Authentication of Cordyceps sinensis by DNA Analyses: Comparison of ITS Sequence Analysis and RAPD-Derived Molecular Markers
Abstract
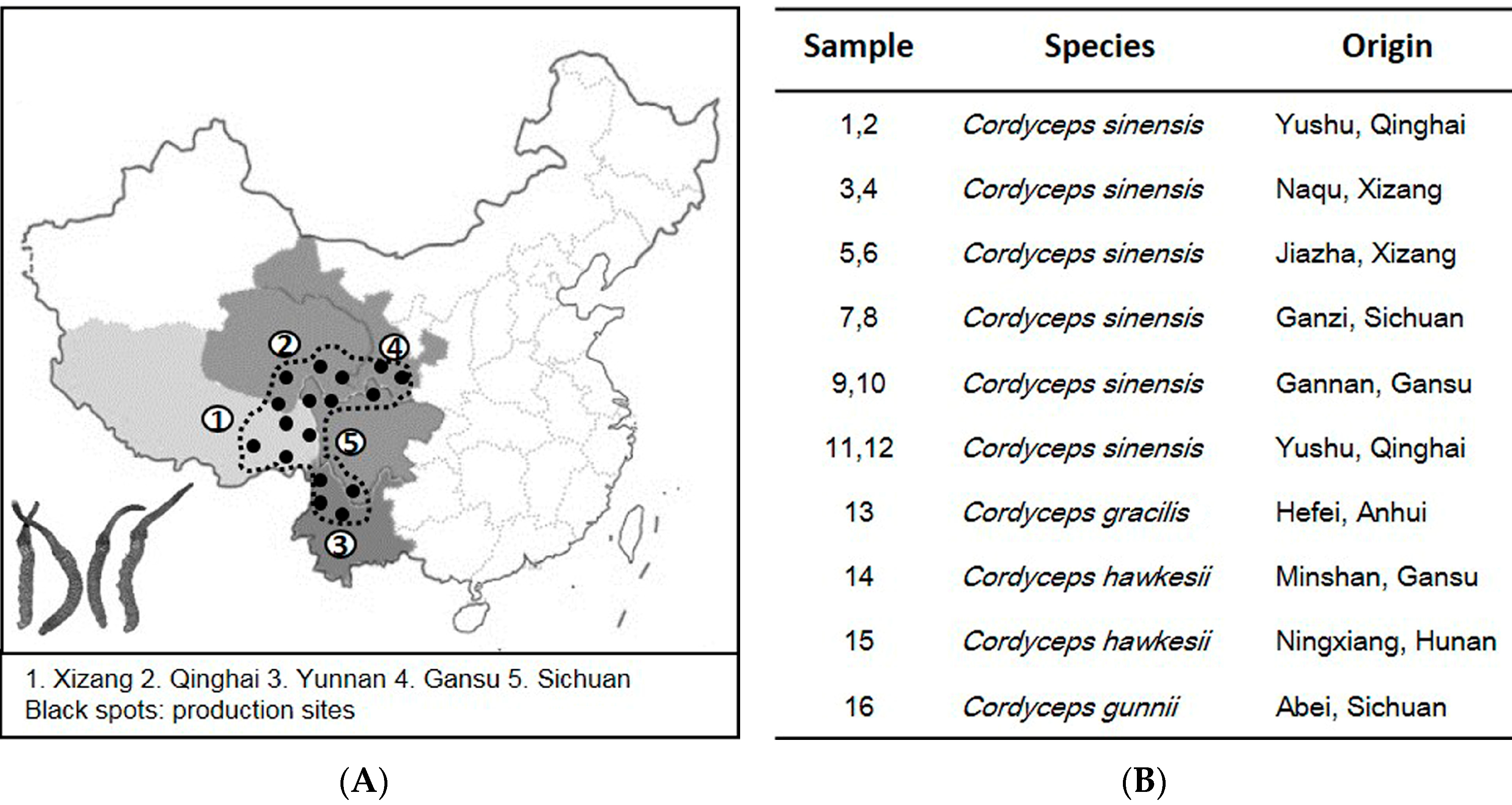
:1. Introduction
2. Results
2.1. ITS Sequence of Cordyceps
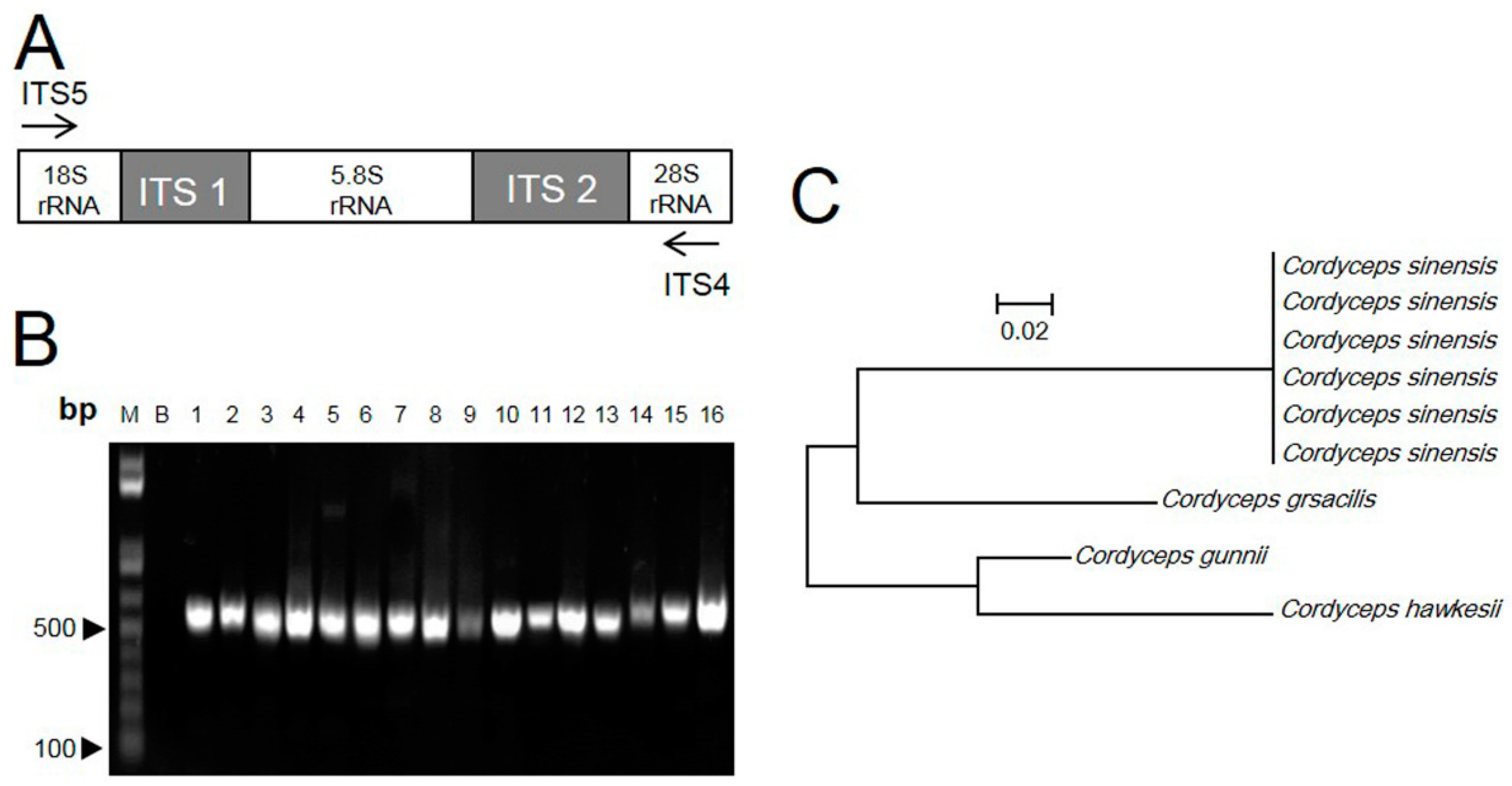
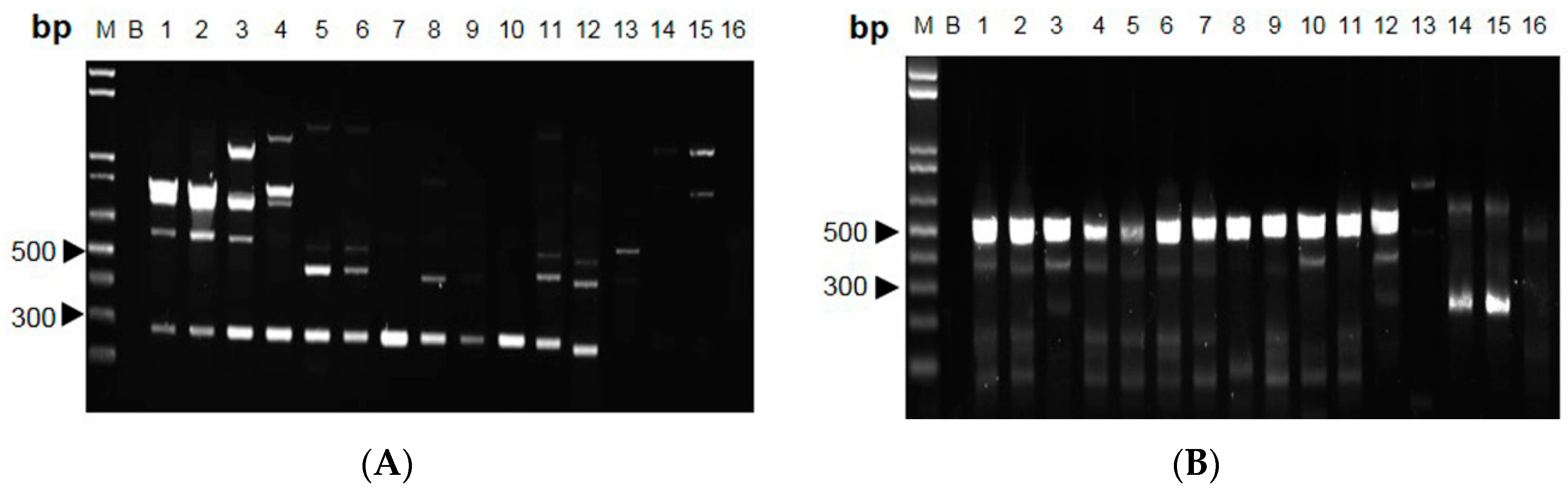
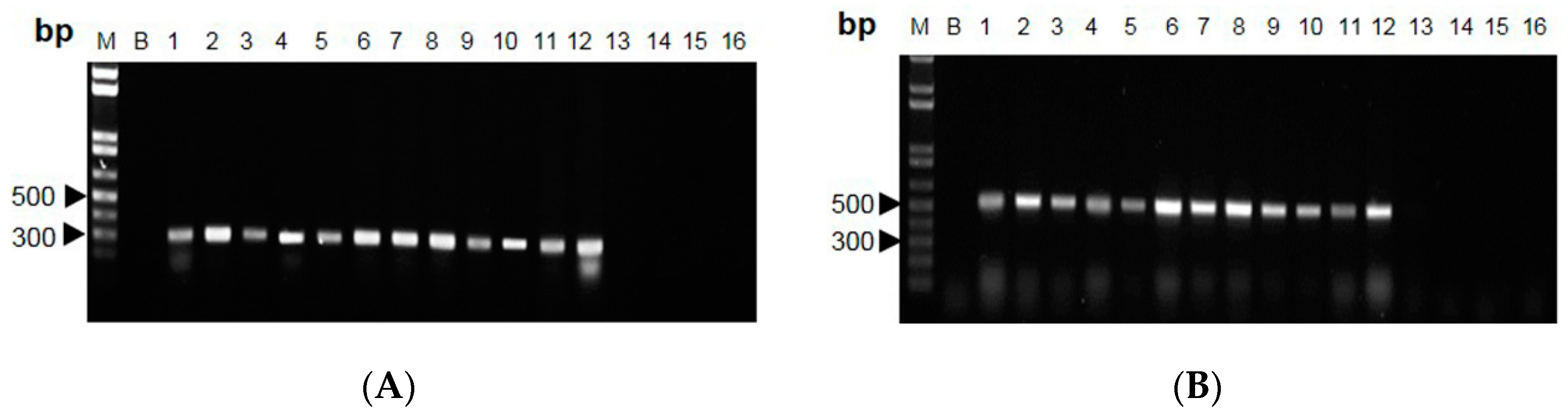
2.2. RAPD-SCAR Application in Cordyceps

3. Discussion
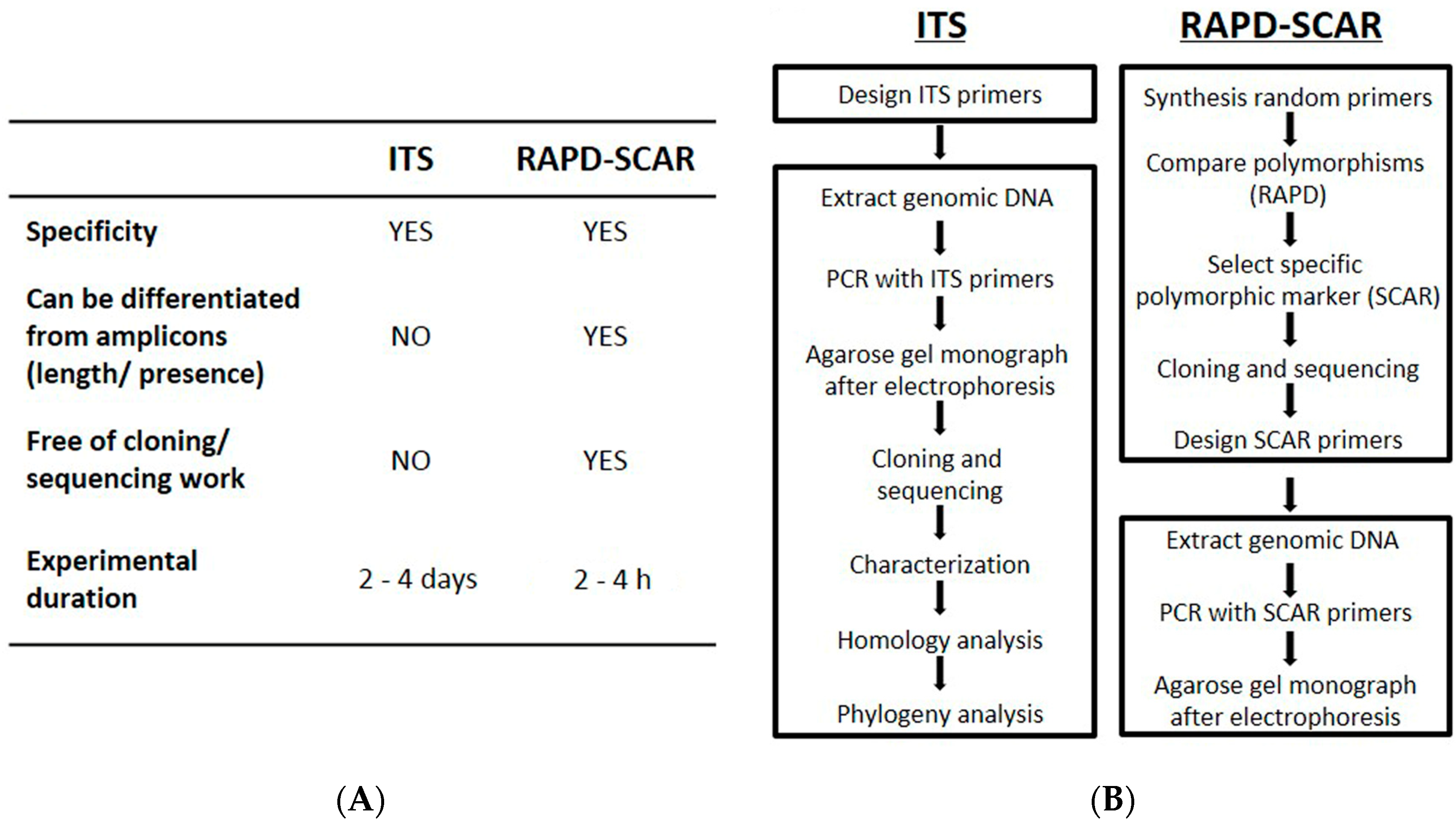
4. Materials and Methods
4.1. DNA Extraction
4.2. ITS Analysis
4.3. RAPD Analysis
4.4. RAPD Amplification and SCAR Design
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sung, G.H.; Hywel-Jones, N.L.; Sung, J.M.; Luangsa-Ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.C.; Su, Y.L.; Yang, H.L.; Chen, T.Y. Identification of Chinese medicinal fungus Cordyceps sinensis by PCR-single-stranded conformation polymorphism and phylogenetic relationship. J. Agric. Food Chem. 2005, 53, 3963–3968. [Google Scholar] [CrossRef] [PubMed]
- Chinese Pharmacopoeia Commission. The Pharmacopoeia of the Peopleʼs Republic of China, 2015 ed.; China Medical Science Press: Beijing, China, 2015. [Google Scholar]
- Chen, J.; Zhang, W.; Lu, T.; Li, J.; Zheng, Y.; Kong, L. Morphological and genetic characterization of a cultivated Cordyceps sinensis fungus and its polysaccharide component possessing antioxidant property in H22 tumor-bearing mice. Life Sci. 2006, 78, 2742–2748. [Google Scholar] [CrossRef] [PubMed]
- Techen, N.; Parveen, I.; Pan, Z.; Khan, I.A. DNA barcoding of medicinal plant material for identification. Curr. Opin. Biotechnol. 2014, 25, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Yue-Qin, C.; Ning, W.; Hui, Z.; Liang-Hu, Q. Differentiation of medicinal Cordyceps species by rDNA ITS sequence analysis. Planta Med. 2002, 68, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.C.; Su, Y.L.; Yang, H.L.; Huang, I.C.; Chen, T.Y. Differentiation of Cordyceps sinensis by a PCR-single-stranded conformation polymorphism-based method and characterization of the fermented products in Taiwan. Food Biotechnol. 2006, 20, 161–170. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, L.; Zhang, S.; Liu, X.; An, Z.; Wang, M.; Guo, Y. Genetic diversity of Ophiocordyceps sinensis, a medicinal fungus endemic to the Tibetan Plateau: Implications for its evolution and conservation. BMC Evol. Biol. 2009, 9, 290. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Wang, S.; Hu, D.J.; Yang, F.Q.; Wang, H.X.; Li, S.P. Random amplified polymorphic DNA (RAPD) analysis and the nucleosides assessment of fungal strains isolated from natural Cordyceps sinensis. J. Pharm. Biomed. Anal. 2009, 50, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; David, B.; Tu, P.; Barbin, Y. Recent analytical approaches in quality control of traditional Chinese medicines—A review. Anal. Chim. Acta 2009, 657, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Balasubramani, S.P.; Murugan, R.; Ravikumar, K.; Venkatasubramanian, P. Development of ITS sequence based molecular marker to distinguish, Tribulus terrestris L. (Zygophyllaceae) from its adulterants. Fitoterapia 2010, 81, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Bertini, L.; Amicucci, A.; Agostini, D.; Polidori, E.; Potenza, L.; Guidi, C.; Stocchi, V. A new pair of primers designed for amplification of the ITS region in Tuber species. FEMS Microbiol. Lett. 1999, 173, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.T.; Ma, X.Q.; Clarke, C.; Song, Z.H.; Ji, Z.N.; Lo, C.K.; Tsim, K.W. Phylogeny of Astragalus in China: Molecular evidence from the DNA sequences of 5S rRNA spacer, ITS, and 18S rRNA. J. Agric. Food Chem. 2003, 51, 6709–6714. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.J.; Dong, T.T.; Cui, X.M.; Tu, P.F.; Tsim, K.W. Genetic distinction of radix adenophorae from its adulterants by the DNA sequence of 5S-rRNA spacer domains. Am. J. Chin. Med. 2003, 31, 919–926. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Hou, P.; Fan, G.; Song, Z.; Arain, S.; Shu, H.; Tang, C.; Yue, Q.; Zhang, Y. Authentication of Angelica anomala Avé-Lall cultivars through DNA barcodes. Mitochondrial DNA 2012, 23, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, D.; Shanmughanandhan, D.; Sarma, R.K.; Joseph, J.C.; Srinivasan, R.V.; Ramalingam, S. DNA barcode ITS effectively distinguishes the medicinal plant Boerhavia diffusa from its adulterants. Genom. Proteom. Bioinform. 2013, 10, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Gao, T.; Yao, H.; Shi, L.; Zhu, Y.; Chen, S. Identification of Lonicera japonica and its related species using the DNA barcoding method. Planta Med. 2010, 77, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Marieschi, M.; Torelli, A.; Bruni, R. Quality control of saffron (Crocus sativus L.): Development of SCAR markers for the detection of plant adulterants used as bulking agents. J. Agric. Food Chem. 2012, 60, 10998–11004. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.M.; Lo, C.K.; Yip, K.L.; Dong, T.T.; Tsim, K.W. Authentication of Panax notoginseng by 5S-rRNA spacer domain and random amplified polymorphic DNA (RAPD) analysis. Planta Med. 2003, 69, 584–586. [Google Scholar] [PubMed]
- Um, J.Y.; Chung, H.S.; Kim, M.S.; Na, H.J.; Kwon, H.J.; Kim, J.J.; Lee, K.M.; Lee, S.J.; Lim, J.P.; Do, K.R.; et al. Molecular authentication of Panax ginseng species by RAPD analysis and PCR-RFLP. Biol. Pharm. Bull. 2001, 24, 872–875. [Google Scholar] [CrossRef] [PubMed]
- Xin, G.Z.; Lam, Y.C.; Maiwulanjiang, M.; Chan, G.K.; Zhu, K.Y.; Tang, W.L.; Dong, T.T.; Shi, Z.Q.; Li, P.; Tsim, K.W. Authentication of Bulbus Fritillariae Cirrhosae by RAPD-derived DNA markers. Molecules 2014, 19, 3450–3459. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.R.D. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- BLAST Algorithm, 2nd ed.; An Algorithm for Comparing Primary Biological Sequence Information. National Center of Biotechnology Information: Bethesda, MD, USA, 2002. Available online: http://www.ncbi.nlm.nih.gov/BLAST/ (accessed on 11 December 2015).
- Sample Availability: Samples of the Cordyceps are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lam, K.Y.C.; Chan, G.K.L.; Xin, G.-Z.; Xu, H.; Ku, C.-F.; Chen, J.-P.; Yao, P.; Lin, H.-Q.; Dong, T.T.X.; Tsim, K.W.K. Authentication of Cordyceps sinensis by DNA Analyses: Comparison of ITS Sequence Analysis and RAPD-Derived Molecular Markers. Molecules 2015, 20, 22454-22462. https://doi.org/10.3390/molecules201219861
Lam KYC, Chan GKL, Xin G-Z, Xu H, Ku C-F, Chen J-P, Yao P, Lin H-Q, Dong TTX, Tsim KWK. Authentication of Cordyceps sinensis by DNA Analyses: Comparison of ITS Sequence Analysis and RAPD-Derived Molecular Markers. Molecules. 2015; 20(12):22454-22462. https://doi.org/10.3390/molecules201219861
Chicago/Turabian StyleLam, Kelly Y. C., Gallant K. L. Chan, Gui-Zhong Xin, Hong Xu, Chuen-Fai Ku, Jian-Ping Chen, Ping Yao, Huang-Quan Lin, Tina T. X. Dong, and Karl W. K. Tsim. 2015. "Authentication of Cordyceps sinensis by DNA Analyses: Comparison of ITS Sequence Analysis and RAPD-Derived Molecular Markers" Molecules 20, no. 12: 22454-22462. https://doi.org/10.3390/molecules201219861





