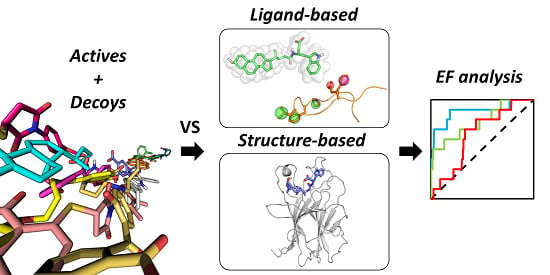
Comparative Analysis of Virtual Screening Approaches in the Search for Novel EphA2 Receptor Antagonists
Abstract
:1. Introduction
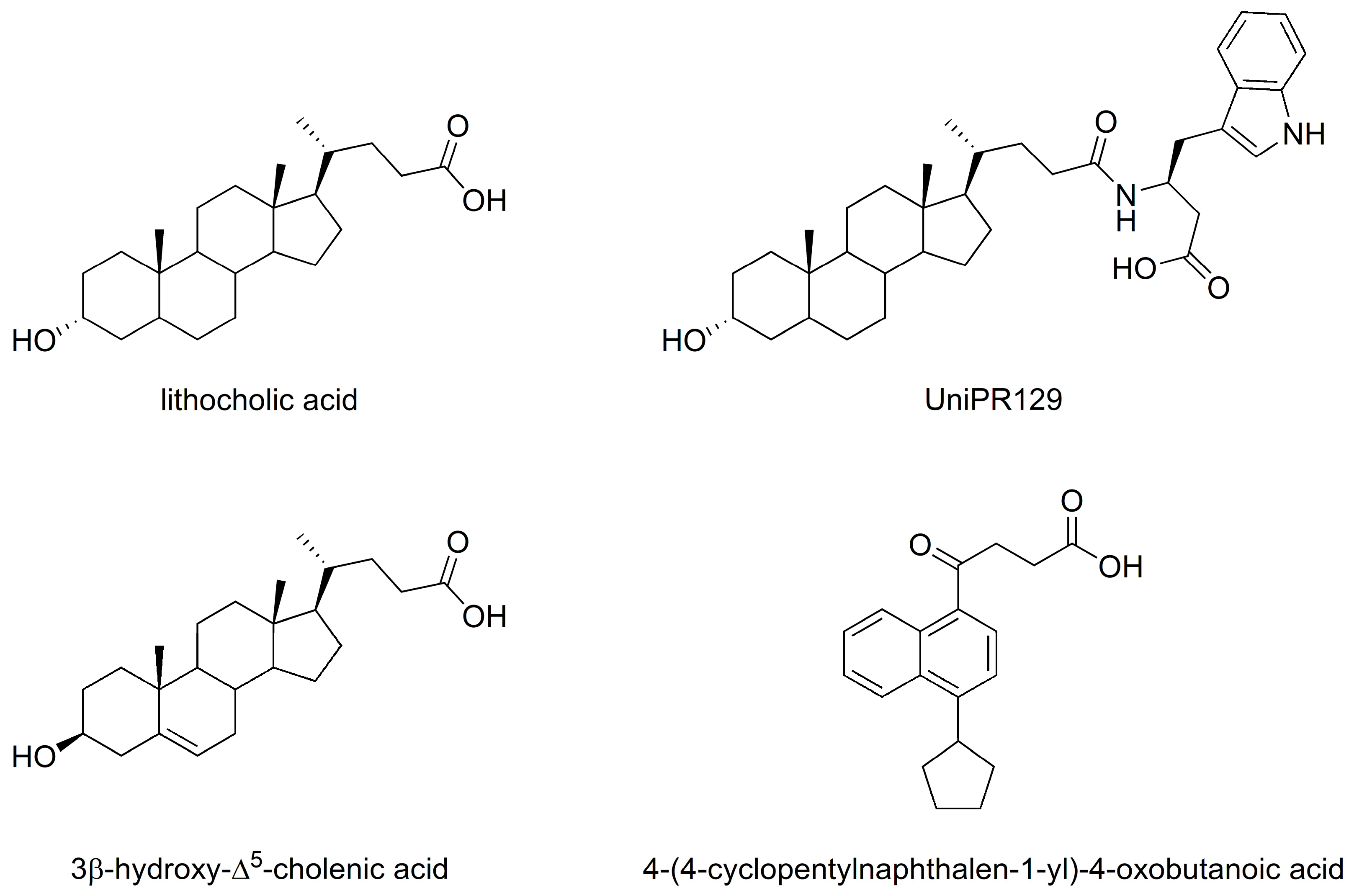
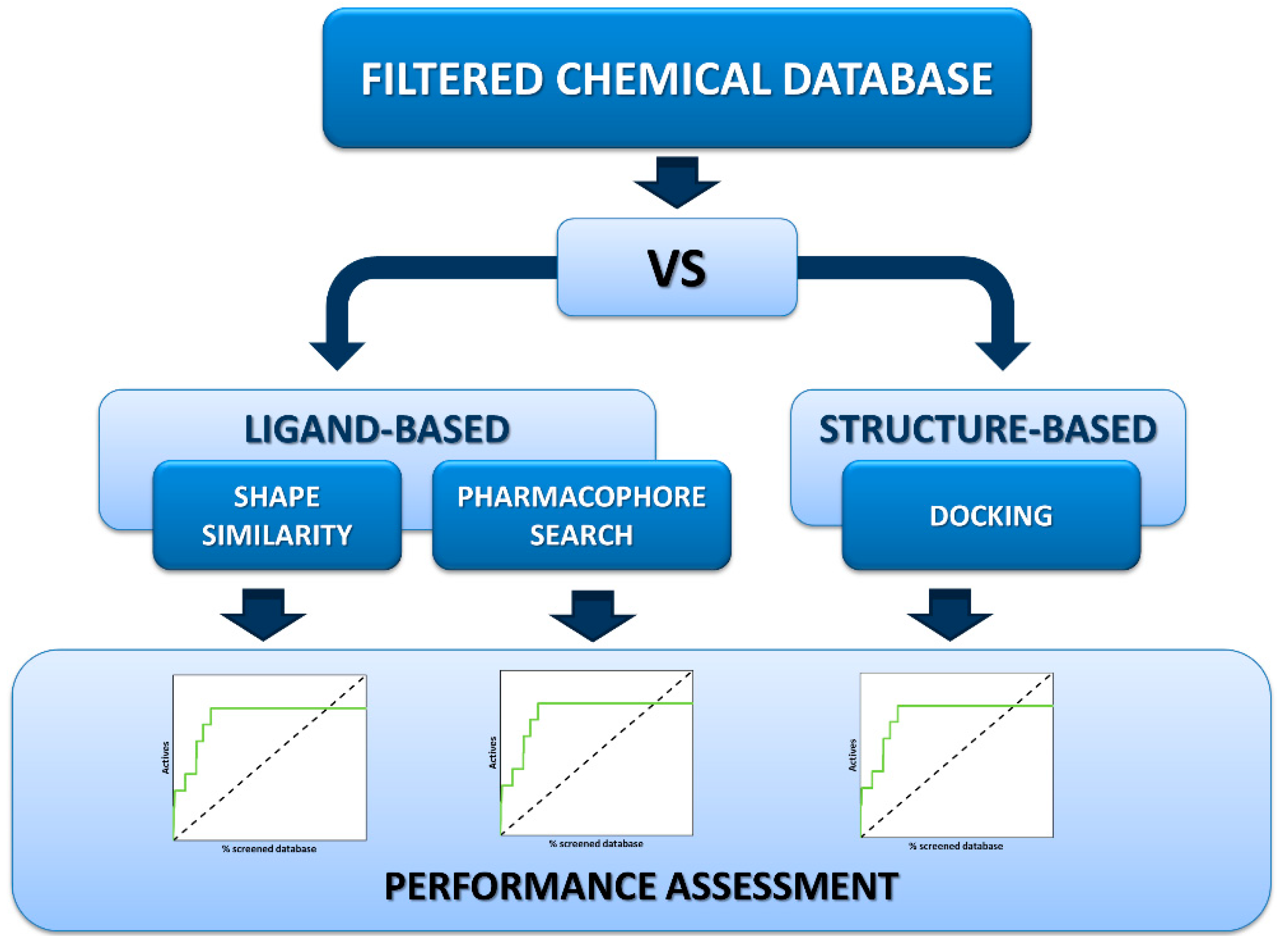
2. Results and Discussion
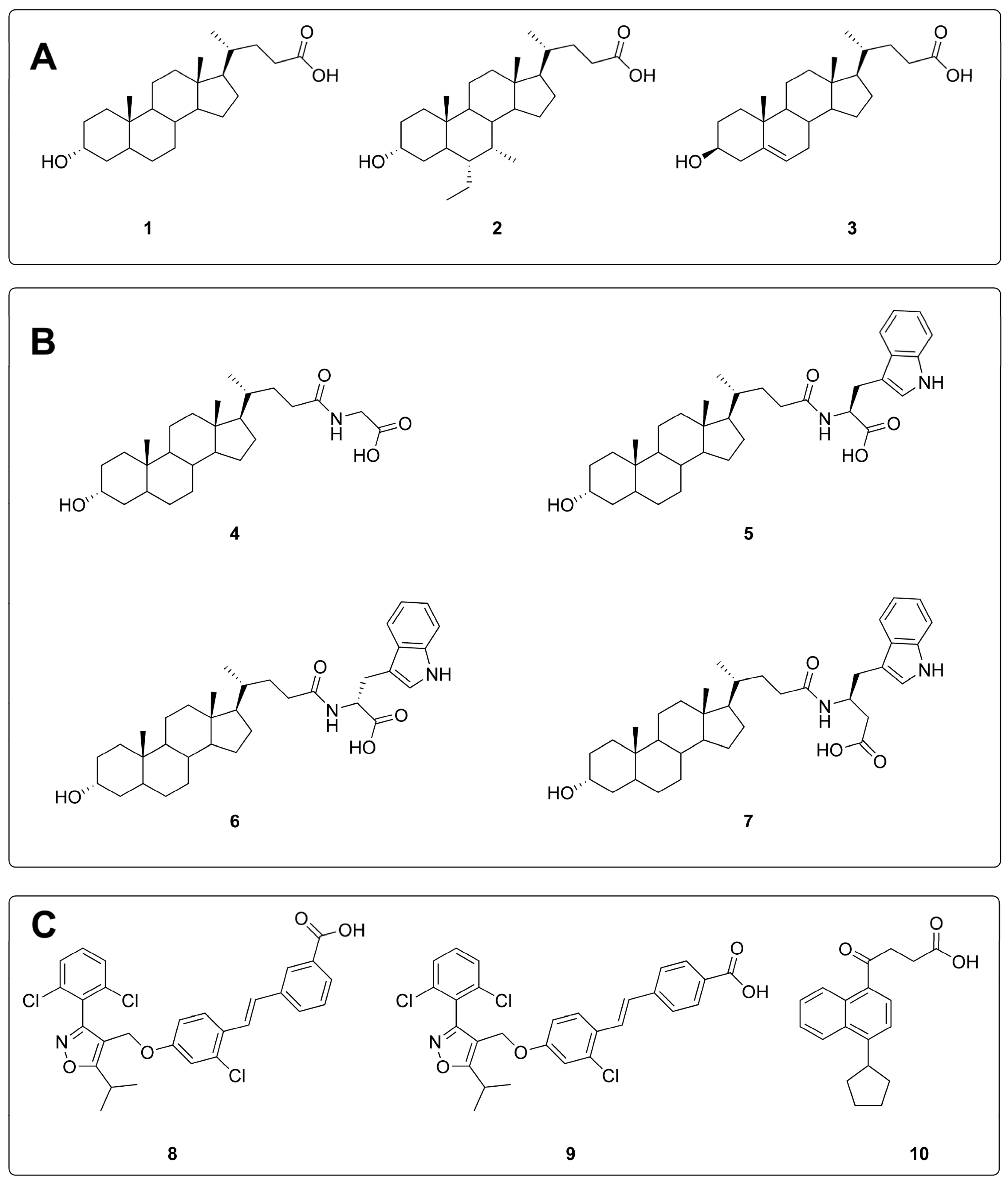

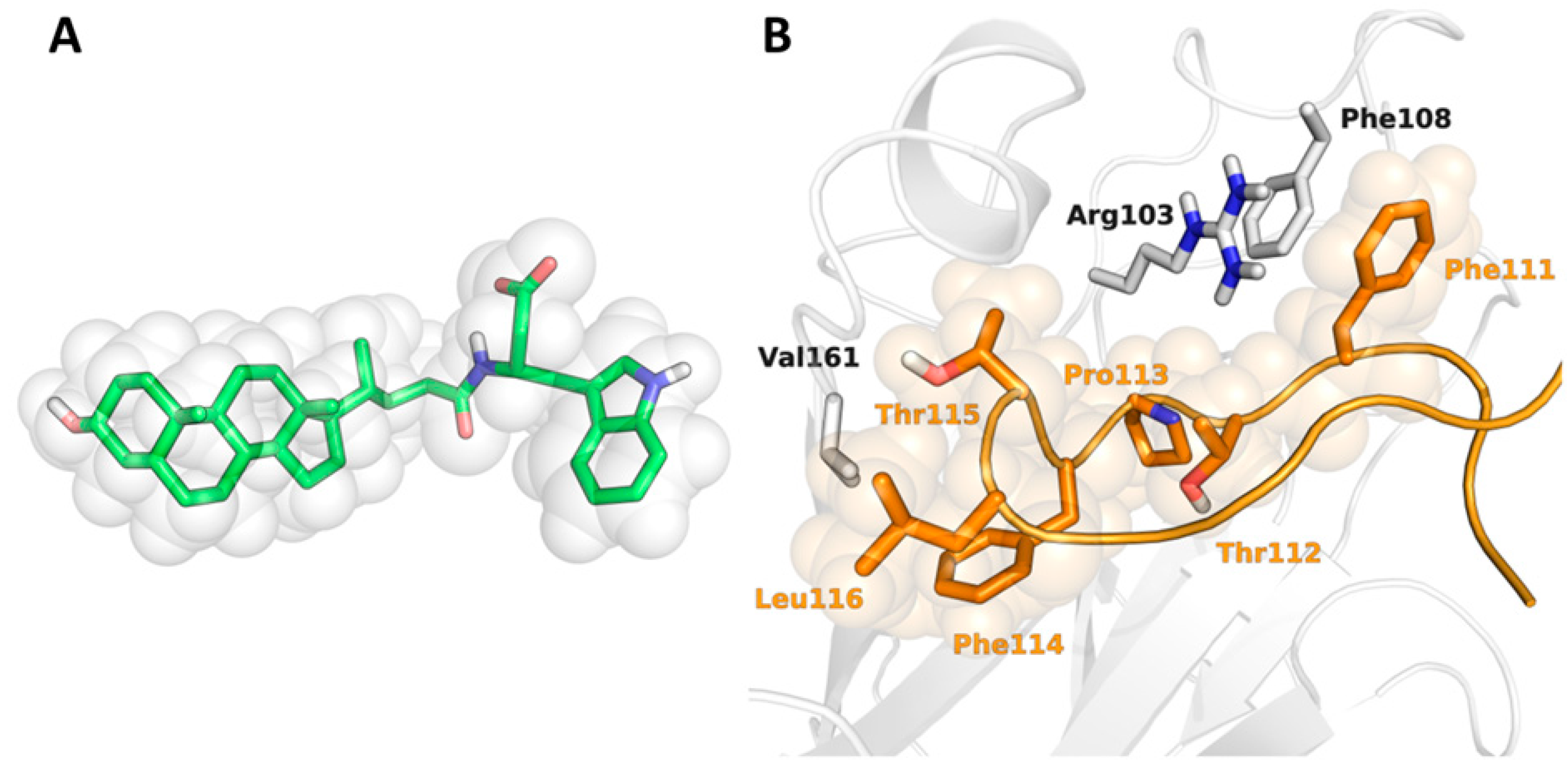
2.1. Shape-Screening
| Template | Method | ChemDiv | ChemBridge | ||
|---|---|---|---|---|---|
| EF2% | EF5% | EF2% | EF5% | ||
| UniPR129 | shape-only | 25 (5) | 14 (7) | 25 (5) | 14 (7) |
| mmod | 35 (7) | 14 (7) | 35 (7) | 14 (7) | |
| Pharm | 35 (7) | 14 (7) | 35 (7) | 14 (7) | |
| G-H loop | shape-only | 20 (4) | 10 (5) | 15 (3) | 6 (3) |
| mmod | 15 (3) | 6 (3) | 15 (3) | 6 (3) | |
| Pharm | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |

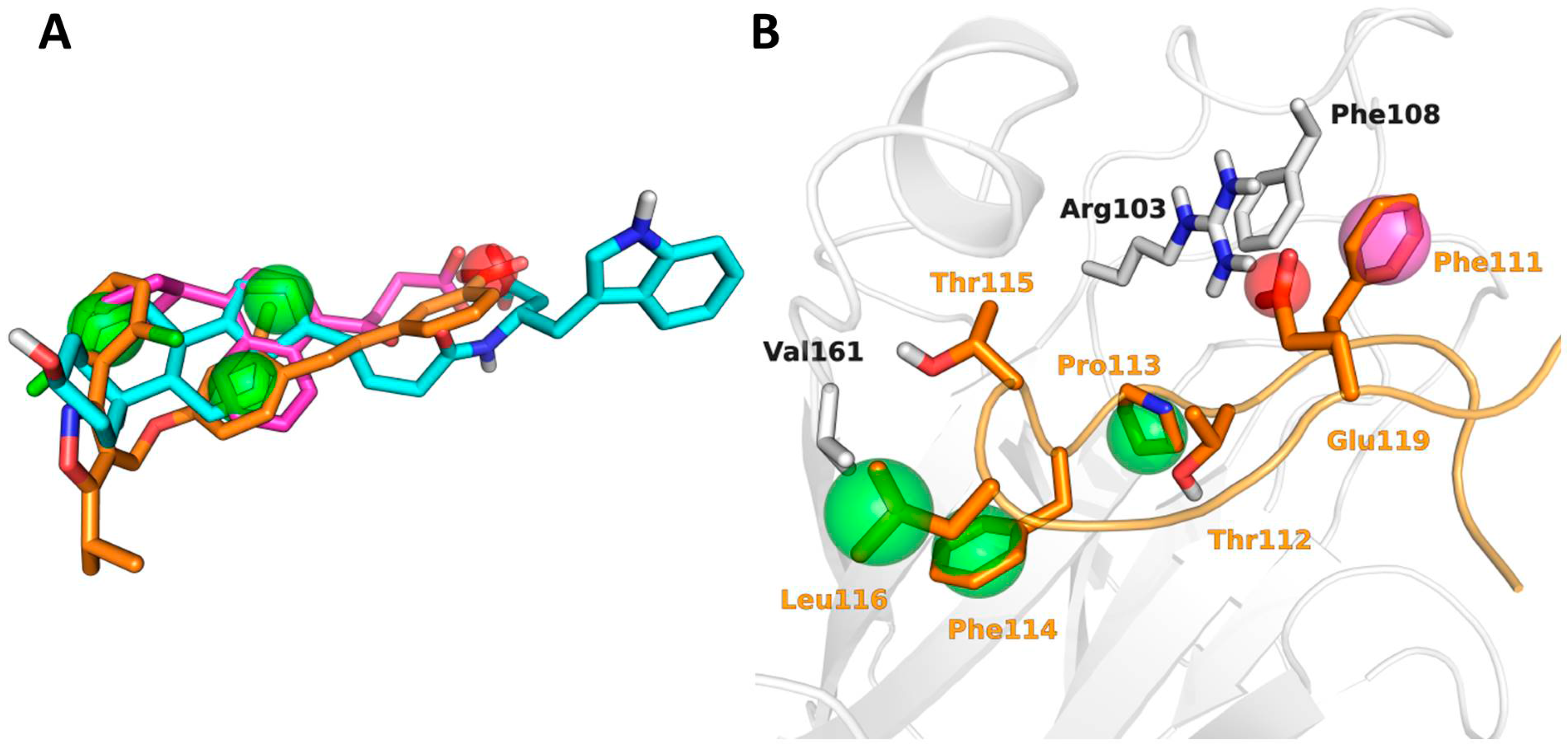
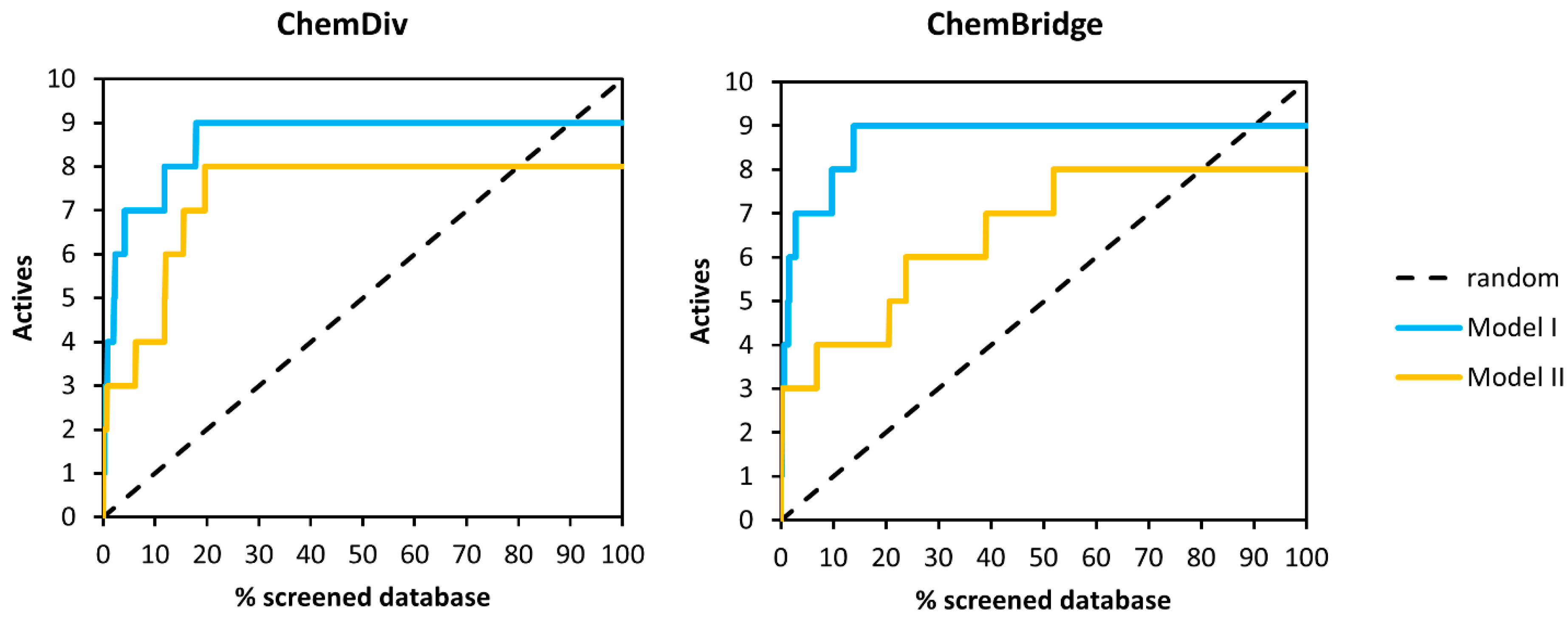
2.2. Pharmacophore Search
| Method | ChemDiv | ChemBridge | ||
|---|---|---|---|---|
| EF2% | EF5% | EF2% | EF5% | |
| Model I | 25(5) | 14 (7) | 30 (6) | 14 (7) |
| Model II | 15 (3) | 6 (3) | 15 (3) | 6 (3) |
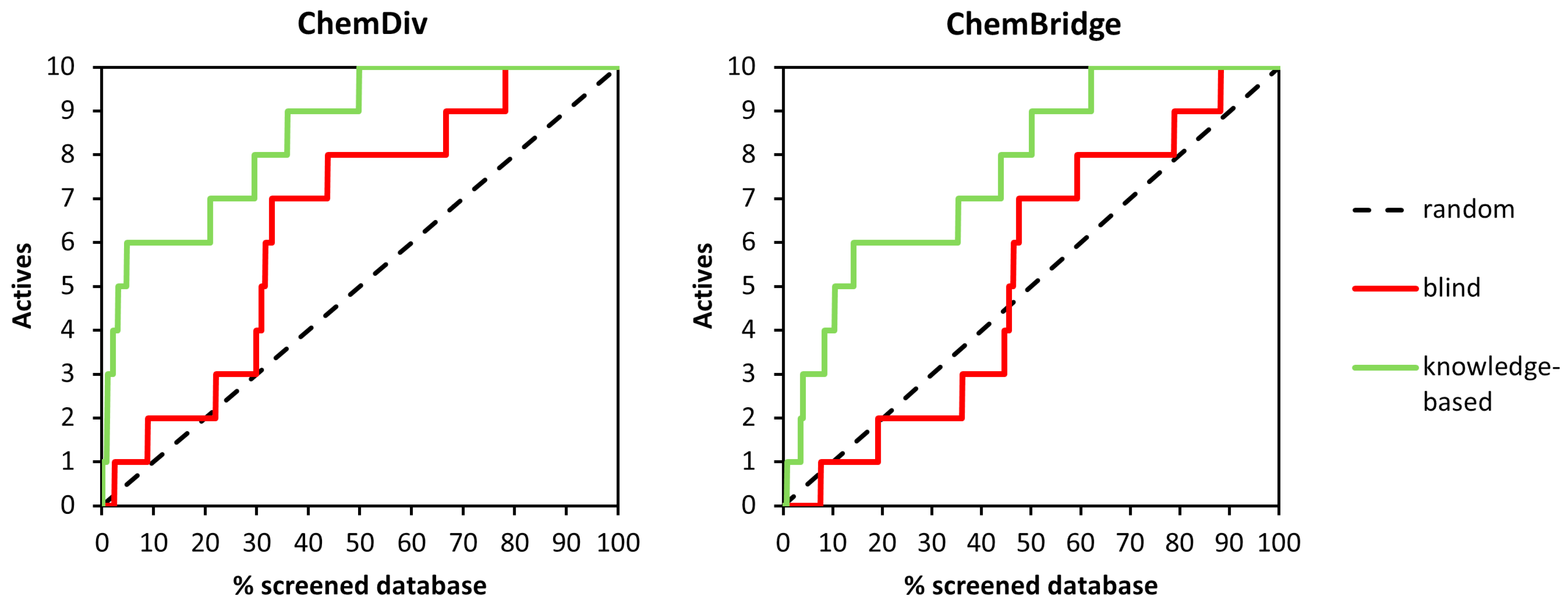
2.3. Docking

| Method | ChemDiv | ChemBridge | ||
|---|---|---|---|---|
| EF2% | EF5% | EF2% | EF5% | |
| blind | 0 (0) | 2 (1) | 0 (0) | 0 (0) |
| knowledge-based | 15 (3) | 12 (6) | 5 (1) | 6 (3) |
3. Materials and Methods
3.1. Database Preparation
3.2. Shape-Screening
3.3. Pharmacophore Models Building
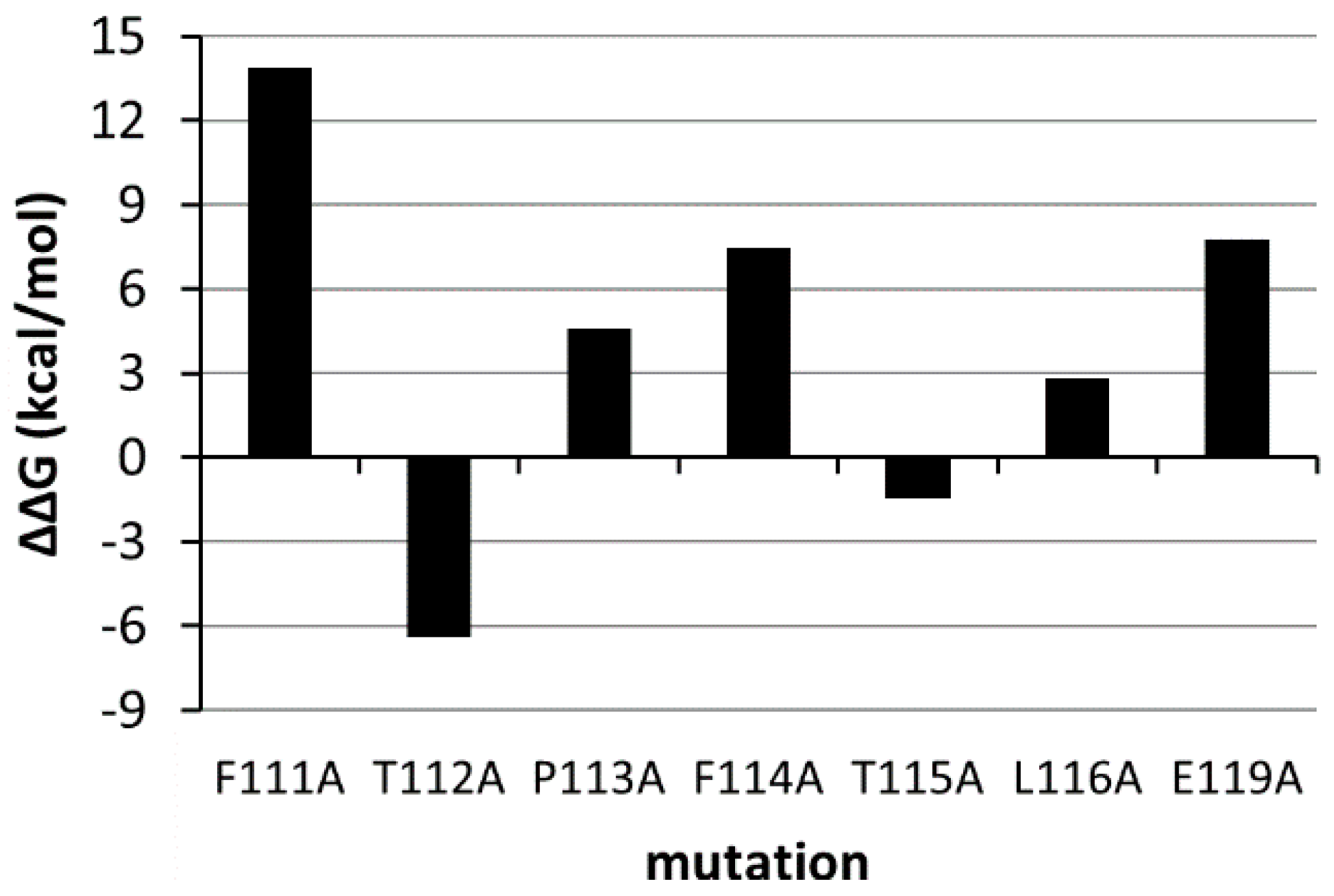
3.4. Pharmacophore Search
3.5. Docking
3.6. Performance Assessment
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Himanen, J.P.; Nikolov, D.B. Eph signaling: A structural view. Trends Neurosci. 2003, 26, 46–51. [Google Scholar] [CrossRef]
- Himanen, J.P.; Chumley, M.J.; Lackmann, M.; Li, C.; Barton, W.A.; Jeffrey, P.D.; Vearing, C.; Geleick, D.; Feldheim, D.A.; Boyd, A.W.; et al. Repelling class discrimination: Ephrin-A5 binds to and activates EphB2 receptor signaling. Nat. Neurosci. 2004, 7, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Pasquale, E.B. Eph-ephrin bidirectional signaling in physiology and disease. Cell 2008, 133, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Pasquale, E.B. Eph receptors and ephrins in cancer: Bidirectional signalling and beyond. Nat. Rev. Cancer 2010, 10, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Tandon, M.; Vemula, S.V.; Mittal, S.K. Emerging strategies for EphA2 receptor targeting for cancer therapeutics. Expert Opin. Ther. Targets 2011, 15, 31–51. [Google Scholar] [CrossRef] [PubMed]
- Landen, C.N.; Lu, C.; Han, L.Y.; Coffman, K.T.; Bruckheimer, E.; Halder, J.; Mangala, L.S.; Merritt, W.M.; Lin, Y.G.; Gao, C.; et al. Efficacy and antivascular effects of EphA2 reduction with an agonistic antibody in ovarian cancer. J. Natl. Cancer Inst. 2006, 98, 1558–1570. [Google Scholar] [CrossRef] [PubMed]
- Dobrzanski, P.; Hunter, K.; Jones-Bolin, S.; Chang, H.; Robinson, C.; Pritchard, S.; Zhao, H.; Ruggeri, B. Antiangiogenic and antitumor efficacy of EphA2 receptor antagonist. Cancer Res. 2004, 64, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Wykosky, J.; Debinski, W. The EphA2 receptor and ephrinA1 ligand in solid tumors: Function and therapeutic targeting. Mol. Cancer Res. 2008, 6, 1795–1806. [Google Scholar] [CrossRef] [PubMed]
- Tognolini, M.; Hassan-Mohamed, I.; Giorgio, C.; Zanotti, I.; Lodola, A. Therapeutic perspectives of Eph-ephrin system modulation. Drug Discov. Today 2014, 19, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Boyd, A.W.; Bartlett, P.F.; Lackmann, M. Therapeutic targeting of EPH receptors and their ligands. Nat. Rev. Drug Discov. 2014, 13, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Barquilla, A.; Pasquale, E.B. Eph receptors and ephrins: Therapeutic opportunities. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 465–487. [Google Scholar] [CrossRef]
- Giorgio, C.; Hassan Mohamed, I.; Flammini, L.; Barocelli, E.; Incerti, M.; Lodola, A.; Tognolini, M. Lithocholic acid is an Eph-ephrin ligand interfering with Eph-kinase activation. PLoS ONE 2011, 6, e18128. [Google Scholar] [CrossRef] [PubMed]
- Tognolini, M.; Incerti, M.; Hassan-Mohamed, I.; Giorgio, C.; Russo, S.; Bruni, R.; Lelli, B.; Bracci, L.; Noberini, R.; Pasquale, E.B. Structure-activity relationships and mechanism of action of Eph-ephrin antagonists: Interaction of cholanic acid with the EphA2 receptor. ChemMedChem 2012, 7, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Russo, S.; Incerti, M.; Tognolini, M.; Castelli, R.; Pala, D.; Hassan-Mohamed, I.; Giorgio, C.; de Franco, F.; Gioiello, A.; Vicini, P.; et al. Synthesis and structure-activity relationships of amino acid conjugates of cholanic acid as antagonists of the EphA2 receptor. Molecules 2013, 18, 13043–13060. [Google Scholar] [PubMed]
- Incerti, M.; Tognolini, M.; Russo, S.; Pala, D.; Giorgio, C.; Hassan-Mohamed, I.; Noberini, R.; Pasquale, E.B.; Vicini, P.; Piersanti, S.; et al. Amino acid conjugates of lithocholic acid as antagonists of the EphA2 receptor. J. Med. Chem. 2013, 56, 2936–2947. [Google Scholar] [CrossRef] [PubMed]
- Hassan-Mohamed, I.; Giorgio, C.; Incerti, M.; Russo, S.; Pala, D.; Pasquale, E.B.; Zanotti, I.; Vicini, P.; Barocelli, E.; Rivara, S.; et al. UniPR129 is a competitive small molecule Eph-ephrin antagonist blocking in vitro angiogenesis at low micromolar concentrations. Br. J. Pharmacol. 2014, 171, 5195–5208. [Google Scholar] [CrossRef] [PubMed]
- Tognolini, M.; Incerti, M.; Lodola, A. Are we using the right pharmacological tools to target EphA4? ACS Chem. Neurosci. 2014, 5, 1146–1147. [Google Scholar] [CrossRef] [PubMed]
- Pala, D.; Castelli, R.; Incerti, M.; Russo, S.; Tognolini, M.; Giorgio, C.; Hassan-Mohamed, I.; Zanotti, I.; Vacondio, F.; Rivara, S.; et al. Combining ligand- and structure-based approaches for the discovery of new inhibitors of the EPHA2-ephrin-A1 interaction. J. Chem. Inf. Model. 2014, 54, 2621–2626. [Google Scholar] [CrossRef] [PubMed]
- Phase; Version 3.7; Schrödinger, LLC: New York, NY, USA, 2013.
- Glide; Version 5.7; Schrödinger, LLC: New York, NY, USA, 2011.
- Himanen, J.P.; Goldgur, Y.; Miao, H.; Myshkin, E.; Guo, H.; Buck, M.; Nguyen, M.; Rajashankar, K.R.; Wang, B.; Nikolov, D.B. Ligand recognition by A-class Eph receptors: Crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex. EMBO Rep. 2009, 10, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Truchon, J.F.; Bayly, C.I. Evaluating virtual screening methods: Good and bad metrics for the “early recognition” problem. J. Chem. Inf. Model. 2007, 47, 488–508. [Google Scholar] [CrossRef] [PubMed]
- McGaughey, G.B.; Sheridan, R.; Bayly, C.I.; Culberson, J.C.; Kreatsoulas, C.; Lindsley, S.; Maiorov, V.; Truchon, J.F.; Cornell, W.D. Comparison of topological, shape, and docking methods in virtual screening. J. Chem. Inf. Model. 2007, 47, 1504–1519. [Google Scholar] [CrossRef] [PubMed]
- Tognolini, M.; Incerti, M.; Pala, D.; Russo, S.; Castelli, R.; Hassan-Mohamed, I.; Giorgio, C.; Lodola, A. Target hopping as a useful tool for the identification of novel EphA2 protein-protein antagonists. ChemMedChem 2014, 9, 67–72. [Google Scholar] [CrossRef] [PubMed]
- ChemDiv—Contract Research Organization. Available online: http://www.chemdiv.com/ (accessed on 10 October 2013).
- ZINC-ChemBridge. Available online: http://zinc.docking.org/catalogs/chbr (accessed on 9 September 2013).
- Verdonk, M.L.; Berdini, V.; Hartshorn, M.J.; Mooij, W.T.M.; Murray, C.W.; Taylor, R.D.; Watson, P. Virtual screening using protein-ligand docking: Avoiding artificial enrichment. J. Chem. Inf. Comput. Sci. 2004, 44, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Scior, T.; Bender, A.; Tresadern, G.; Medina-Franco, J.L.; Martínez-Mayorga, K.; Langer, T.; Cuanalo-Contreras, K.; Agrafiotis, D.K. Recognizing pitfalls in virtual screening: A critical review. J. Chem. Inf. Model. 2012, 52, 867–881. [Google Scholar] [CrossRef] [PubMed]
- Labbé, C.M.; Laconde, G.; Kuenemann, M.A.; Villoutreix, B.O.; Sperandio, O. iPPI-DB: A manually curated and interactive database of small non-peptide inhibitors of protein-protein interactions. Drug Discov. Today 2013, 18, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Morelli, X.; Bourgeas, R.; Roche, P. Chemical and structural lessons from recent successes in protein-protein interaction inhibition (2P2I). Curr. Opin. Chem. Biol. 2011, 15, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Betzi, S.; Morelli, X.; Roche, P. Focused chemical libraries—Design and enrichment: An example of protein-protein interaction chemical space. Future Med. Chem. 2014, 6, 1291–1307. [Google Scholar] [CrossRef] [PubMed]
- Reynès, C.; Host, H.; Camproux, A.C.; Laconde, G.; Leroux, F.; Mazars, A.; Deprez, B.; Fahraeus, R.; Villoutreix, B.O.; Sperandio, O. Designing focused chemical libraries enriched in protein-protein interaction inhibitors using machine-learning methods. PLoS Comput. Biol. 2010, 6, e1000695. [Google Scholar] [CrossRef] [PubMed]
- Lavecchia, A.; di Giovanni, C. Virtual screening strategies in drug discovery: A critical review. Curr. Med. Chem. 2013, 20, 2839–2860. [Google Scholar] [CrossRef] [PubMed]
- Krüger, D.M.; Evers, A. Comparison of structure- and ligand-based virtual screening protocols considering hit list complementarity and enrichment factors. ChemMedChem 2010, 5, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Dixon, S.L.; Sherman, W. Rapid shape-based ligand alignment and virtual screening method based on atom/feature-pair similarities and volume overlap scoring. J. Chem. Inf. Model. 2011, 51, 2455–2466. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.L.; Smondyrev, A.M.; Rao, S.N. PHASE: A Novel Approach to Pharmacophore Modeling and 3D Database Searching. Chem. Biol. Drug Des. 2006, 67, 370–372. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.L.; Smondyrev, A.M.; Knoll, E.H.; Rao, S.N.; Shaw, D.E.; Friesner, R.A. PHASE: A new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J. Comput. Aided Mol. Des. 2006, 20, 647–671. [Google Scholar] [CrossRef] [PubMed]
- Brogi, S.; Papazafiri, P.; Roussis, V.; Tafi, A. 3D-QSAR using pharmacophore-based alignment and virtual screening for discovery of novel MCF-7 cell line inhibitors. Eur. J. Med. Chem. 2013, 67, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.G.; dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Duggineni, S.; Koolpe, M.; Zhu, X.; Huang, Z.; Pasquale, E.B. Structure-activity relationship analysis of peptides targeting the EphA2 receptor. Biochemistry 2010, 49, 6687–6695. [Google Scholar] [CrossRef] [PubMed]
- Mor, M.; Rivara, S.; Lodola, A.; Lorenzi, S.; Bordi, F.; Plazzi, P.V.; Spadoni, G.; Bedini, A.; Duranti, A.; Tontini, A.; et al. Application of 3D-QSAR in the rational design of receptor ligands and enzyme inhibitors. Chem. Biodivers. 2005, 2, 1438–1451. [Google Scholar] [CrossRef] [PubMed]
- Braga, R.C.; Andrade, C.H. Assessing the performance of 3D pharmacophore models in virtual screening: How good are they? Curr. Top. Med. Chem. 2013, 13, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, C.R.; Moitessier, N. Docking ligands into flexible and solvated macromolecules. 3. Impact of input ligand conformation, protein flexibility, and water molecules on the accuracy of docking programs. J. Chem. Inf. Model. 2009, 49, 997–1009. [Google Scholar] [CrossRef] [PubMed]
- Tubert-Brohman, I.; Sherman, W.; Repasky, M.; Beuming, T. Improved docking of polypeptides with Glide. J. Chem. Inf. Model. 2013, 53, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Maestro; Version 9.6; Schrödinger, LLC: New York, NY, USA, 2013.
- LigPrep; Version 2.5; Schrödinger, LLC: New York, NY, USA, 2011.
- Epik; Version 2.2; Schrödinger, LLC: New York, NY, USA, 2011.
- MacroModel; Version 10.2; Schrödinger, LLC: New York, NY, USA, 2013.
- ConfGen; Version 2.6; Schrödinger, LLC: New York, NY, USA, 2013.
- Beard, H.; Cholleti, A.; Pearlman, D.; Sherman, W.; Loving, K.A. Applying physics-based scoring to calculate free energies of binding for single amino acid mutations in protein-protein complexes. PLoS ONE 2013, 8, e82849. [Google Scholar] [CrossRef] [PubMed]
- Prime; Version 3.4; Schrödinger, LLC: New York, NY, USA, 2013.
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Triballeau, N.; Acher, F.; Brabet, I.; Pin, J.P.; Bertrand, H.O. Virtual screening workflow development guided by the “receiver operating characteristic” curve approach. Application to high-throughput docking on metabotropic glutamate receptor subtype 4. J. Med. Chem. 2005, 48, 2534–2547. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Pala, D.; Beuming, T.; Sherman, W.; Lodola, A.; Rivara, S.; Mor, M. Structure-based virtual screening of MT2 melatonin receptor: Influence of template choice and structural refinement. J. Chem. Inf. Model. 2013, 53, 821–835. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comp. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Inakollu, V.S.S.; Sherman, W. Boosting virtual screening enrichments with data fusion: Coalescing hits from two-dimensional fingerprints, shape, and docking. J. Chem. Inf. Model. 2013, 53, 1531–1542. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of compounds 1–10 are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Callegari, D.; Pala, D.; Scalvini, L.; Tognolini, M.; Incerti, M.; Rivara, S.; Mor, M.; Lodola, A. Comparative Analysis of Virtual Screening Approaches in the Search for Novel EphA2 Receptor Antagonists. Molecules 2015, 20, 17132-17151. https://doi.org/10.3390/molecules200917132
Callegari D, Pala D, Scalvini L, Tognolini M, Incerti M, Rivara S, Mor M, Lodola A. Comparative Analysis of Virtual Screening Approaches in the Search for Novel EphA2 Receptor Antagonists. Molecules. 2015; 20(9):17132-17151. https://doi.org/10.3390/molecules200917132
Chicago/Turabian StyleCallegari, Donatella, Daniele Pala, Laura Scalvini, Massimiliano Tognolini, Matteo Incerti, Silvia Rivara, Marco Mor, and Alessio Lodola. 2015. "Comparative Analysis of Virtual Screening Approaches in the Search for Novel EphA2 Receptor Antagonists" Molecules 20, no. 9: 17132-17151. https://doi.org/10.3390/molecules200917132