2.1. The Metabolic Rates of Flavonoids by Intestinal Enzyme and Intestinal Flora of Rats
Five prenylated flavonoids were incubated with rat intestinal enzyme and intestinal flora solution and the degradation products were analyzed with time by HPLC-UV and LC/LC/MS. The HPLC-UV chromatograms of five prenylated flavonoids are shown in
Figure 2. We can see that there were two metabolites and one metabolite created from icariin after hydrolysis by intestinal enzyme and intestinal flora, respectively, whereas epimedin A, epimedin B and epimedin C gave only one metabolite with both intestinal enzyme and intestinal flora. As to baohuoside I, no hydrolysis products were found with either intestinal enzyme or intestinal flora.
To intuitively calculate the drug metabolic rates, the logarithmic concentrations of flavonoids (Y) and time (X/h) were used to get the corresponding regression equations.
Figure 3 shows the metabolic results for intestinal enzyme and
Figure 4 illustrates the metabolic results for intestinal flora. A lower slope value indicates a higher metabolic rate. Epimedin A, epimedin B, epimedin C and icariin all had higher metabolic rates with intestinal enzyme (the slope values were −0.0706 ± 0.00010, −0.0248 ± 0.00021, −0.0438 ± 0.00015, −0.2551 ± 0.00025, respectively) than with intestinal flora (the slope values were −0.0098 ± 0.00025, −0.0158 ± 0.00011, −0.0085 ± 0.00050, −0.0176 ± 0.00015, respectively). Baohuoside I had a similar metabolic rate with intestinal enzyme and flora, and the slope values were −0.0019 ± 0.00015 and −0.0018 ± 0.00011, respectively. Except for baohuoside I, the slope values for epimedin A, epimedin B, epimedin C and icariin showed significant differences between intestinal enzyme and intestinal flora (
p < 0.05). The sequence of metabolic rates for intestinal enzyme was icariin > epimedin A > epimedin C > epimedin B > baohuoside I, and the order of metabolic rates for intestinal flora was icariin > epimedin B > epimedin A > epimedin C > baohuoside I.
Figure 2.
HPLC-UV elution profiles of five prenylfalvonoids and their intestinal metabolites. (1) icariin; (2) M1; (3) baohuoside I; (4) testosterone; (5) epimedin A; (6) M3; (7) epimedin B; (8) M4; (9) epimedin C; (10) M5. Testosterone was used as an internal standard (IS). M1, M3, M4, M5 were defined as the metabolites.
Figure 2.
HPLC-UV elution profiles of five prenylfalvonoids and their intestinal metabolites. (1) icariin; (2) M1; (3) baohuoside I; (4) testosterone; (5) epimedin A; (6) M3; (7) epimedin B; (8) M4; (9) epimedin C; (10) M5. Testosterone was used as an internal standard (IS). M1, M3, M4, M5 were defined as the metabolites.
Figure 3.
The metabolic rates of flavonoids by intestinal enzyme of rats.
Figure 3.
The metabolic rates of flavonoids by intestinal enzyme of rats.
Figure 4.
The metabolic rates of flavonoids by intestinal flora of rats.
Figure 4.
The metabolic rates of flavonoids by intestinal flora of rats.
Our data suggest that intestinal hydrolysis of glycosides by intestinal enzymes is rapid. Even icariin was completely metabolized in 6 h and the epimedin A was totally metabolized in 12 h in incubations with intestinal enzyme. These results inform that intestinal glycosidases are brush-border enzymes. LPH is the only mammalian brush border-glucosidase, thus, we hypothesize that LPH is responsible for the observed hydrolysis of prenylated flavonoids in YYH. LPH has two distinct catalytic active sites, one for the hydrolysis of lactose and flavonoid glucosides and another, phlorizin hydrolase, for the hydrolysis of phlorizin and –glucosylceramides [
16]. As reported by Wilkinson
et al. [
17], LPH plays a major role in the deglycosylation of daidzin. In our previous research, we used gluconolactone, a LPH enzyme inhibitor, to see if LPH was involved in the hydrolysis of the flavonoids. If LPH plays an important role in absorption of icariin, epimedin A, epimdin B, epimedin C, and baohuoside I, then low activity of LPH would result in a reduced rate of metabolism.
The data of HPLC-UV show that flavonoid metabolic rates with rat intestinal enzyme were higher than those with intestinal flora. Moreover, LPH is located in the mammalian small intestine, and the intestinal flora usually exists in the large intestine. After oral administration, the drug lagged in the small intestine for a long time before it reached the large intestine, so if this situation occurs as fast in humans as we observed in the rats, the role played by intestinal flora in hydrolyzing glycosides would be significantly diminished.
2.2. Identification of Flavonoid Metabolites with Intestinal Flora and Intestinal Enzyme of Rats
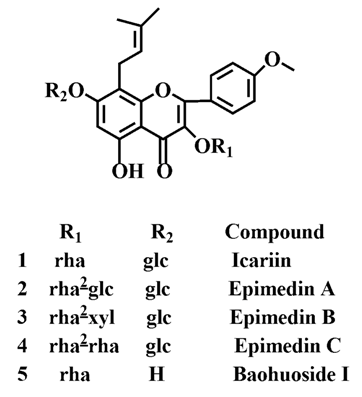
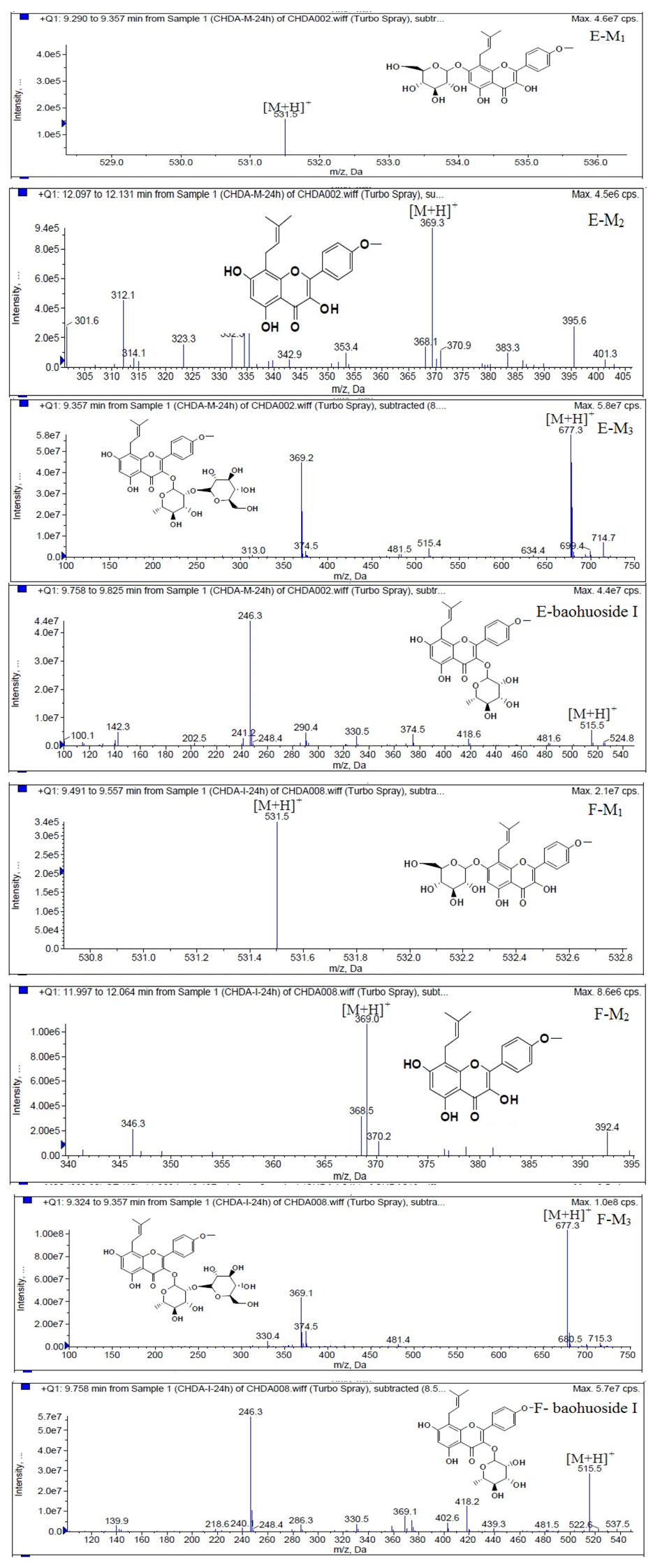
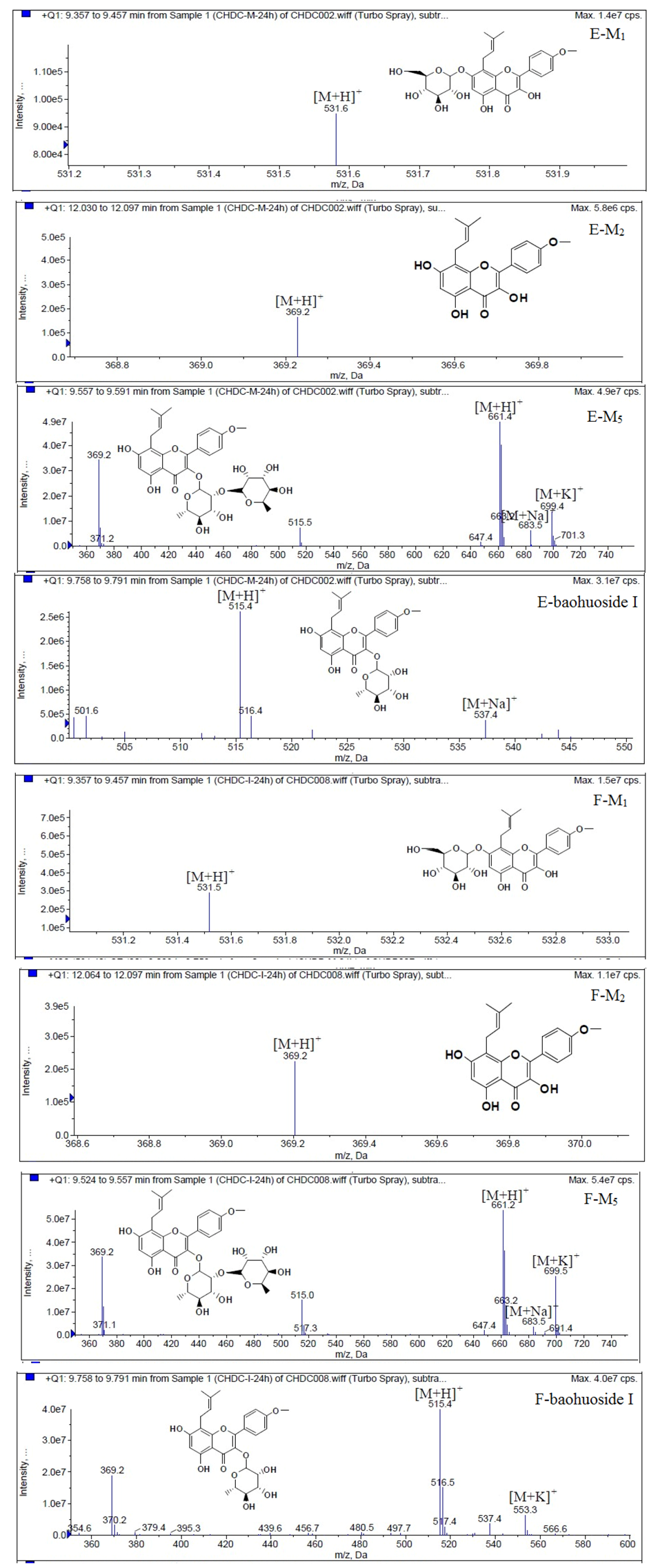
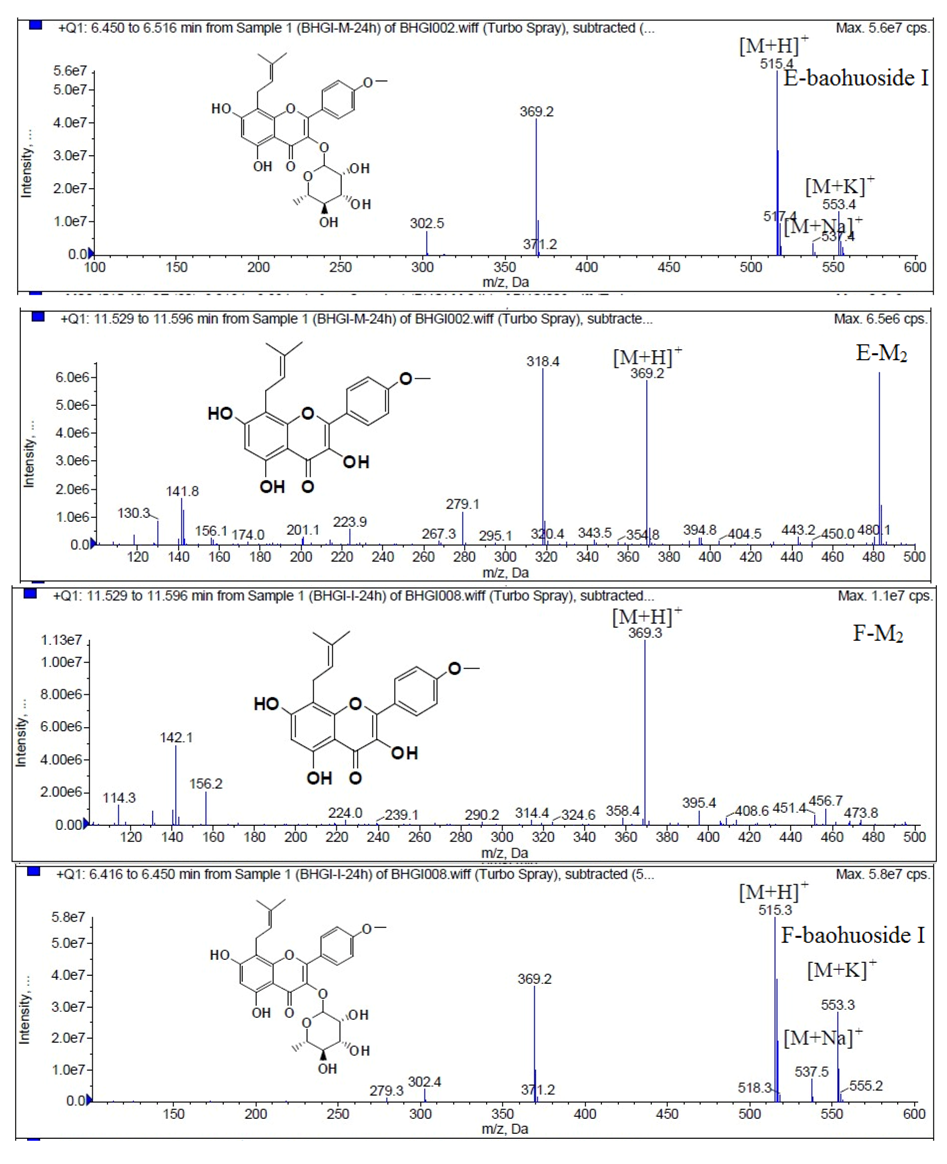
LC/MS/MS was used for the purpose of identifying the metabolites of the flavonoids and finding if there any other metabolites not being detected in HPLC-UV. Positive mode ESI mass spectroscopy was used in this study, [M+H]
+ ions of sufficient abundance could be subjected to MS
n analysis and provided much structural information. LC-MS/MS spectra of the metabolites of icariin, epimedin A, epimedin B, epimedin C and baohuoside I were obtained via fragmentation of molecular ions were used for more precise structural identification of metabolites. The chromatographic and mass spectrometry conditions were optimized for icariin, epimedin A, epimedin B, epimedin C and baohuoside I. Full scan mass spectrum analysis of icariin, epimedin A, epimedin B, epimedin C and baohuoside I showed the presence of [M+H]
+ at
m/
z 677, 839, 809, 823, 515 and [M+K]
+ at
m/
z 715, 877, 847, 862, 553 respectively. The MS
2 daughter ions spectrum of icariin, epimedin A, epimedin B, epimedin C and baohuoside I are shown in
Figure 5. Fragmentation of icariin in the ion trap leaded to four product ions at
m/
z 313, 369, 515, 531. The most abundant product ion at
m/
z 531 were formed by the loss of rhamnose (146 Da). The product ion at
m/
z 369 was produced by the loss of rahmnose and glucose (308 Da). The product ion at
m/
z 515 was generated by the loss of glucose. The MS
2 spectra of epimedin A shows three identical fragment ions at
m/
z 369, 531, 677. The fragment ion at
m/
z 369 was attributed to the loss of rahmnose and bimolecular glucose. The product ion at
m/
z 531 was generated through the reduction of bimolecular glucose and the ion at
m/
z 677 was produced via the loss of glucose. The daughter ions appearing at
m/
z 369, 531, 677 in the MS
2 spectra of epimedin B were produced by the neutral loss of 440, 278, 132 Da, corresponding to the losses of glucose, rhamnose and xylose, glucose and xylose, xylose, respectively. Epimedin C yielded five main daughter ions at
m/
z 313, 369, 515, 531, 677. The fragment ions at
m/
z 369, 515, 531 and 677 were generated by the reduction of glucose and bimolecular rhamnose, glucose and rhamnose, bimolecular rhamnose, rhamnose respectively. Baohuoside I gave two product ions at
m/
z 313 and 369, in which the latter was attributed to the loss of rhamnose. All the product ions at
m/
z 313 were 56 Da less than
m/
z 369 by the loss of C
4H
7 (56 Da), which are produced by the rearrangement of the isopentene group at the position 8 of the A-ring. Above all, these characterisitic product ions and neutral losses were a sound basis to identify the metabolites of icariin, epimedin A, epimedin B, epimedin C and baohuoside I (
Figure 6,
Figure 7,
Figure 8,
Figure 9 and
Figure 10).
Figure 5.
Mass spectra corresponding to prenylflavonoids. (A) MS and MS2 spectrum of icariin; (B) MS and MS2 spectrum of epimedin A; (C) MS and MS2 spectrum of epimedin B; (D) MS and MS2 spectrum of epimedin C; (E) MS and MS2 spectrum of baohuoside I.
Figure 5.
Mass spectra corresponding to prenylflavonoids. (A) MS and MS2 spectrum of icariin; (B) MS and MS2 spectrum of epimedin A; (C) MS and MS2 spectrum of epimedin B; (D) MS and MS2 spectrum of epimedin C; (E) MS and MS2 spectrum of baohuoside I.
Figure 6.
Mass spectra of metabolite of icariin. E for metabolites in intestinal enzyme; F for metabolites in intestinal flora.
Figure 6.
Mass spectra of metabolite of icariin. E for metabolites in intestinal enzyme; F for metabolites in intestinal flora.
Figure 7.
Mass spectra of metabolite of epimedin A. E for metabolites in intestinal enzyme; F for metabolites in intestinal flora.
Figure 7.
Mass spectra of metabolite of epimedin A. E for metabolites in intestinal enzyme; F for metabolites in intestinal flora.
Figure 8.
Mass spectra of metabolite of epimedin B. E for metabolites in intestinal enzyme; F for metabolites in intestinal flora.
Figure 8.
Mass spectra of metabolite of epimedin B. E for metabolites in intestinal enzyme; F for metabolites in intestinal flora.
Figure 9.
Mass spectra of metabolite of epimedin C. E for metabolites in intestinal enzyme; F for metabolites in intestinal flora.
Figure 9.
Mass spectra of metabolite of epimedin C. E for metabolites in intestinal enzyme; F for metabolites in intestinal flora.
Figure 10.
Mass spectra of metabolite of baohuoside I. E for metabolites in intestinal enzyme; F for metabolites in intestinal flora.
Figure 10.
Mass spectra of metabolite of baohuoside I. E for metabolites in intestinal enzyme; F for metabolites in intestinal flora.
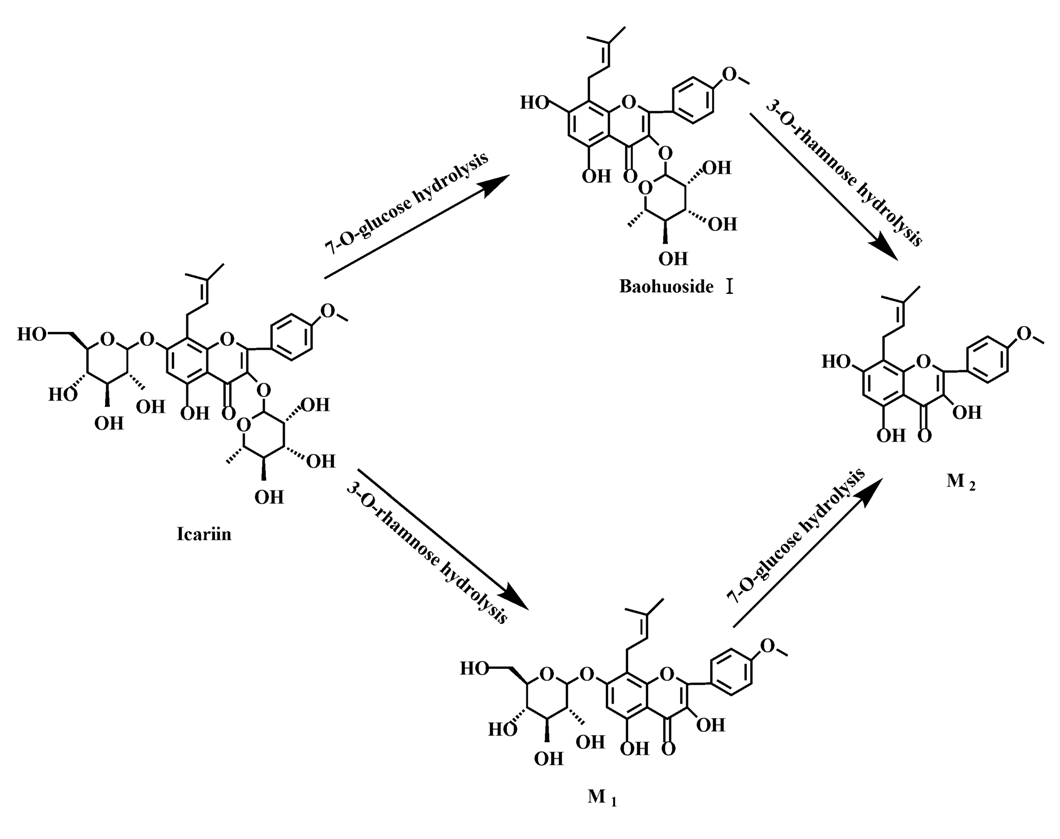
Based on our analysis, the metabolic pathways of icariin by rat intestinal flora and enzyme solution were basically the same, including 3-
O-rhamnose, 7-
O-glucose hydrolysis or dual 3, 7-hydrolysis (
Figure 11). Compared to 3-
O-rhamnose and dual hydrolysis of 3, 7-, the 7-
O-glucose hydrolysis was easier. The yielded metabolites contained M
1 (icariside I), M
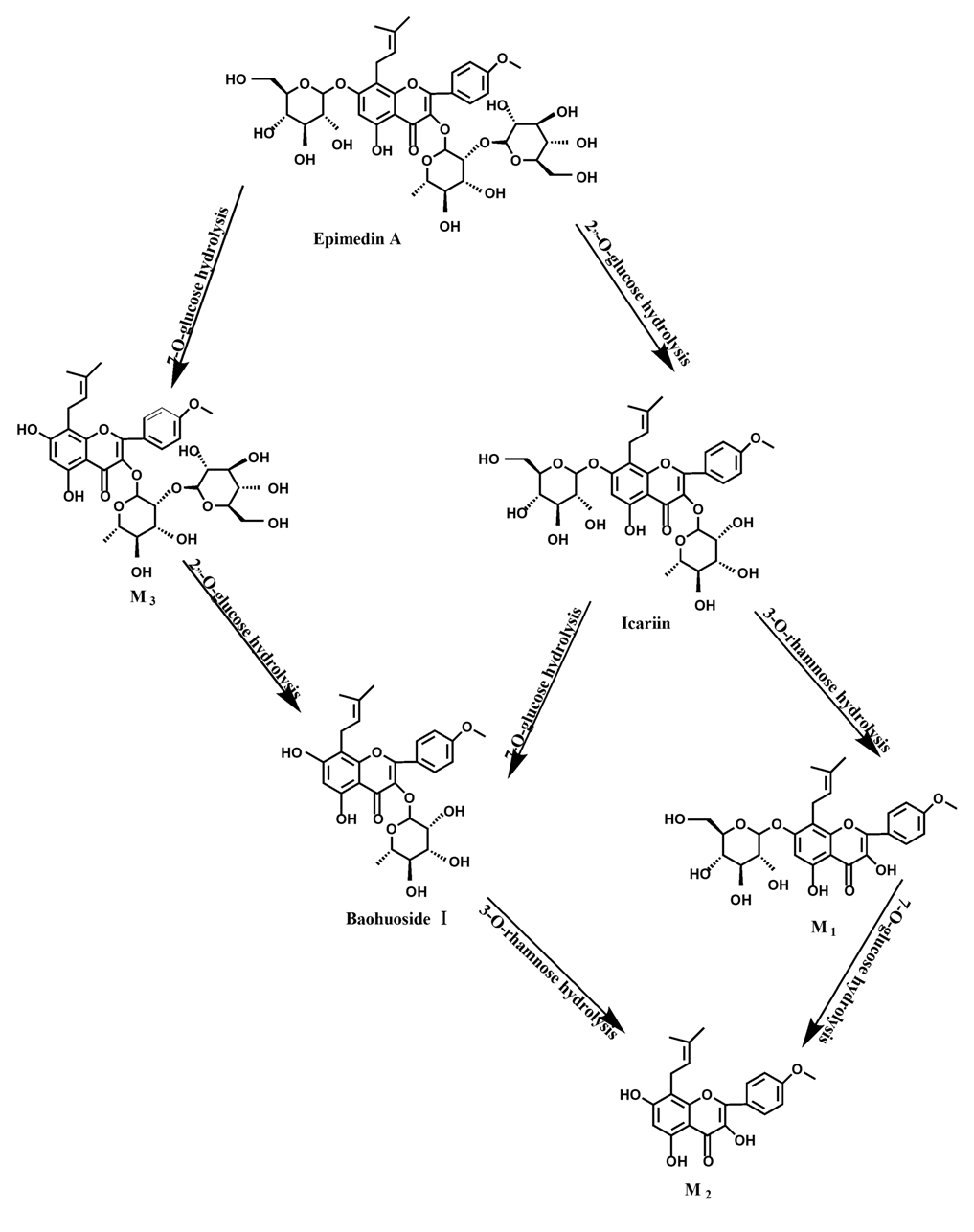
2 (icaritin), and baohuoside I. After incubation for 24 h, the parent drug, icariin, could not be found in the intestinal enzyme solution, but it was still visible in intestinal flora solution at the same time point, which demonstrated that the intestinal enzyme could more easily hydrolyze icariin. The results concluded from LC/MS/MS told that the metabolism of epimedin A by intestinal enzyme and intestinal flora were similar to each other. The metabolites included icariin, M
1, M
2, M
3 (sagittatoside A) and baohuoside I, which were generated by 3-
O-glucose-(1→2)-rhamnose hydrolysis and 7-
O-glucose hydrolysis (
Figure 12). Comparatively speaking, the 2"-
O-glucose and 7-
O-glucose could be removed more easily. The medicine precursor could be found in the intestinal flora of rats but not in the intestinal enzyme after incubation for 24 h. This phenomenon indicated that epimedin A was quickly metabolized by intestinal enzyme. M
1 and M
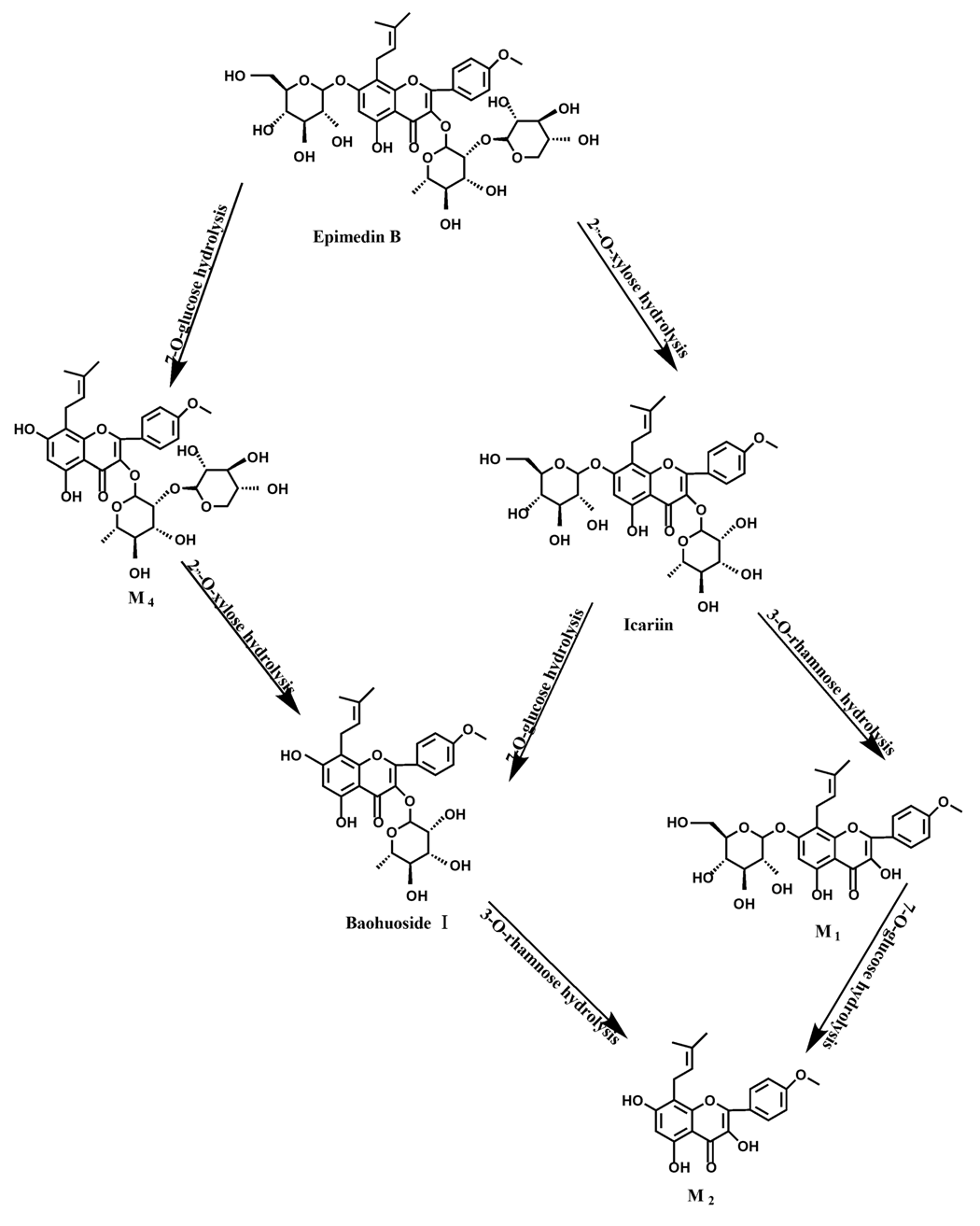
2 instead of icariin were detected in the incubated sample of 24 h, which demonstrated that icariin was quite easily hydrolyzed further. The sample of epimedin B incubated in intestinal enzyme and intestinal flora was determined by LC/MS/MS. Metabolic pathways by intestinal enzyme and intestinal flora were the same, including 3-
O-xylose-(1→2)-rhamnose hydrolysis and 7-
O-glucose hydrolysis (
Figure 13). Icariin, M
1, M
2, M
4 (sagittatoside B) and baohuoside I were the metabolites. The order of hydrolysis from easy to difficult is 7-
O-glucose > 2"-
O-xylose > 3-
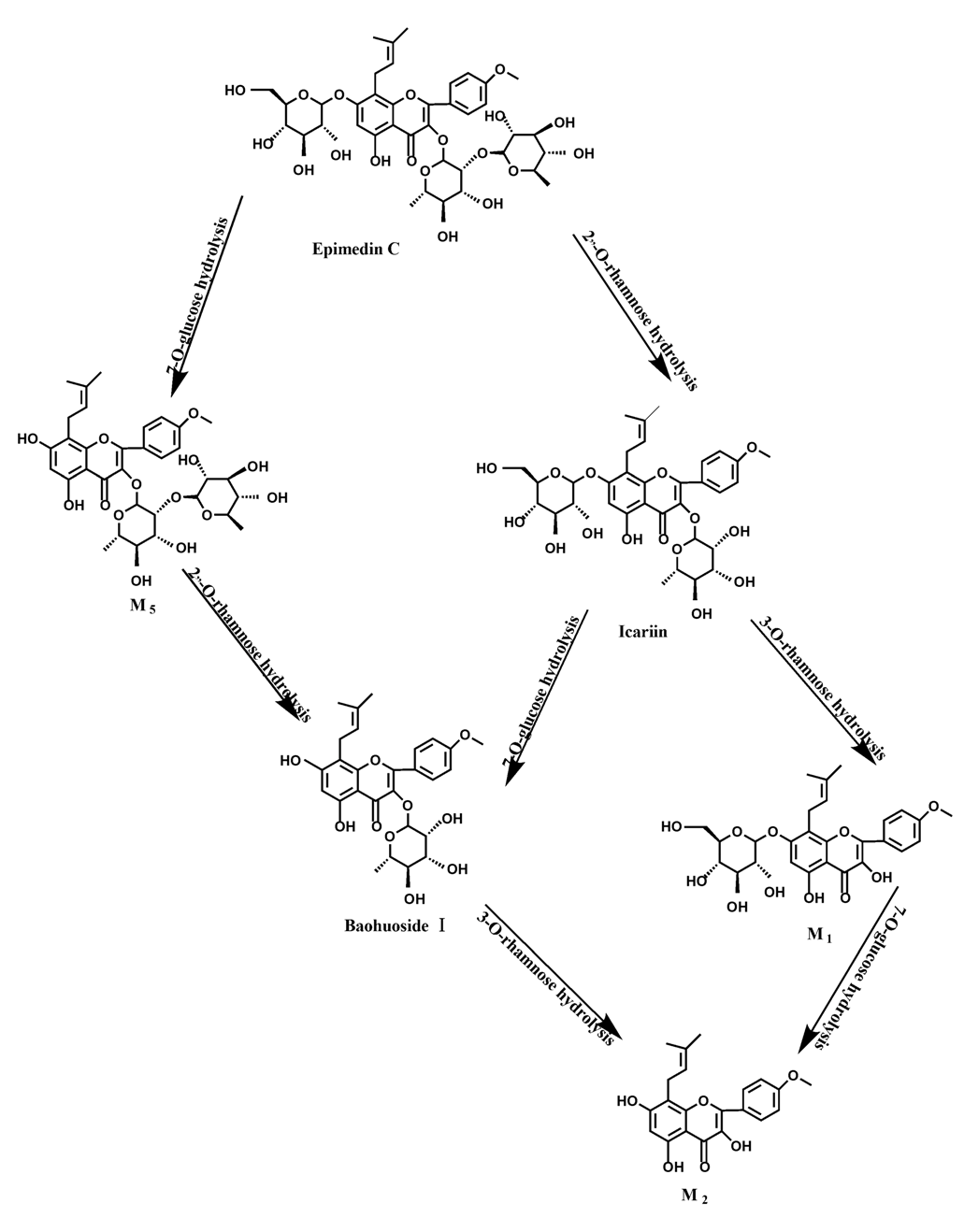
O-rhamnose. Like epimedin A, M1 and M2 replaced icariin were found in the incubated sample of epimedin B at 24 h. The conclusions summarized from LC/MS/MS data show that the metabolism of epimedin C by intestinal enzyme and intestinal flora were similar to each other. The metabolites contained icariin, M
1, M
2, M
5 (2"-
O-rhamonosylicariside) and baohuoside I, which were generated by 3-
O-rhamnose-(1→2)-rhamnose hydrolysis and 7-
O-glucose hydrolysis (
Figure 14). Comparatively speaking, the 7-
O-glucose could be removed more easily and 3-
O-rhamnose was difficult to hydrolyze. M
1 and M
2 replacing icariin could be detected in the incubated sample at 24 h demonstrating that icariin was quite easily hydrolyzed further. This phenomenon also ocurred in epimedin A and B. Simultaneously, the content of baohuoside I in samples after incubation of epimedin C was significantly higher than that of epimedin A and B. However, the contents of M
1 and M
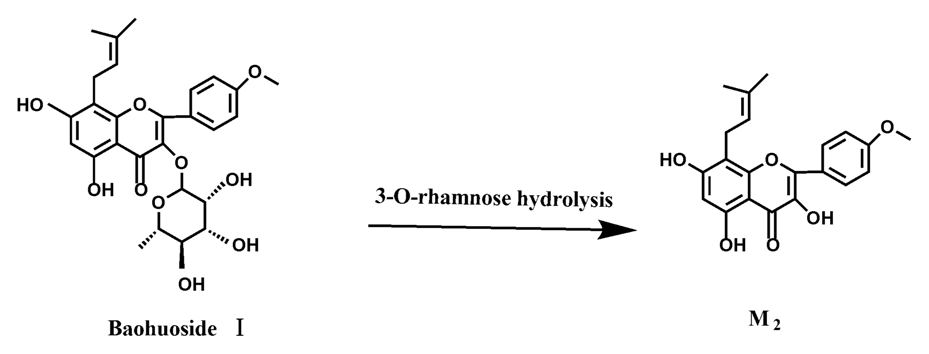
2 were so low that they could be barely be measured. Presumably these were two rhamnosyls in the molecular structure of epimedin C which made epimedin C hydrolyze more difficultly in the intestinal enzyme and intestinal flora incubations. The metabolic pathway of baohuoside I by intestinal flora was consistent with that by intestinal enzyme, which was only 3-
O-rhamnose hydrolysis (
Figure 15). After incubation for 24 h with both intestinal flora and intestinal enzyme, a high content of baohuoside I could be detected. It indicated that 3-
O-rhamnose was hard to hydrolyze by either intestinal flora or intestinal enzyme.
Figure 11.
The metabolic pathway of icariin in intestinal flora and enzyme of rats.
Figure 11.
The metabolic pathway of icariin in intestinal flora and enzyme of rats.
Figure 12.
The metabolic pathway of epimedin A in intestinal flora and enzyme of rats.
Figure 12.
The metabolic pathway of epimedin A in intestinal flora and enzyme of rats.
Figure 13.
The metabolic pathway of epimedin B in intestinal flora and enzyme of rats.
Figure 13.
The metabolic pathway of epimedin B in intestinal flora and enzyme of rats.
Figure 14.
The metabolic pathway of epimedin C in intestinal flora and enzyme of rats.
Figure 14.
The metabolic pathway of epimedin C in intestinal flora and enzyme of rats.
Figure 15.
The metabolic pathway of baohuoside I in intestinal flora and enzyme of rats.
Figure 15.
The metabolic pathway of baohuoside I in intestinal flora and enzyme of rats.
Icariin is a dual glucoside, it was thought to be metabolized by hydrolysis to first produce baohuoside I after removal of 7-O-glucose and then baohuoside I was metabolized to produce M2 after further removal of 3-O-rhamnose. Meanwhile, icariin could be metabolized to generate M1 after only removal of 3-O-rahamnose. Epimedin A, B, C were triple glucosides with similar structures, and besides the M3 for epimedin A, M4 for epimedin B, M5 for epimedin C, they had other four common metabolites: icariin, M1, M2 and baohuoside I. Epimedin A was considered to be metabolized by hydrolysis to first produce M3 after removal of the 2"-O-glucose and then M3 was metabolized to produce baohuoside I after further removal of 3-O-rhamnose. Epimedin A also can be metabolized by removal of 7-O-glucose to then produce icariin. The generated icariin had the same metabolic pathway as above. Epimedin B was thought to be metabolized by hydrolysis to first produce icariin which had the same metabolic pathway as above after removal of 7-O-glucose. Epimedin B also can be metabolized by removal of 2"-O-oxylose to next produce M4 and then M4 was metabolized to produce baohuoside I after further removal of 3-O-rhamnose. Epimedin C was thought to be metabolized by hydrolysis to first produce icarrin which had the same metabolic pathway as above after removal of 7-O-glucose. Epimedin C also can be metabolized by removal of 3-O-rahmnose to secondly produce M5 and then M5 was metabolized to produce baohuoside I after further removal of 3-O-rhamnose. Presumably it was the presence of two rhamnosyls in the molecular structure of epimedin C which made epimedin C difficultly hydrolyzed by intestinal flora and intestinal enzyme. It can be deduced from the above data that icariin, epimedin A, B, C were generally absorbed as metabolites. On the other hand, as a single glucoside, it was difficult for baohuoside I to hydrolyze in the intestine and usually it was absorbed as the precursor.
Most of the previous studies consider that the deglycosylation of flavonoids may be caused by intestinal flora and hepatic biotransformation enzymes [
18,
19]. However, our study indicated that intestinal enzymes play an important role in the metabolism of the prenylated flavonoids present in YYH. Meanwhile, based on our analysis and literature data, the metabolic pathways of icariin, epimedin A, epimedin B, epimedin C and baohuoside I in intestinal flora and enzyme solution of rats were basically the same, and the absorption pathways of
Epimedium flavonoids in rat intestine initially involved deglycosylation of flavonoid glycosides, yielding the major hydrolytic metabolites. It demonstrated that the enzyme existing in flora was a β-enzyme which had the same function as LPH in hydrolyzing
Epimedium flavonoids. The metabolic effects in intestinal enzyme were far higher than those in intestinal flora. It might because of the expression and distribution of LPH in the upper gastrointestinal tract were comparatively large and there were no bacterial growth in the upper gastrointestinal tract.