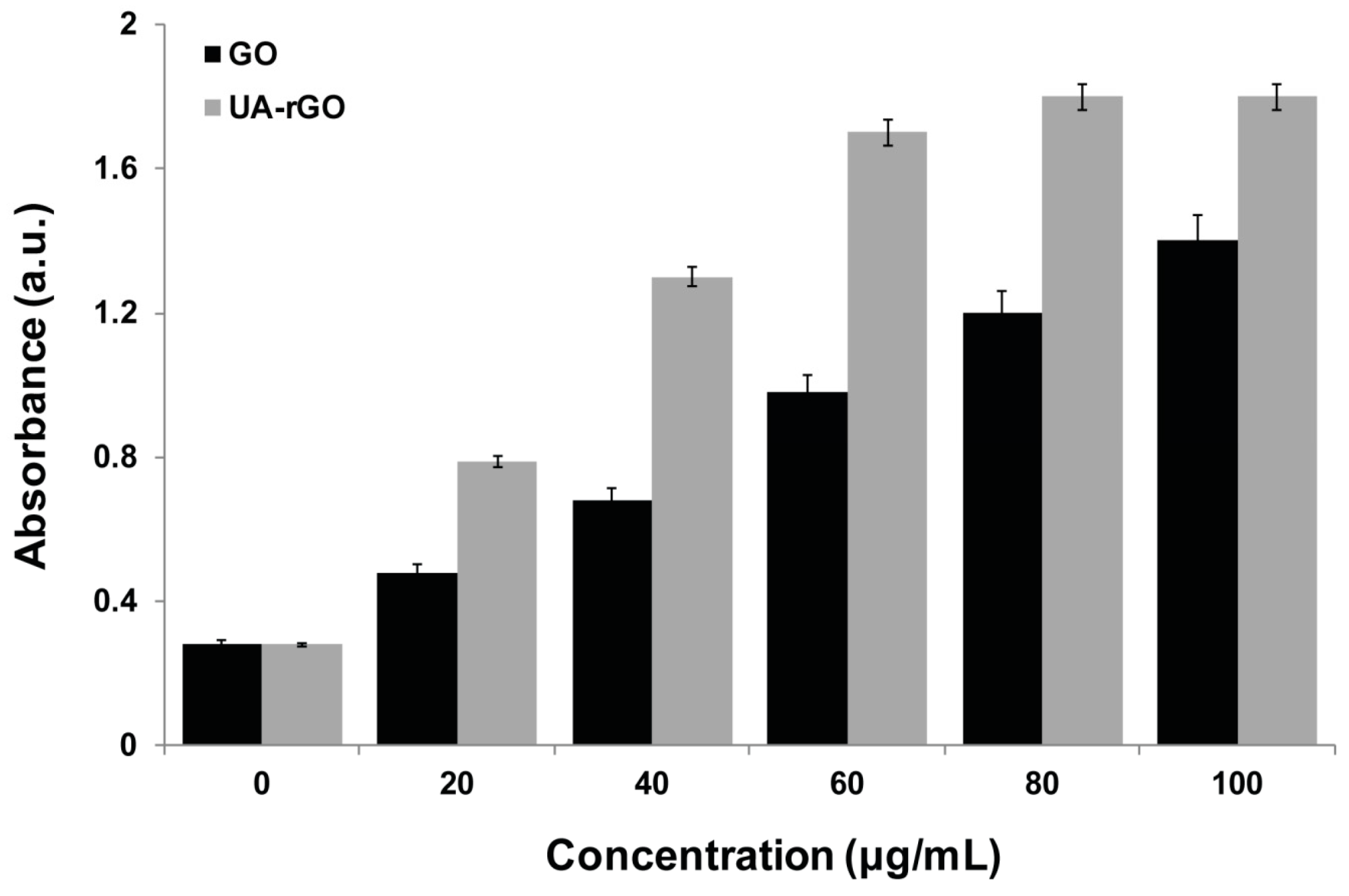
2.7. UA-rGO Inhibits Cell Viability of Ovarian Cancer Cells
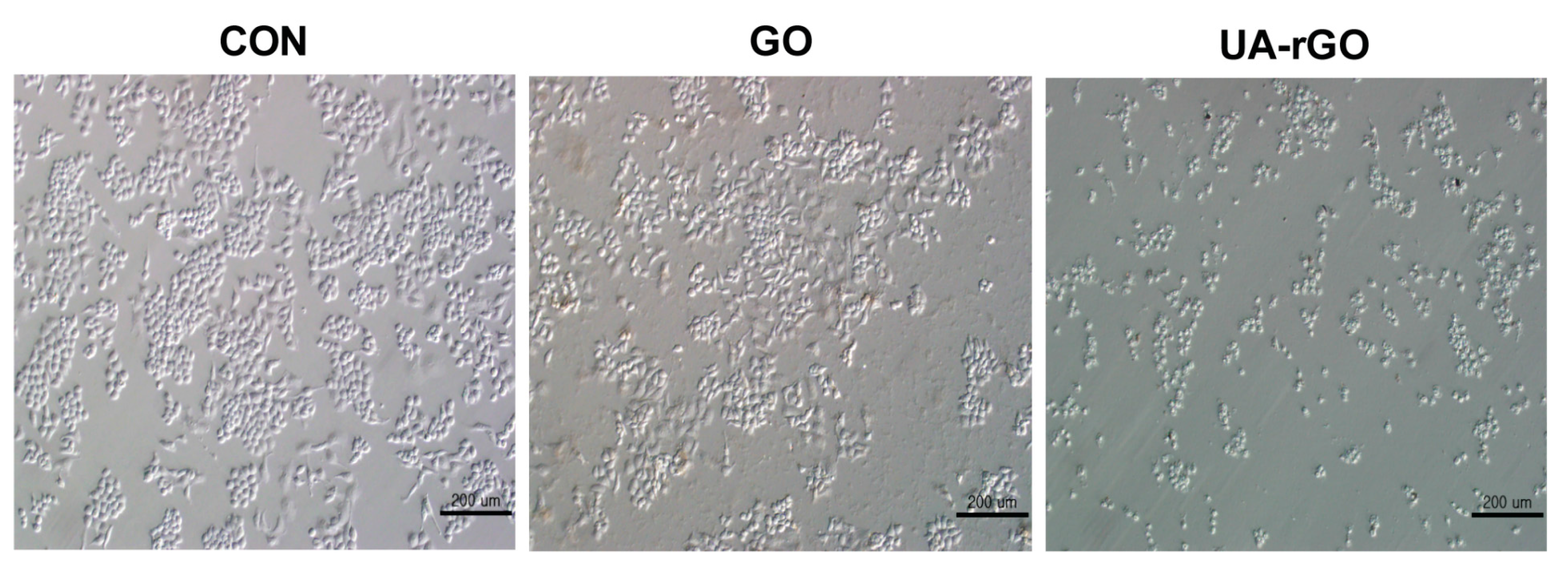
2.8. Effect of UA-rGO on Cell Morphology
2.9. Effect of UA-rGO on Membrane Integrity
2.10. UA-rGO Induces ROS Generation
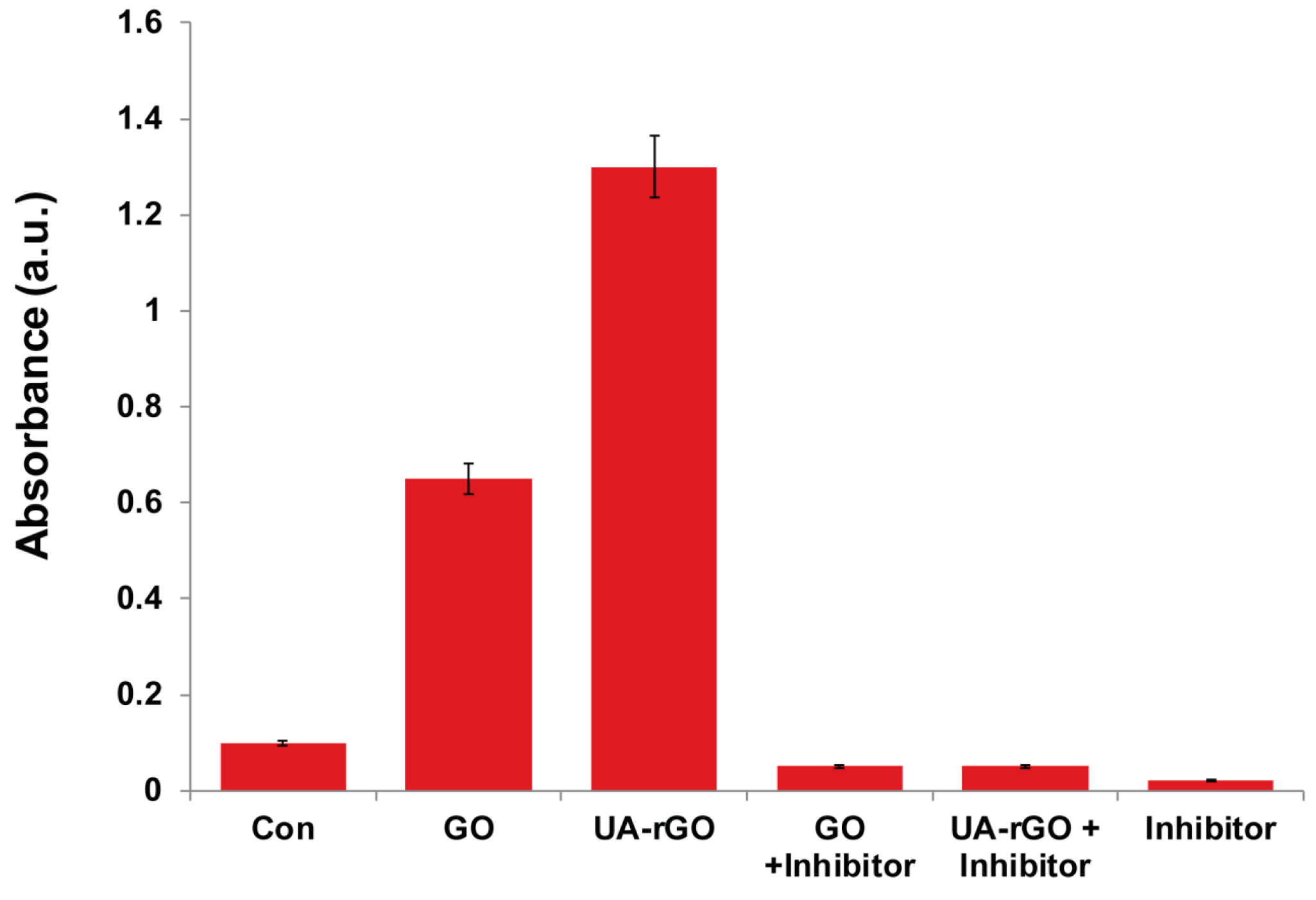
2.11. UA-rGO Activates Caspase-3 Activity
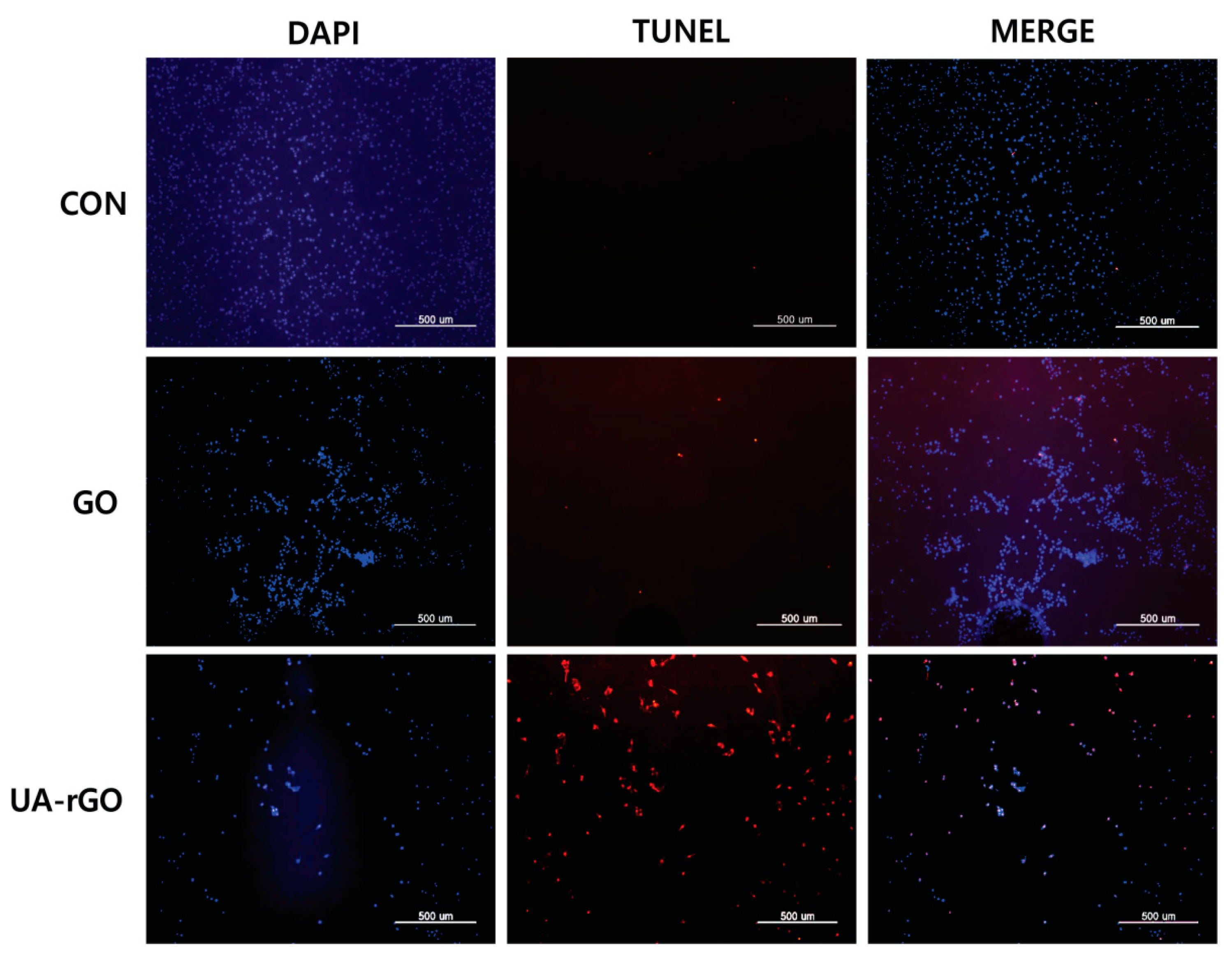
2.12. UA-rGO Activates DNA Fragmentation
3. Materials and Methods
3.1. Materials
3.2. GO Synthesis
3.3. Preparation of UA-rGO
3.4. Characterization of GO and UA-rGO
3.5. Cell Culture and Exposure to GO and UA-rGO
3.6. Cell Viability Assay
3.7. Cell Morphology
3.8. Membrane Integrity
3.9. Determination of ROS
3.10. Measurement of Caspase-3 Activity
3.11. TUNEL Assay
3.12. Statistical Analyses
4. Conclusions
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Novoselov, K.S.; Geim, A.K.; Morozov, S.V.; Jiang, D.; Zhang, Y.; Dubonos, S.V.; Grigorieva, I.V.; Firsov, A.A. Electric field effect in atomically thin carbon films. Science 2004, 306, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, D.R.; Park, S.; Bielawski, C.W.; Ruoff, R.S. The chemistry of graphene oxide. Chem Soc. Rev. 2010, 39, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Compton, O.C.; Nguyen, S.T. Graphene oxide, highly reduced graphene oxide, and graphene: Versatile building blocks for carbon-based materials. Small 2010, 6, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.Z.; Guo, S.J.; Fang, Y.X.; Dong, S.J. Reducing sugar: New functional molecules for the green synthesis of graphene nanosheets. ACS Nano 2010, 4, 2429–2437. [Google Scholar] [CrossRef] [PubMed]
- Balandin, A.A.; Ghosh, S.; Bao, W.; Calizo, I.; Teweldebrhan, D.; Miao, F.; Lau, C.N. Superior thermal conductivity of single-layer graphene. Nano Lett. 2008, 8, 902–907. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.N.; Sood, A.K.; Subrahmanyam, K.S.; Govindaraj, A. Graphene: The new two-dimensional nanomaterial. Angew. Chem. Int. Ed. Engl. 2009, 48, 7752–7777. [Google Scholar] [CrossRef] [PubMed]
- Emtsev, K.V.; Bostwick, A.; Horn, K.; Jobst, J.; Kellogg, G.L.; Ley, L.; McChesney, J.L.; Ohta, T.; Reshanov, S.A.; Rohrl, J.; et al. Towards wafer-size graphene layers by atmospheric pressure graphitization of silicon carbide. Nat. Mater. 2009, 8, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.Y.; Laurent, S.; Chen, W.; Akhavan, O.; Imani, M.; Ashkarran, A.A.; Mahmoudi, M. Graphene: Promises, facts, opportunities, and challenges in nanomedicine. Chem Rev. 2013, 113, 3407–3424. [Google Scholar] [CrossRef] [PubMed]
- Stankovich, S.; Piner, R.D.; Chen, X.Q.; Wu, N.Q.; Nguyen, S.T.; Ruoff, R.S. Stable aqueous dispersions of graphitic nanoplatelets via the reduction of exfoliated graphite oxide in the presence of poly(sodium 4-styrenesulfonate). J. Mater. Chem. 2006, 16, 155–158. [Google Scholar] [CrossRef]
- Kim, H.; Abdala, A.A.; Macosko, C.W. Graphene/polymer nanocomposites. Macromolecules 2010, 43, 6515–6530. [Google Scholar] [CrossRef]
- Fernandez-Merino, M.J.; Guardia, L.; Paredes, J.I.; Villar-Rodil, S.; Solis-Fernandez, P.; Martinez-Alonso, A.; Tascon, J.M.D. Vitamin C is an ideal substitute for hydrazine in the reduction of graphene oxide suspensions. J. Phys. Chem. C 2010, 114, 6426–6432. [Google Scholar] [CrossRef]
- Gao, J.; Liu, F.; Liu, Y.L.; Ma, N.; Wang, Z.Q.; Zhang, X. Environment-friendly method to produce graphene that employs vitamin C and amino acid. Chem. Mater. 2010, 22, 2213–2218. [Google Scholar] [CrossRef]
- Akhavan, O.; Ghaderi, E.; Aghayee, S.; Fereydooni, Y.; Talebi, A. The use of a glucose-reduced graphene oxide suspension for photothermal cancer therapy. J. Mater. Chem. 2012, 22, 13773–13781. [Google Scholar] [CrossRef]
- Liu, J.B.; Fu, S.H.; Yuan, B.; Li, Y.L.; Deng, Z.X. Toward a universal “adhesive nanosheet” for the assembly of multiple nanoparticles based on a protein-induced reduction/decoration of graphene oxide. J. Am. Chem. Soc. 2010, 132, 7279–7281. [Google Scholar] [CrossRef] [PubMed]
- Esfandiar, A.; Akhavan, O.; Irajizad, A. Melatonin as a powerful bio-antioxidant for reduction of graphene oxide. J. Mater. Chem. 2011, 21, 10907–10914. [Google Scholar] [CrossRef]
- Gurunathan, S.; Han, J.; Kim, J.H. Humanin: A novel functional molecule for the green synthesis of graphene. Colloid Surface B 2013, 111, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Kim, E.; Kwon, D.N.; Park, J.K.; Kim, J.H. Enhanced green fluorescent protein-mediated synthesis of biocompatible graphene. J. Nanobiotechnol. 2014, 12. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Kim, E.S.; Park, J.H.; Kim, J.H. Reduction of graphene oxide by resveratrol: A novel and simple biological method for the synthesis of an effective anticancer nanotherapeutic molecule. Int. J. Nanomed. 2015, 10, 2951–2969. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, G.; Vila, M.; Portoles, M.T.; Vallet-Regi, M.; Gracio, J.; Marques, P.A.A.P. Nano-graphene oxide: A potential multifunctional platform for cancer therapy. Adv. Healthc. Mater. 2013, 2, 1072–1090. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, Y.J.; Tan, X.F.; Peng, R.; Liu, Z. Behavior and toxicity of graphene and its functionalized derivatives in biological systems. Small 2013, 9, 1492–1503. [Google Scholar] [CrossRef] [PubMed]
- Akhavan, O.; Ghaderi, E. Toxicity of graphene and graphene oxide nanowalls against bacteria. ACS Nano 2010, 4, 5731–5736. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.B.; Peng, C.; Luo, W.J.; Lv, M.; Li, X.M.; Li, D.; Huang, Q.; Fan, C.H. Graphene-based antibacterial paper. ACS Nano 2010, 4, 4317–4323. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.B.; Zeng, T.H.; Hofmann, M.; Burcombe, E.; Wei, J.; Jiang, R.R.; Kong, J.; Chen, Y. Antibacterial activity of graphite, graphite oxide, graphene oxide, and reduced graphene oxide: Membrane and oxidative stress. ACS Nano 2011, 5, 6971–6980. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Dayem, A.A.; Eppakayala, V.; Kim, J.H. Oxidative stress-mediated antibacterial activity of graphene oxide and reduced graphene oxide in Pseudomonas aeruginosa. Int. J. Nanomed. 2012, 7, 5901–5914. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Dayem, A.A.; Eppakayala, V.; Park, M.R.; Kwon, D.N.; Kim, J.H. Antibacterial activity of dithiothreitol reduced graphene oxide. J. Ind. Eng. Chem. 2013, 19, 1280–1288. [Google Scholar] [CrossRef]
- Chang, Y.L.; Yang, S.T.; Liu, J.H.; Dong, E.; Wang, Y.W.; Cao, A.N.; Liu, Y.F.; Wang, H.F. In vitro toxicity evaluation of graphene oxide on A549 cells. Toxicol. Lett. 2011, 200, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Hu, W.B.; Li, J.; Tao, L.; Wei, Y. A comparative study of cellular uptake and cytotoxicity of multi-walled carbon nanotubes, graphene oxide, and nanodiamond. Toxicol. Res. 2012, 1, 62–68. [Google Scholar] [CrossRef]
- Gurunathan, S.; Han, J.W.; Eppakayala, V.; Kim, J.H. Green synthesis of graphene and its cytotoxic effects in human breast cancer cells. Int. J. Nanomed. 2013, 8, 1015–1027. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.; Park, J.H.; Kim, J.H. An in vitro evaluation of graphene oxide reduced by Ganoderma spp. In human breast cancer cells (MDA-MB-231). Int. J. Nanomed. 2014, 9, 1783–1797. [Google Scholar] [CrossRef] [PubMed]
- Akhavan, O.; Ghaderi, E.; Akhavan, A. Size-dependent genotoxicity of graphene nanoplatelets in human stem cells. Biomaterials 2012, 33, 8017–8025. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.M.; Qian, F.; Saltikov, C.; Jiao, Y.Q.; Li, Y. Microbial reduction of graphene oxide by shewanella. Nano Res. 2011, 4, 563–570. [Google Scholar] [CrossRef]
- Yang, K.; Zhang, S.; Zhang, G.; Sun, X.; Lee, S.T.; Liu, Z. Graphene in mice: Ultrahigh in vivo tumor uptake and efficient photothermal therapy. Nano Lett 2010, 10, 3318–3323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Guo, Z.Y.; Huang, D.Q.; Liu, Z.M.; Guo, X.; Zhong, H.Q. Synergistic effect of chemo-photothermal therapy using pegylated graphene oxide. Biomaterials 2011, 32, 8555–8561. [Google Scholar] [CrossRef] [PubMed]
- Kurman, R.J.; Shih, I.M. The origin and pathogenesis of epithelial ovarian cancer: A proposed unifying theory. Am. J. Surg. Pathol. 2010, 34, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Park, J.H.; Kim, E.; Choi, Y.J.; Kwon, D.N.; Kim, J.H. Reduced graphene oxide-silver nanoparticle nanocomposite: A potential anticancer nanotherapy. Int. J. Nanomed. 2015, 10, 6257–6276. [Google Scholar] [CrossRef] [PubMed]
- Sautin, Y.Y.; Johnson, R.J. Uric acid: The oxidant-antioxidant paradox. Nucleosides Nucleotides Nucl. 2008, 27, 608–619. [Google Scholar] [CrossRef] [PubMed]
- Amaro, S.; Soy, D.; Obach, V.; Cervera, A.; Planas, A.M.; Chamorro, A. A pilot study of dual treatment with recombinant tissue plasminogen activator and uric acid in acute ischemic stroke. Stroke 2007, 38, 2173–2175. [Google Scholar] [CrossRef] [PubMed]
- Titelman, G.I.; Gelman, V.; Bron, S.; Khalfin, R.L.; Cohen, Y.; Bianco-Peled, H. Characteristics and microstructure of aqueous colloidal dispersions of graphite oxide. Carbon 2005, 43, 641–649. [Google Scholar] [CrossRef]
- Szabo, T.; Tombacz, E.; Illes, E.; Dekany, I. Enhanced acidity and pH-dependent surface charge characterization of successively oxidized graphite oxides. Carbon 2006, 44, 537–545. [Google Scholar] [CrossRef]
- Thema, F.T.; Moloto, M.J.; Dikio, E.D.; Nyangiwe, N.N.; Kotsedi, L.; Maaza, M.; Khenfouch, M. Synthesis and characterization of graphene thin films by chemical reduction of exfoliated and intercalated graphite oxide. J. Chem. 2013, 2013, 6. [Google Scholar] [CrossRef]
- Kaniyoor, A.; Baby, T.T.; Ramaprabhu, S. Graphene synthesis via hydrogen induced low temperature exfoliation of graphite oxide. J. Mater. Chem. 2010, 20, 8467–8469. [Google Scholar] [CrossRef]
- Konkena, B.; Vasudevan, S. Understanding aqueous dispersibility of graphene oxide and reduced graphene oxide through pKa measurements. J. Phys. Chem. Lett. 2012, 3, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Andryushina, N.S.; Stroyuk, O.L.; Yanchuk, I.B.; Yefanov, A.V. A dynamic light scattering study of photochemically reduced colloidal graphene oxide. Colloid. Polym. Sci. 2014, 292, 539–546. [Google Scholar] [CrossRef]
- Gurunathan, S.; Han, J.W.; Kim, J.H. Green chemistry approach for the synthesis of biocompatible graphene. Int. J. Nanomed. 2013, 8, 2719–2732. [Google Scholar] [CrossRef] [PubMed]
- Stankovich, S.; Dikin, D.A.; Piner, R.D.; Kohlhaas, K.A.; Kleinhammes, A.; Jia, Y.; Wu, Y.; Nguyen, S.T.; Ruoff, R.S. Synthesis of graphene-based nanosheets via chemical reduction of exfoliated graphite oxide. Carbon 2007, 45, 1558–1565. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, H.; Shen, G.; Cheng, P.; Zhang, J.; Guo, S. Reduction of graphene oxide via l-ascorbic acid. Chem. Commun. 2010, 46, 1112–1114. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.Z.; Li, L.D.; Guo, L. An environment-friendly preparation of reduced graphene oxide nanosheets via amino acid. Nanotechnology 2011, 22. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Eppakayala, V.; Dayem, A.A.; Kwon, D.N.; Kim, J.H. Biocompatibility effects of biologically synthesized graphene in primary mouse embryonic fibroblast cells. Nanoscale Res. Lett. 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Park, J.H.; Eppakayala, V.; Kim, J.H. Ginkgo biloba: A natural reducing agent for the synthesis of cytocompatible graphene. Int. J. Nanomed. 2014, 9, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Mhamane, D.; Ramadan, W.; Fawzy, M.; Rana, A.; Dubey, M.; Rode, C.; Lefez, B.; Hannoyer, B.; Ogale, S. From graphite oxide to highly water dispersible functionalized graphene by single step plant extract-induced deoxygenation. Green Chem. 2011, 13, 1990–1996. [Google Scholar] [CrossRef]
- Kuila, T.; Bose, S.; Khanra, P.; Mishra, A.K.; Kim, N.H.; Lee, J.H. A green approach for the reduction of graphene oxide by wild carrot root. Carbon 2012, 50, 914–921. [Google Scholar] [CrossRef]
- Khanra, P.; Kuila, T.; Kim, N.H.; Bae, S.H.; Yu, D.S.; Lee, J.H. Simultaneous bio-functionalization and reduction of graphene oxide by baker’s yeast. Chem. Eng. J. 2012, 183, 526–533. [Google Scholar] [CrossRef]
- Gurunathan, S.; Han, J.W.; Eppakayala, V.; Kim, J.H. Microbial reduction of graphene oxide by Escherichia coli: A green chemistry approach. Colloid. Surface B 2013, 102, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Liang, S.; Ma, L.; Ding, S.J.; Yu, X.F.; Zhou, L.; Wang, Q.Q. One-pot synthesis of CdS-reduced graphene oxide 3D composites with enhanced photocatalytic properties. CrystEngComm 2014, 16, 399–405. [Google Scholar] [CrossRef]
- Bourlinos, A.B.; Gournis, D.; Petridis, D.; Szabo, T.; Szeri, A.; Dekany, I. Graphite oxide: Chemical reduction to graphite and surface modification with primary aliphatic amines and amino acids. Langmuir 2003, 19, 6050–6055. [Google Scholar] [CrossRef]
- Murdock, R.C.; Braydich-Stolle, L.; Schrand, A.M.; Schlager, J.J.; Hussain, S.M. Characterization of nanomaterial dispersion in solution prior to in vitro exposure using dynamic light scattering technique. Toxicol. Sci. 2008, 101, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Ang, B.T.; Zhang, L.L.; Zhao, X.S.; Wu, J.S. Pyrolyzed graphene oxide/resorcinol-formaldehyde resin composites as high-performance supercapacitor electrodes. J. Mater. Chem. 2011, 21, 2663–2670. [Google Scholar] [CrossRef]
- Fu, C.J.; Zhao, G.G.; Zhang, H.J.; Li, S. Evaluation and characterization of reduced graphene oxide nanosheets as anode materials for lithium-ion batteries. Int J. Electrochem. Sc. 2013, 8, 6269–6280. [Google Scholar]
- Meyer, J.C.; Geim, A.K.; Katsnelson, M.I.; Novoselov, K.S.; Booth, T.J.; Roth, S. The structure of suspended graphene sheets. Nature 2007, 446, 60–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.L.; Yu, C.Z.; Wang, M.S.; Zhang, Y.P.; Pan, C.X. Synthesis of nitrogen doped graphene from graphene oxide within an ammonia flame for high performance supercapacitors. RSC Adv. 2014, 4, 55394–55399. [Google Scholar] [CrossRef]
- Zhang, H.X.; Feng, P.X. Fabrication and characterization of few-layer graphene. Carbon 2010, 48, 359–364. [Google Scholar] [CrossRef]
- Wang, Z.L.; Xu, D.; Huang, Y.; Wu, Z.; Wang, L.M.; Zhang, X.B. Facile, mild and fast thermal-decomposition reduction of graphene oxide in air and its application in high-performance lithium batteries. Chem. Commun. 2012, 48, 976–978. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.C.; Meyer, J.C.; Scardaci, V.; Casiraghi, C.; Lazzeri, M.; Mauri, F.; Piscanec, S.; Jiang, D.; Novoselov, K.S.; Roth, S.; et al. Raman spectrum of graphene and graphene layers. Phys. Rev. Lett. 2006, 97. [Google Scholar] [CrossRef] [PubMed]
- Jorio, A. Raman Spectroscopy in Graphene Related Systems; Wiley-VCH: Weinheim, Germany, 2011. [Google Scholar]
- Ferrari, A.C.; Robertson, J. Interpretation of raman spectra of disordered and amorphous carbon. Phys. Rev. B 2000, 61, 14095–14107. [Google Scholar] [CrossRef]
- Guo, Y.Q.; Sun, X.Y.; Liu, Y.; Wang, W.; Qiu, H.X.; Gao, J.P. One pot preparation of reduced graphene oxide (RGO) or Au (Ag) nanoparticle-RGO hybrids using chitosan as a reducing and stabilizing agent and their use in methanol electrooxidation. Carbon 2012, 50, 2513–2523. [Google Scholar] [CrossRef]
- Akhavan, O.; Ghaderi, E. Escherichia coli bacteria reduce graphene oxide to bactericidal graphene in a self-limiting manner. Carbon 2012, 50, 1853–1860. [Google Scholar] [CrossRef]
- Lammel, T.; Boisseaux, P.; Fernandez-Cruz, M.L.; Navas, J.M. Internalization and cytotoxicity of graphene oxide and carboxyl graphene nanoplatelets in the human hepatocellular carcinoma cell line HepG2. Part. Fibre Toxicol. 2013, 10. [Google Scholar] [CrossRef] [PubMed]
- Akhavan, O.; Ghaderi, E.; Emamy, H.; Akhavan, F. Genotoxicity of graphene nanoribbons in human mesenchymal stem cells. Carbon 2013, 54, 419–431. [Google Scholar] [CrossRef]
- Jaworski, S.; Sawosz, E.; Grodzik, M.; Winnicka, A.; Prasek, M.; Wierzbicki, M.; Chwalibog, A. In vitro evaluation of the effects of graphene platelets on glioblastoma multiforme cells. Int. J. Nanomed. 2013, 8, 413–420. [Google Scholar]
- Jaworski, S.; Sawosz, E.; Kutwin, M.; Wierzbicki, M.; Hinzmann, M.; Grodzik, M.; Winnicka, A.; Lipinska, L.; Wlodyga, K.; Chwalibog, A. In vitro and in vivo effects of graphene oxide and reduced graphene oxide on glioblastoma. In. J. Nanomed. 2015, 10, 1585–1596. [Google Scholar]
- Hatamie, S.; Akhavan, O.; Sadrnezhaad, S.K.; Ahadian, M.M.; Shirolkar, M.M.; Wang, H.Q.Q. Curcumin-reduced graphene oxide sheets and their effects on human breast cancer cells. Mater. Sci. Eng. C Mater. 2015, 55, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.B.; Ali, S.F.; Dervishi, E.; Xu, Y.; Li, Z.R.; Casciano, D.; Biris, A.S. Cytotoxicity effects of graphene and single-wall carbon nanotubes in neural phaeochromocytoma-derived pc12 cells. ACS Nano 2010, 4, 3181–3186. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Wang, Y.P.; Xu, X.; Zeng, C.; Hou, J.P.; Lin, M.M.; Xu, J.Z.; Sun, F.; Huang, X.J.; Dai, L.M.; et al. Can graphene oxide cause damage to eyesight? Chem. Res. Toxicol. 2012, 25, 1265–1270. [Google Scholar] [CrossRef] [PubMed]
- Schinwald, A.; Murphy, F.A.; Jones, A.; MacNee, W.; Donaldson, K. Graphene-based nanoplatelets: A new risk to the respiratory system as a consequence of their unusual aerodynamic properties. ACS Nano 2012, 6, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.X.; Pu, K.F.; Dong, B.; Liu, Y.; Zhang, L.M.; Zhang, Z.J.; Duan, W.; Zhu, Y.M. Role of surface charge and oxidative stress in cytotoxicity and genotoxicity of graphene oxide towards human lung fibroblast cells. J. Appl. Toxicol. 2013, 33, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.T.; Desikan, R.; Neill, S.J. Role of reactive oxygen species in cell signalling pathways. Biochem. Soc. Trans. 2001, 29, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Andreyev, A.I.; Kushnareva, Y.E.; Starkov, A.A. Mitochondrial metabolism of reactive oxygen species. Biochemistry 2005, 70, 200–214. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Eppakayala, V.; Jeyaraj, M.; Kim, J.H. Cytotoxicity of biologically synthesized silver nanoparticles in mda-mb-231 human breast cancer cells. Biomed. Res. Int. 2013, 2013, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Fu, Y.J.; Wei, T.T.; Le Guyader, L.; Gao, G.; Liu, R.S.; Chang, Y.Z.; Chen, C.Y. The triggering of apoptosis in macrophages by pristine graphene through the MAPK and TGF-β signaling pathways. Biomaterials 2012, 33, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, A.; Panchakarla, L.S.; Sadanandan, A.R.; Ashokan, A.; Chandran, P.; Girish, C.M.; Menon, D.; Nair, S.V.; Rao, C.N.R.; Koyakutty, M. Hemocompatibility and macrophage response of pristine and functionalized graphene. Small 2012, 8, 1251–1263. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.R.N.; Reddy, Y.N.; Krishna, D.R.; Himabindu, V. Multi wall carbon nanotubes induce oxidative stress and cytotoxicity in human embryonic kidney (HEK293) cells. Toxicology 2010, 272, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.R.; Jing, L.; Valladares, A.; Mehta, S.L.; Wang, Z.Z.; Li, P.A.; Bang, J.J. Silver nanoparticle exposure induced mitochondrial stress, caspase-3 activation and cell death: Amelioration by sodium selenite. Int J. Biol. Sci. 2015, 11, 860–867. [Google Scholar] [CrossRef] [PubMed]
- AshaRani, P.V.; Mun, G.L.K.; Hande, M.P.; Valiyaveettil, S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano 2009, 3, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Nhan, T.Q.; Liles, W.C.; Schwartz, S.M. Physiological functions of caspases beyond cell death. Am. J. Pathol. 2006, 169, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Cohen, G.M. Caspases: The executioners of apoptosis. Biochem. J. 1997, 326, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, R.M. Mechanisms of disease: Apoptosis and caspases in neurodegenerative diseases. New Eng. J. Med. 2003, 348, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zou, H.; Slaughter, C.; Wang, X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell 1997, 89, 175–184. [Google Scholar] [CrossRef]
- Sakahira, H.; Enari, M.; Nagata, S. Cleavage of cad inhibitor in cad activation and DNA degradation during apoptosis. Nature 1998, 391, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Henkels, K.M.; Turchi, J.J. Cisplatin-induced apoptosis proceeds by caspase-3-dependent and -independent pathways in cisplatin-resistant and -sensitive human ovarian cancer cell lines. Cancer Res. 1999, 59, 3077–3083. [Google Scholar] [PubMed]
- Liu, Y.Y.; Luo, Y.; Wu, J.; Wang, Y.S.; Yang, X.Y.; Yang, R.; Wang, B.Q.; Yang, J.R.; Zhang, N. Graphene oxide can induce in vitro and in vivo mutagenesis. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Xiong, W.; Zhu, L.; Osawa, E.; Hussin, S.; Dai, L.M. DNA damage in embryonic stem cells caused by nanodiamonds. ACS Nano 2011, 5, 2376–2384. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Chang, D.W.; Dai, L.M.; Hong, Y.L. DNA damage induced by multiwalled carbon nanotubes in mouse embryonic stem cells. Nano Lett 2007, 7, 3592–3597. [Google Scholar] [CrossRef] [PubMed]
- Hinzmann, M.; Jaworski, S.; Kutwin, M.; Jagiello, J.; Kozinski, R.; Wierzbicki, M.; Grodzik, M.; Lipinska, L.; Sawosz, E.; Chwalibog, A. Nanoparticles containing allotropes of carbon have genotoxic effects on glioblastoma multiforme cells. Int. J. Nanomed. 2014, 9, 2409–2417. [Google Scholar]
- Zhang, G.; Chang, H.C.; Amatore, C.; Chen, Y.; Jiang, H.; Wang, X.M. Apoptosis induction and inhibition of drug resistant tumor growth in vivo involving daunorubicin-loaded graphene-gold composites. J. Mater. Chem. B 2013, 1, 493–499. [Google Scholar] [CrossRef]
- Hummers, W.S.; Offeman, R.E. Preparation of graphitic oxide. J. Am. Chem. Soc. 1958, 80, 1339. [Google Scholar] [CrossRef]
- Liao, K.H.; Lin, Y.S.; Macosko, C.W.; Haynes, C.L. Cytotoxicity of graphene oxide and graphene in human erythrocytes and skin fibroblasts. ACS Appl. Mater. Interf. 2011, 3, 2607–2615. [Google Scholar] [CrossRef] [PubMed]
- Sriram, M.I.; Kanth, S.B.M.; Kalishwaralal, K.; Gurunathan, S. Antitumor activity of silver nanoparticles in dalton's lymphoma ascites tumor model. Int. J. Nanomed. 2010, 5, 753–762. [Google Scholar]
- Sample Availability: Samples of the compounds are not available from the authors.






© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, Y.-J.; Kim, E.; Han, J.; Kim, J.-H.; Gurunathan, S. A Novel Biomolecule-Mediated Reduction of Graphene Oxide: A Multifunctional Anti-Cancer Agent. Molecules 2016, 21, 375. https://doi.org/10.3390/molecules21030375
Choi Y-J, Kim E, Han J, Kim J-H, Gurunathan S. A Novel Biomolecule-Mediated Reduction of Graphene Oxide: A Multifunctional Anti-Cancer Agent. Molecules. 2016; 21(3):375. https://doi.org/10.3390/molecules21030375
Chicago/Turabian StyleChoi, Yun-Jung, Eunsu Kim, JaeWoong Han, Jin-Hoi Kim, and Sangiliyandi Gurunathan. 2016. "A Novel Biomolecule-Mediated Reduction of Graphene Oxide: A Multifunctional Anti-Cancer Agent" Molecules 21, no. 3: 375. https://doi.org/10.3390/molecules21030375





