A Family of Fitness Landscapes Modeled through Gene Regulatory Networks
Abstract
:1. Introduction
2. Methods
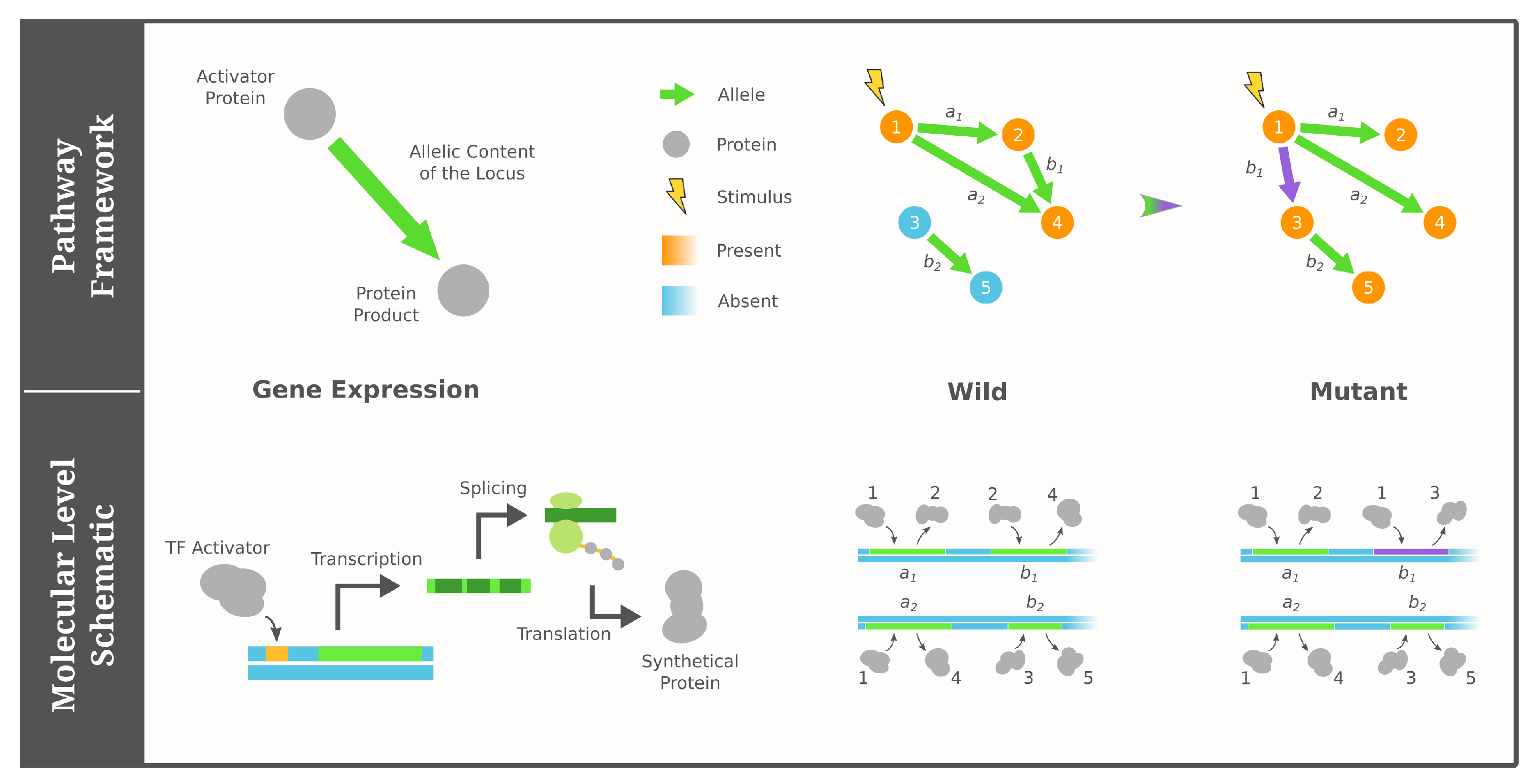
2.1. Pathway Framework of GRNs
2.2. Fitness Landscape of GRNs under the Pathway Framework
3. Results
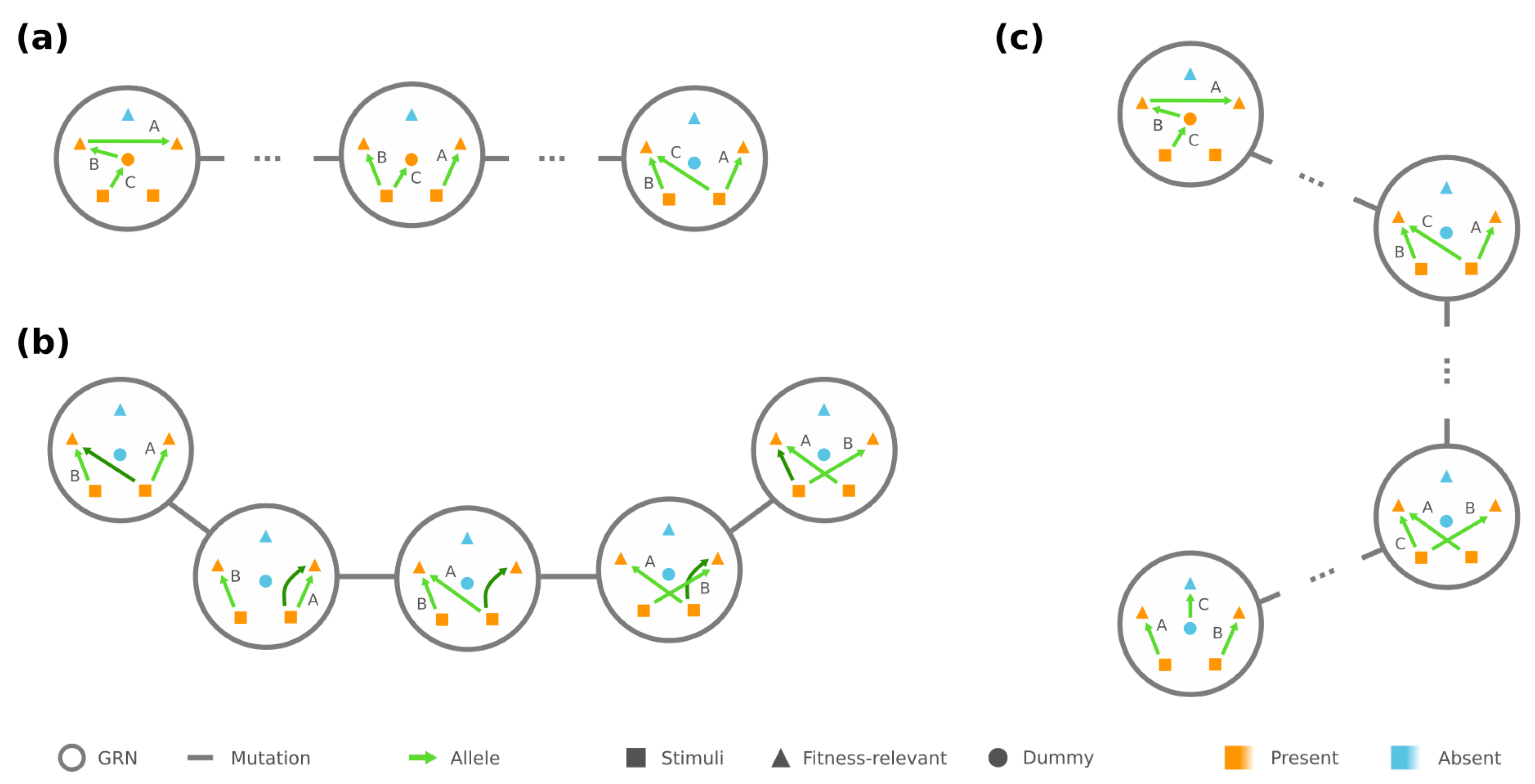
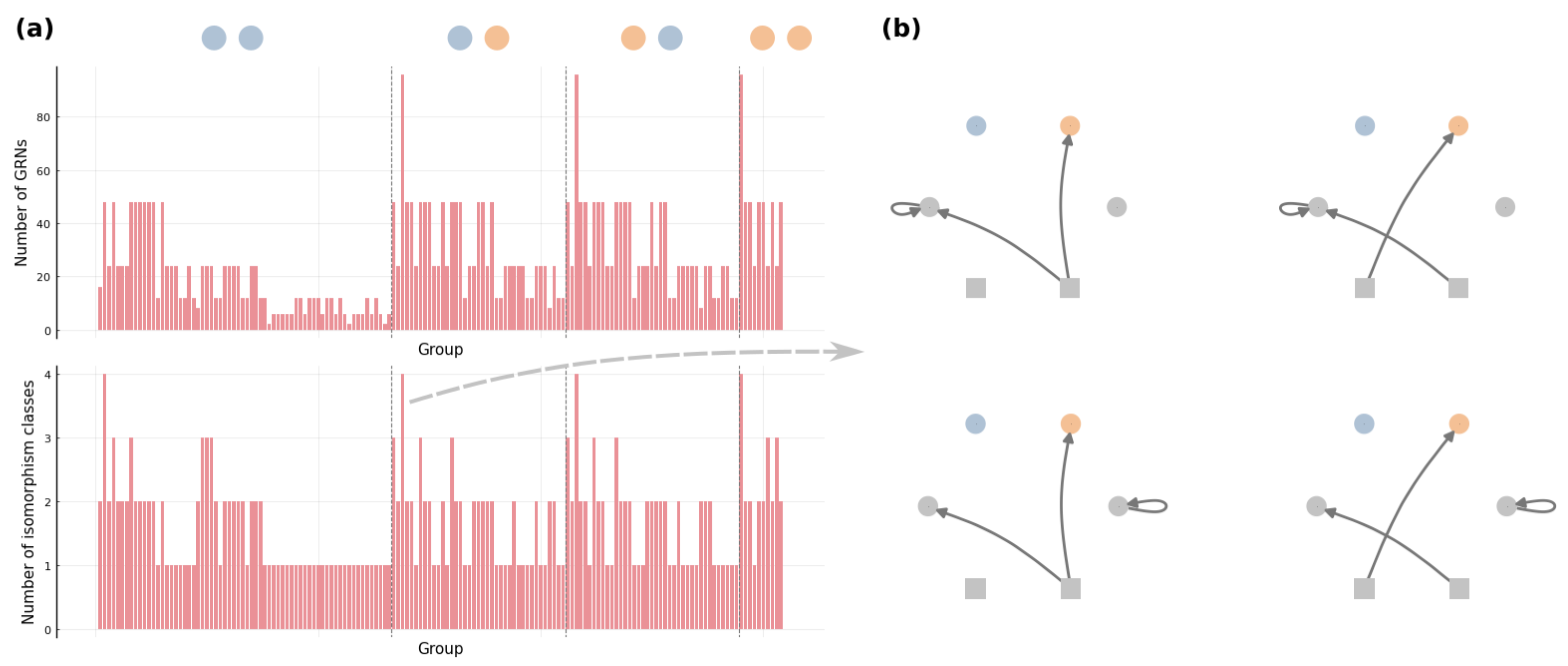
3.1. Connectivity and Accessibility in a Fitness Landscape of GRNs
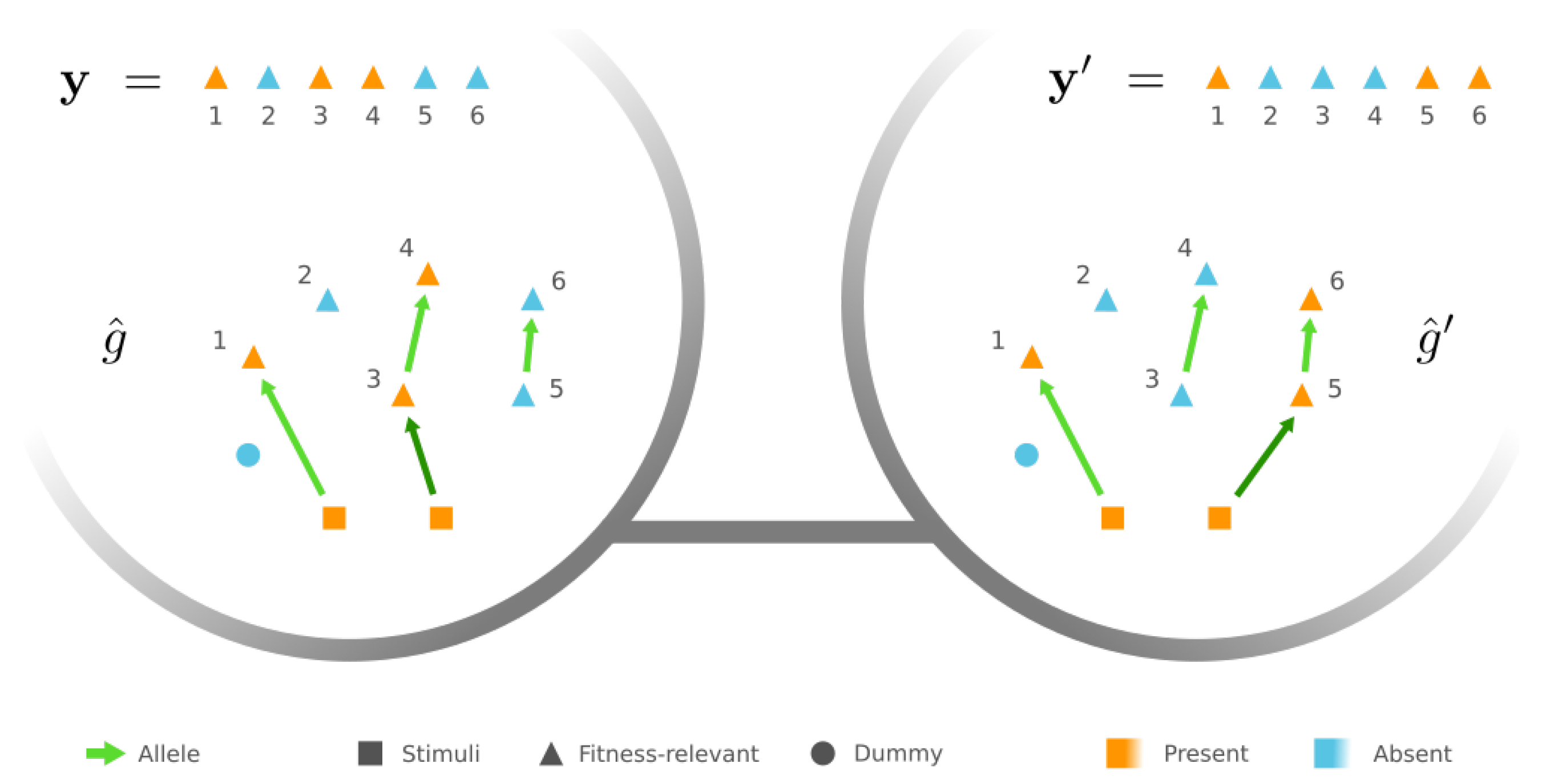
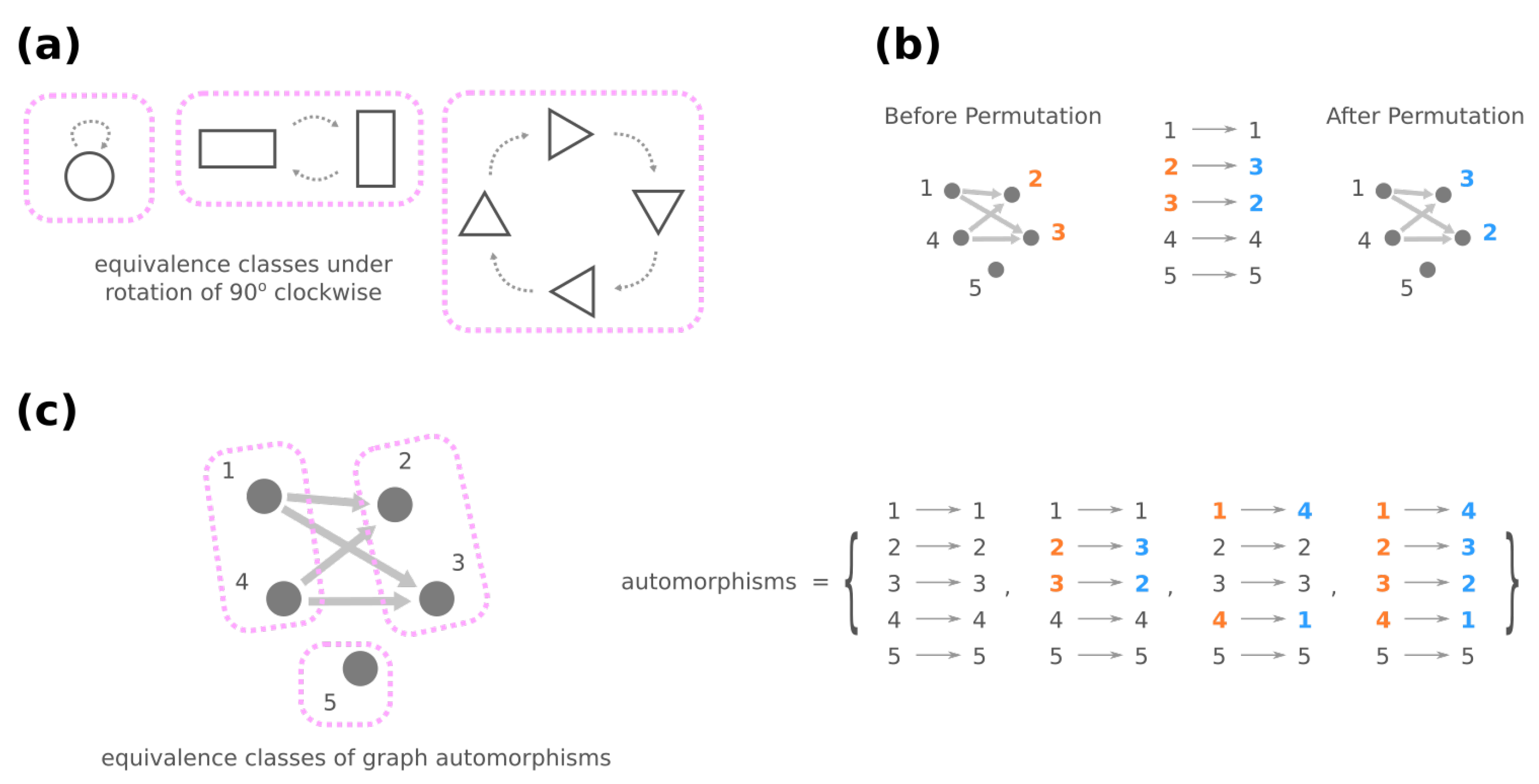
3.2. Mesoscopic Skeleton Derived from “Symmetries” in the Genotype Network of GRNs
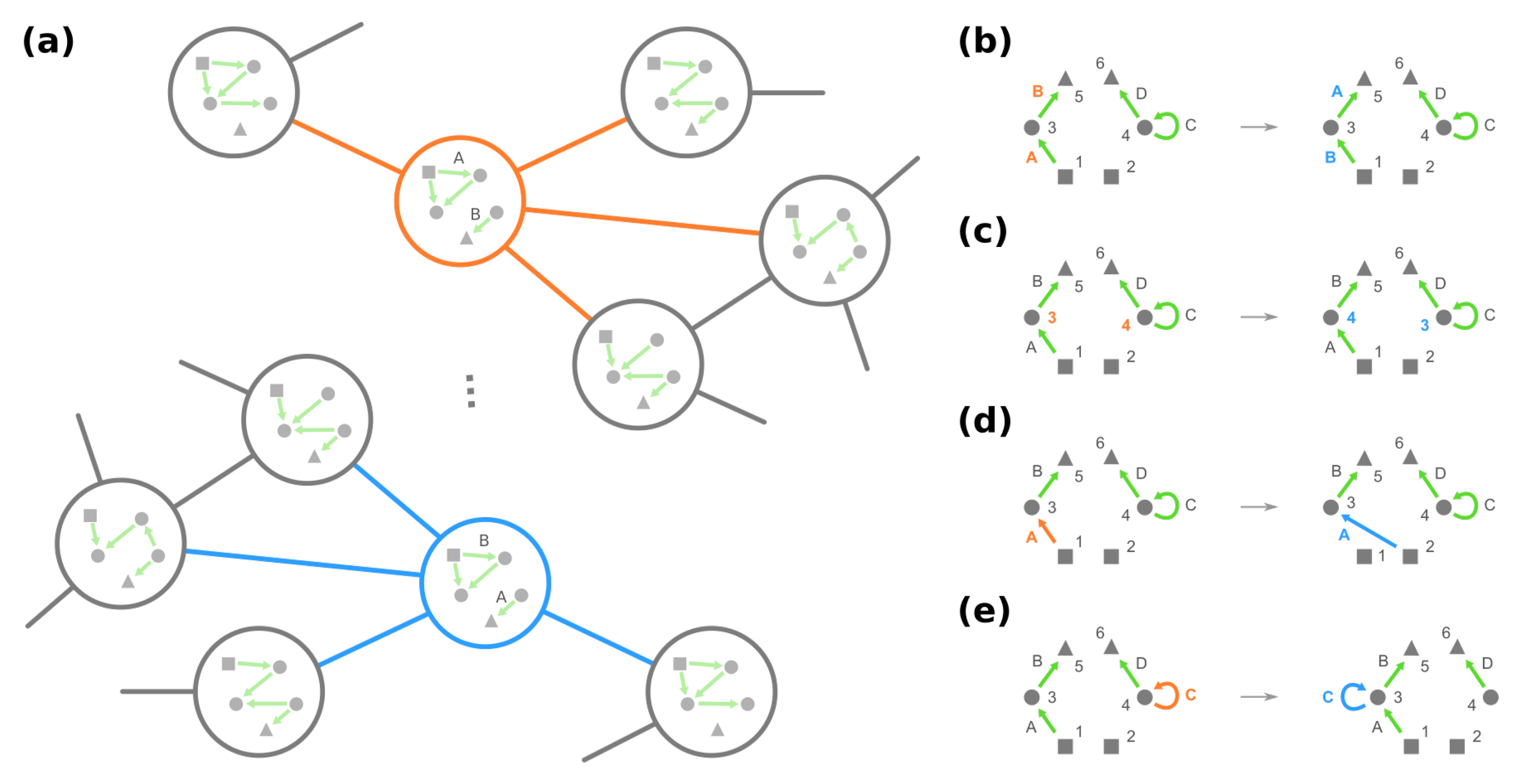
- (iii)
- Change the source node of an edge from one stimulus to another stimulus and vice versa, e.g., in Figure 4d, moving an edge pointing from node 1 to node 3 to pointing from node 2. (Note that this operation is not necessarily equivalent to permuting the identities of stimuli since at most only the single focal edge will be affected.)
- (iv)
- Move a self-loop at one node to another node and vice versa, for example, re-allocating a self-loop at node 3 to node 4 in Figure 4e.
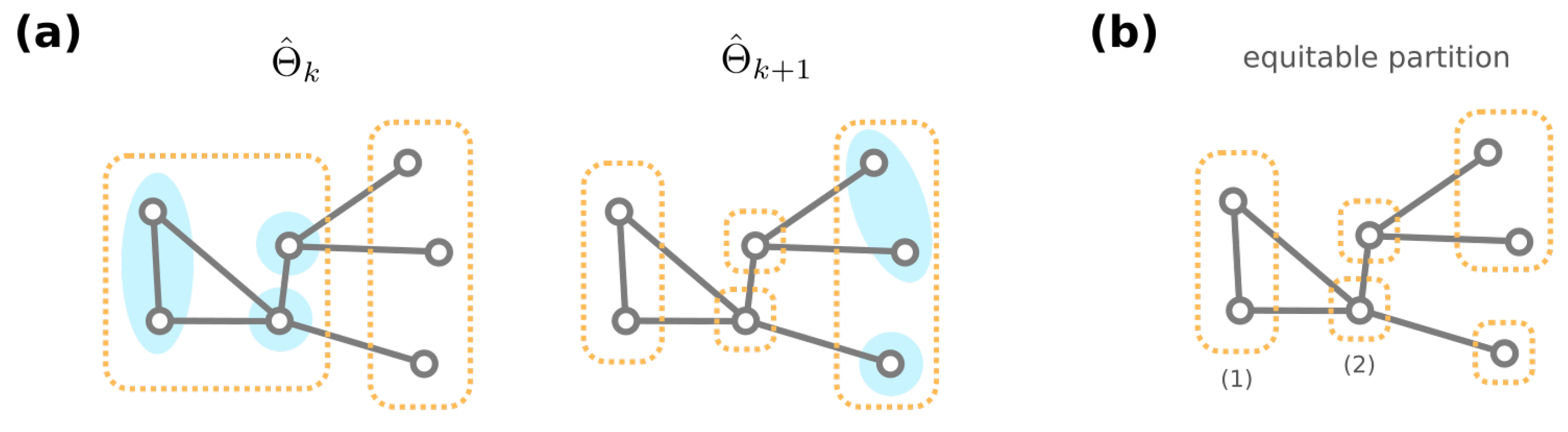
3.3. Algorithmic Construction of the Mesoscopic Backbone of GRN Fitness Landscape
- If has one more non-self-loop edge than g, then ;
- If has one less non-self-loop edge than g, then we have ;
- If has the same number of non-self-loop edges as g, and then they share a common mutational neighbor , where the only different edge between g and is rewired to a self-loop and thus .
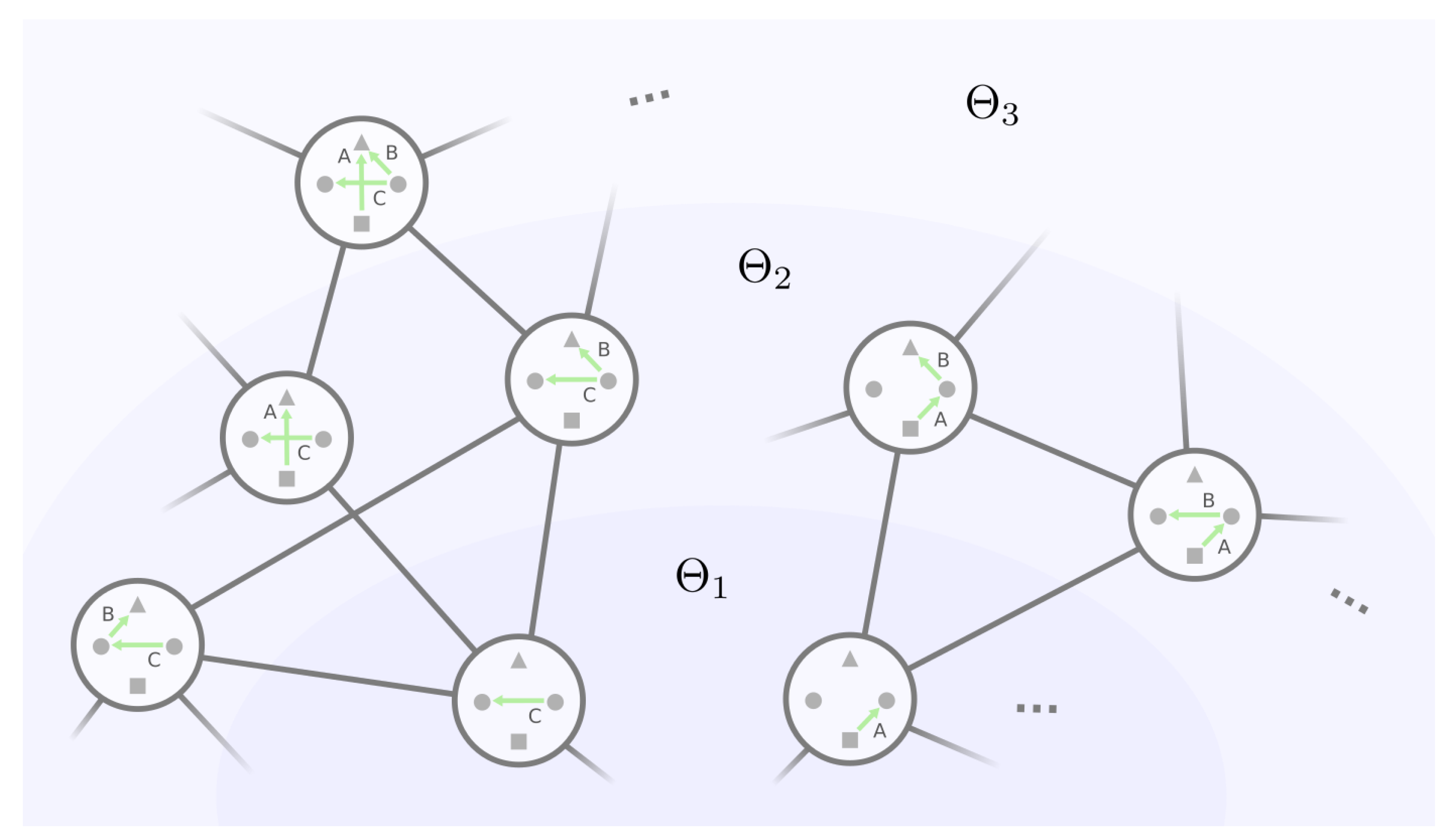
- (A)
- For an equivalence class and its representative GRN , under what condition will belong to the same equivalence class in layer ?
- (B)
- For two distinct equivalence classes and their representative GRNs and , under what condition will and belong to the same equivalence class in layer ?
- There is an integer p such that and ;
- There is another integer such that and ;
- for ;
- for ;
- For any locus and non-self-loop source–target pair such that for , we have if and only if .
- (I)
- For every representative GRN g in and every phenotype-preserving automorphism of g, there is an operation that joins together the groups of and , where and ;
- (II)
- For every representative GRN g in and every phenotype-preserving automorphism of each subgraph of g such that the edge differences are sequentially connected via , there is an operation that joins together the groups of and , where automorphism consecutively transforms edge into through ;
- (III)
- For every representative GRN in and each and in two different equivalence classes and , such that we have phenotype-preserving isomorphisms / from / to the representative GRN / after self-loop removal, there is an operation that joins together the groups of , and .
| Algorithm 1 Constructing the underlying space of a fitness landscape of GRNs |
| Require: The fixed underlying collections of loci and proteins of GRNs Ensure: The representative GRN of each equivalence class , and its number of mutational neighbors in any equivalence class
|
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Central and Peripheral GRNs Where No Regulation Presents
Appendix B. Phenotype-Preserving Automorphisms of the Genotype Network of GRNs
Appendix C. Combining Mutational Neighbors into Equivalence Classes
- (A)
- For an equivalence class and its representative GRN , under what condition will belong to the same equivalence class in layer ?
- (B)
- For two distinct equivalence classes and their representative GRNs and , under what condition will and belong to the same equivalence class in layer ?
- i.
- ;
- ii.
- ;
- iii.
- and for ;
- iv.
- for .
- i.
- and belong to the same equivalence class;
- ii.
- ;
- iii.
- .
Appendix D. Size of an Equivalence Class of GRNs
References
- Wright, S. The roles of mutation, inbreeding, crossbreeding, and selection in evolution. In Proceedings of the Sixth International Congress on Genetics, Ithaca, NY, USA, 24–31 August 1932; pp. 356–366. [Google Scholar]
- De Visser, J.A.G.M.; Krug, J. Empirical fitness landscapes and the predictability of evolution. Nat. Rev. Genet. 2014, 15, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Fragata, I.; Blanckaert, A.; Louro, M.A.D.; Liberles, D.A.; Bank, C. Evolution in the light of fitness landscape theory. Trends Ecol. Evol. 2019, 34, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Szendro, I.G.; Schenk, M.F.; Franke, J.; Krug, J.; De Visser, J.A.G. Quantitative analyses of empirical fitness landscapes. J. Stat. Mech. Theory Exp. 2013, 2013, P01005. [Google Scholar] [CrossRef] [Green Version]
- Wagner, A. The Origins of Evolutionary Innovations: A Theory of Transformative Change in Living Systems; Oxford University Press: Oxford, UK, 2011. [Google Scholar]
- Jain, K.; Krug, J. Deterministic and stochastic regimes of asexual evolution on rugged fitness landscapes. Genetics 2007, 175, 1275–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kryazhimskiy, S.; Tkačik, G.; Plotkin, J.B. The dynamics of adaptation on correlated fitness landscapes. Proc. Natl. Acad. Sci. USA 2009, 106, 18638–18643. [Google Scholar] [CrossRef] [Green Version]
- Draghi, J.A.; Parsons, T.L.; Wagner, G.P.; Plotkin, J.B. Mutational robustness can facilitate adaptation. Nature 2010, 463, 353–355. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.C.; Dai, L.; Olson, C.A.; Lloyd-Smith, J.O.; Sun, R. Adaptation in protein fitness landscapes is facilitated by indirect paths. eLife 2016, 5, e16965. [Google Scholar] [CrossRef]
- Gavrilets, S. Fitness Landscapes and the Origin of Species; Princeton University Press: Princeton, NY, USA, 2004. [Google Scholar]
- Fraïsse, C.; Gunnarsson, P.A.; Roze, D.; Bierne, N.; Welch, J.J. The genetics of speciation: Insights from fisher’s geometric model. Evolution 2016, 70, 1450–1464. [Google Scholar] [CrossRef] [Green Version]
- de Visser, J.A.G.; Park, S.C.; Krug, J. Exploring the effect of sex on empirical fitness landscapes. Am. Nat. 2009, 174, S15–S30. [Google Scholar] [CrossRef] [Green Version]
- Otto, S.P. The evolutionary enigma of sex. Am. Nat. 2009, 174, S1–S14. [Google Scholar] [CrossRef] [Green Version]
- Watson, R.A.; Weinreich, D.M.; Wakeley, J. Genome structure and the benefit of sex. Evol. Int. J. Org. Evol. 2011, 65, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Orr, H.A. The genetic theory of adaptation: A brief history. Nat. Rev. Genet. 2005, 6, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Lobkovsky, A.E.; Wolf, Y.I.; Koonin, E.V. Predictability of evolutionary trajectories in fitness landscapes. PLoS Comput. Biol. 2011, 7, e1002302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salverda, M.L.; Dellus, E.; Gorter, F.A.; Debets, A.J.; Van Der Oost, J.; Hoekstra, R.F.; Taw, D.S.; de Visser, J.A.G. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 2011, 7, e1001321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bank, C.; Matuszewski, S.; Hietpas, R.T.; Jensen, J.D. On the (un) predictability of a large intragenic fitness landscape. Proc. Natl. Acad. Sci. USA 2016, 113, 14085–14090. [Google Scholar] [CrossRef] [Green Version]
- Van Nimwegen, E.; Crutchfield, J.P.; Huynen, M. Neutral evolution of mutational robustness. Proc. Natl. Acad. Sci. USA 1999, 96, 9716–9720. [Google Scholar] [CrossRef] [Green Version]
- Van Nimwegen, E.; Crutchfield, J.P. Metastable evolutionary dynamics: Crossing fitness barriers or escaping via neutral paths? Bull. Math. Biol. 2000, 62, 799–848. [Google Scholar] [CrossRef] [Green Version]
- Wilke, C.O. Adaptive evolution on neutral networks. Bull. Math. Biol. 2001, 63, 715–730. [Google Scholar] [CrossRef] [Green Version]
- Smith, T.; Husbands, P.; O’Shea, M. Neutral networks and evolvability with complex genotype-phenotype mapping. In European Conference on Artificial Life; Springer: Berlin/Heidelberg, Germany, 2001; pp. 272–281. [Google Scholar]
- Aguirre, J.; Catalán, P.; Cuesta, J.A.; Manrubia, S. On the networked architecture of genotype spaces and its critical effects on molecular evolution. Open Biol. 2018, 8, 180069. [Google Scholar] [CrossRef] [Green Version]
- Manrubia, S.; Cuesta, J.A.; Aguirre, J.; Ahnert, S.E.; Altenberg, L.; Cano, A.V.; Catalán, P.; Diaz-Uriarte, R.; Elena, S.F.; García-Martín, J.A.; et al. From genotypes to organisms: State-of-the-art and perspectives of a cornerstone in evolutionary dynamics. Phys. Life Rev. 2021, 38, 55–106. [Google Scholar] [CrossRef]
- Tenaillon, O. The utility of fisher’s geometric model in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 179–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, R.A. The Genetical Theory of Natural Selection; Oxford University Press: Oxford, UK, 1930. [Google Scholar]
- Kingman, J.F.C. A simple model for the balance between selection and mutation. J. Appl. Probab. 1978, 15, 1–12. [Google Scholar] [CrossRef]
- Kauffman, S.; Levin, S. Towards a general theory of adaptive walks on rugged landscapes. J. Theor. Biol. 1987, 128, 11–45. [Google Scholar] [CrossRef]
- Kauffman, S.A.; Weinberger, E.D. The nk model of rugged fitness landscapes and its application to maturation of the immune response. J. Theor. Biol. 1989, 141, 211–245. [Google Scholar] [CrossRef]
- Aita, T.; Uchiyama, H.; Inaoka, T.; Nakajima, M.; Kokubo, T.; Husimi, Y. Analysis of a local fitness landscape with a model of the rough mt. fuji-type landscape: Application to prolyl endopeptidase and thermolysin. Biopolym. Orig. Res. Biomol. 2000, 54, 64–79. [Google Scholar] [CrossRef]
- Neidhart, J.; Szendro, I.G.; Krug, J. Adaptation in tunably rugged fitness landscapes: The rough mount fuji model. Genetics 2014, 198, 699–721. [Google Scholar] [CrossRef] [Green Version]
- Weinreich, D.M.; Delaney, N.F.; DePristo, M.A.; Hartl, D.L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 2006, 312, 111–114. [Google Scholar] [CrossRef] [Green Version]
- Chou, H.-H.; Chiu, H.-C.; NDelaney, N.F.; Segrè, D.; Marx, C.J. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 2011, 332, 1190–1192. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.I.; Dinh, D.M.; Schneider, D.; Lenski, R.E.; Cooper, T.F. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 2011, 332, 1193–1196. [Google Scholar] [CrossRef]
- De Visser, J.A.G.M.; Hoekstra, R.F.; van den Ende, H. Test of interaction between genetic markers that affect fitness in aspergillus niger. Evolution 1997, 51, 1499–1505. [Google Scholar] [CrossRef] [Green Version]
- Hall, D.W.; Agan, M.; Pope, S.C. Fitness epistasis among 6 biosynthetic loci in the budding yeast saccharomyces cerevisiae. J. Hered. 2010, 101 (Suppl. S1), S75–S84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitlock, M.C.; Bourguet, D. Factors affecting the genetic load in drosophila: Synergistic epistasis and correlations among fitness components. Evolution 2000, 54, 1654–1660. [Google Scholar] [CrossRef] [PubMed]
- Hinkley, T.; Martins, J.; Chappey, C.; Haddad, M.; Stawiski, E.; Whitcomb, J.M.; Petropoulos, C.J.; Bonhoeffer, S. A systems analysis of mutational effects in hiv-1 protease and reverse transcriptase. Nat. Genet. 2011, 43, 487–489. [Google Scholar] [CrossRef] [PubMed]
- Kouyos, R.D.; Leventhal, G.E.; Hinkley, T.; Haddad, M.; Whitcomb, J.M.; Petropoulos, C.J.; Bonhoeffer, S. Exploring the complexity of the hiv-1 fitness landscape. PLoS Genet. 2012, 8, e1002551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hietpas, R.T.; Jensen, J.D.; Bolon, D.N.A. Experimental illumination of a fitness landscape. Proc. Natl. Acad. Sci. USA 2011, 108, 7896–7901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otwinowski, J.; Nemenman, I. Genotype to phenotype mapping and the fitness landscape of the E. coli lac promoter. PLoS ONE 2013, 8, e61570. [Google Scholar] [CrossRef] [Green Version]
- Sarkisyan, K.S.; Bolotin, D.A.; Meer, M.V.; Usmanova, D.R.; Mishin, A.S.; Sharonov, G.V.; Ivankov, D.N.; Bozhanova, N.G.; Baranov, M.S.; Soylemez, O.; et al. Local fitness landscape of the green fluorescent protein. Nature 2016, 533, 397–401. [Google Scholar] [CrossRef] [Green Version]
- Rogers, Z.N.; McFarland, C.D.; Winters, I.P.; Seoane, J.A.; Brady, J.J.; Yoon, S.; Curtis, C.; Petrov, D.A.; Winslow, M.M. Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice. Nat. Genet. 2018, 50, 483–486. [Google Scholar] [CrossRef]
- Watson, C.J.; Papula, A.L.; Poon, G.Y.P.; Wong, W.H.; Young, A.L.; Druley, T.E.; Fisher, D.S.; Blundell, J.R. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science 2020, 367, 1449–1454. [Google Scholar] [CrossRef]
- Rowe, W.; Platt, M.; Wedge, D.C.; Day, P.J.; Kell, D.B.; Knowles, J. Analysis of a complete DNA–protein affinity landscape. J. R. Soc. Interface 2010, 7, 397–408. [Google Scholar] [CrossRef]
- Pitt, J.N.; Ferré-D’Amaré, A.R. Rapid construction of empirical rna fitness landscapes. Science 2010, 330, 376–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez, J.I.; Xulvi-Brunet, R.; Campbell, G.W.; Turk-MacLeod, R.; Chen, I.A. Comprehensive experimental fitness landscape and evolutionary network for small rna. Proc. Natl. Acad. Sci. USA 2013, 110, 14984–14989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Qian, W.; Maclean, C.J.; Zhang, J. The fitness landscape of a trna gene. Science 2016, 352, 837–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendixsen, D.P.; Østman, B.; Hayden, E.J. Negative epistasis in experimental rna fitness landscapes. J. Mol. Evol. 2017, 85, 159–168. [Google Scholar] [CrossRef]
- Aguilar-Rodríguez, J.; Payne, J.L.; Wagner, A. A thousand empirical adaptive landscapes and their navigability. Nat. Ecol. Evol. 2017, 1, 0045. [Google Scholar] [CrossRef] [Green Version]
- Martin, C.H. Context dependence in complex adaptive landscapes: Frequency and trait-dependent selection surfaces within an adaptive radiation of caribbean pupfishes. Evolution 2016, 70, 1265–1282. [Google Scholar] [CrossRef]
- Boucher, J.I.; Cote, P.; Flynn, J.; Jiang, L.; Laban, A.; Mishra, P.; Roscoe, B.P.; Bolon, D.N.A. Viewing protein fitness landscapes through a next-gen lens. Genetics 2014, 198, 461–471. [Google Scholar] [CrossRef]
- Aita, T.; Iwakura, M.; Husimi, Y. A cross-section of the fitness landscape of dihydrofolate reductase. Protein Eng. 2001, 14, 633–638. [Google Scholar] [CrossRef] [Green Version]
- Carneiro, M.; Hartl, D.L. Adaptive landscapes and protein evolution. Proc. Natl. Acad. Sci. USA 2010, 107 (Suppl. S1), 1747–1751. [Google Scholar] [CrossRef] [Green Version]
- Poelwijk, F.J.; Tănase-Nicola, S.; Kiviet, D.J.; Tans, S.J. Reciprocal sign epistasis is a necessary condition for multi-peaked fitness landscapes. J. Theor. Biol. 2011, 272, 141–144. [Google Scholar] [CrossRef] [Green Version]
- Lozovsky, E.R.; Chookajorn, T.; Brown, K.M.; Imwong, M.; Shaw, P.J.; Kamchonwongpaisan, S.; Neafsey, D.E.; Weinreich, D.M.; Hartl, D.L. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc. Natl. Acad. Sci. USA 2009, 106, 12025–12030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunzer, M.; Miller, S.P.; Felsheim, R.; Dean, A.M. The biochemical architecture of an ancient adaptive landscape. Science 2005, 310, 499–501. [Google Scholar] [CrossRef] [PubMed]
- Bridgham, J.T.; Carroll, S.M.; Thornton, J.W. Evolution of hormone-receptor complexity by molecular exploitation. Science 2006, 312, 97–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poelwijk, F.J.; Kiviet, D.J.; Tans, S.J. Evolutionary potential of a duplicated repressor-operator pair: Simulating pathways using mutation data. PLoS Comput. Biol. 2006, 2, e58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poelwijk, F.J.; Kiviet, D.J.; Weinreich, D.M.; Tans, S.J. Empirical fitness landscapes reveal accessible evolutionary paths. Nature 2007, 445, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Franke, J.; Klözer, A.; de Visser, J.A.G.; Krug, J. Evolutionary accessibility of mutational pathways. PLoS Comput. Biol. 2011, 7, e1002134. [Google Scholar] [CrossRef] [Green Version]
- Hegarty, P.; Martinsson, A. On the existence of accessible paths in various models of fitness landscapes. Ann. Appl. Probab. 2014, 24, 1375–1395. [Google Scholar] [CrossRef]
- Zagorski, M.; Burda, Z.; Waclaw, B. Beyond the hypercube: Evolutionary accessibility of fitness landscapes with realistic mutational networks. PLoS Comput. Biol. 2016, 12, e1005218. [Google Scholar] [CrossRef] [Green Version]
- Kell, D.B. Genotype–phenotype mapping: Genes as computer programs. Trends Genet. 2002, 18, 555–559. [Google Scholar] [CrossRef]
- Kondrashov, D.A.; Kondrashov, F.A. Topological features of rugged fitness landscapes in sequence space. Trends Genet. 2015, 31, 24–33. [Google Scholar] [CrossRef]
- Chan, H.S.; Bornberg-Bauer, E. Perspectives on protein evolution from simple exact models. Appl. Bioinform. 2002, 50, 121–144. [Google Scholar]
- Cowperthwaite, M.C.; Economo, E.P.; Harcombe, W.R.; Miller, E.L.; Meyers, L.A. The ascent of the abundant: How mutational networks constrain evolution. PLoS Comput. Biol. 2008, 4, e1000110. [Google Scholar] [CrossRef] [PubMed]
- Stich, M.; Lázaro, E.; Manrubia, S.C. Phenotypic effect of mutations in evolving populations of rna molecules. BMC Evol. Biol. 2010, 10, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, M.E.; Moudgil, A.; Feldman, M.W. Long-term evolution is surprisingly predictable in lattice proteins. J. R. Soc. Interface 2013, 10, 20130026. [Google Scholar] [CrossRef] [Green Version]
- Bershtein, S.; Serohijos, A.W.R.; Shakhnovich, E.I. Bridging the physical scales in evolutionary biology: From protein sequence space to fitness of organisms and populations. Curr. Opin. Struct. Biol. 2017, 42, 31–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perfeito, L.; Ghozzi, S.; Berg, J.; Schnetz, K.; Lässig, M. Nonlinear fitness landscape of a molecular pathway. PLoS Genet. 2011, 7, e1002160. [Google Scholar] [CrossRef] [Green Version]
- Chou, H.-H.; Delaney, N.F.; Draghi, J.A.; Marx, C.J. Mapping the fitness landscape of gene expression uncovers the cause of antagonism and sign epistasis between adaptive mutations. PLoS Genet. 2014, 10, e1004149. [Google Scholar] [CrossRef] [Green Version]
- Friedlander, T.; Prizak, R.; Barton, N.H.; Tkačik, G. Evolution of new regulatory functions on biophysically realistic fitness landscapes. Nat. Commun. 2017, 8, 216. [Google Scholar] [CrossRef] [Green Version]
- Cuypers, T.D.; Rutten, J.P.; Hogeweg, P. Evolution of evolvability and phenotypic plasticity in virtual cells. BMC Evol. Biol. 2017, 17, 60. [Google Scholar] [CrossRef] [Green Version]
- Yubero, P.; Manrubia, S.; Aguirre, J. The space of genotypes is a network of networks: Implications for evolutionary and extinction dynamics. Sci. Rep. 2017, 7, 13813. [Google Scholar] [CrossRef] [Green Version]
- Harmand, N.; Gallet, R.; Jabbour-Zahab, R.; Martin, G.; Lenormand, T. Fisher’s geometrical model and the mutational patterns of antibiotic resistance across dose gradients. Evolution 2017, 71, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-H.; Scarpino, S.V. Reproductive barriers as a byproduct of gene network evolution. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yang, C.-H.; Scarpino, S.V. The ensemble of gene regulatory networks at mutation-selection balance. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ciliberti, S.; Martin, O.C.; Wagner, A. Innovation and robustness in complex regulatory gene networks. Proc. Natl. Acad. Sci. USA 2007, 104, 13591–13596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payne, J.L.; Wagner, A. Latent phenotypes pervade gene regulatory circuits. BMC Syst. Biol. 2014, 8, 64. [Google Scholar] [CrossRef] [Green Version]
- Godsil, C.D. Compact graphs and equitable partitions. Linear Algebra Its Appl. 1997, 255, 259–266. [Google Scholar] [CrossRef] [Green Version]
- Das, S.G.; Direito, S.O.L.; Waclaw, B.; Allen, R.J.; Krug, J. Predictable properties of fitness landscapes induced by adaptational tradeoffs. eLife 2020, 9, e55155. [Google Scholar] [CrossRef]
- Wagner, A. Neutralism and selectionism: A network-based reconciliation. Nat. Rev. Genet. 2008, 9, 965–974. [Google Scholar] [CrossRef] [Green Version]
- Bendixsen, D.P.; Collet, J.; Østman, B.; Hayden, E.J. Genotype network intersections promote evolutionary innovation. PLoS Biol. 2019, 17, e3000300. [Google Scholar] [CrossRef] [Green Version]
- Hunt, G.; Hopkins, M.J.; Lidgard, S. Simple versus complex models of trait evolution and stasis as a response to environmental change. Proc. Natl. Acad. Sci. USA 2015, 112, 4885–4890. [Google Scholar] [CrossRef] [Green Version]
- Heasley, L.R.; Sampaio, N.M.V.; Argueso, J.L. Systemic and rapid restructuring of the genome: A new perspective on punctuated equilibrium. Curr. Genet. 2020, 67, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Rodríguez, J.; Peel, L.; Stella, M.; Wagner, A.; Payne, J.L. The architecture of an empirical genotype-phenotype map. Evolution 2018, 72, 1242–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Presa, J.L.; Chiroque, L.F.; Anta, A.F. Novel techniques to speed up the computation of the automorphism group of a graph. J. Appl. Math. 2014, 2014, 934637. [Google Scholar] [CrossRef]
- Stoichev, S.D. New exact and heuristic algorithms for graph automorphism group and graph isomorphism. J. Exp. Algorithmics (JEA) 2019, 24, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Battiston, F.; Cencetti, G.; Iacopini, I.; Latora, V.; Lucas, M.; Patania, A.; Young, J.-G.; Petri, G. Networks beyond pairwise interactions: Structure and dynamics. Phys. Rep. 2020, 874, 1–92. [Google Scholar] [CrossRef]
- Maheshwari, P.; Albert, R. A framework to find the logic backbone of a biological network. BMC Syst. Biol. 2017, 11, 122. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Gómez Tejeda Zañudo, J.; Albert, R. Target control in logical models using the domain of influence of nodes. Front. Physiol. 2018, 9, 454. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Tomassini, M.; Banzhaf, W. A network perspective on genotype–phenotype mapping in genetic programming. Genet. Program. Evolvable Mach. 2020, 21, 375–397. [Google Scholar] [CrossRef]
- Greenbury, S.F.; Louis, A.A.; Ahnert, S.E. The structure of genotype-phenotype maps makes fitness landscapes navigable. bioRxiv 2021. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.-H.; Scarpino, S.V. A Family of Fitness Landscapes Modeled through Gene Regulatory Networks. Entropy 2022, 24, 622. https://doi.org/10.3390/e24050622
Yang C-H, Scarpino SV. A Family of Fitness Landscapes Modeled through Gene Regulatory Networks. Entropy. 2022; 24(5):622. https://doi.org/10.3390/e24050622
Chicago/Turabian StyleYang, Chia-Hung, and Samuel V. Scarpino. 2022. "A Family of Fitness Landscapes Modeled through Gene Regulatory Networks" Entropy 24, no. 5: 622. https://doi.org/10.3390/e24050622





