Information Theoretic-Based Interpretation of a Deep Neural Network Approach in Diagnosing Psychogenic Non-Epileptic Seizures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects and Electrophysiological Recordings
2.2. Time-Frequency Feature Extraction
2.3. Deep Learning (DL) Approach
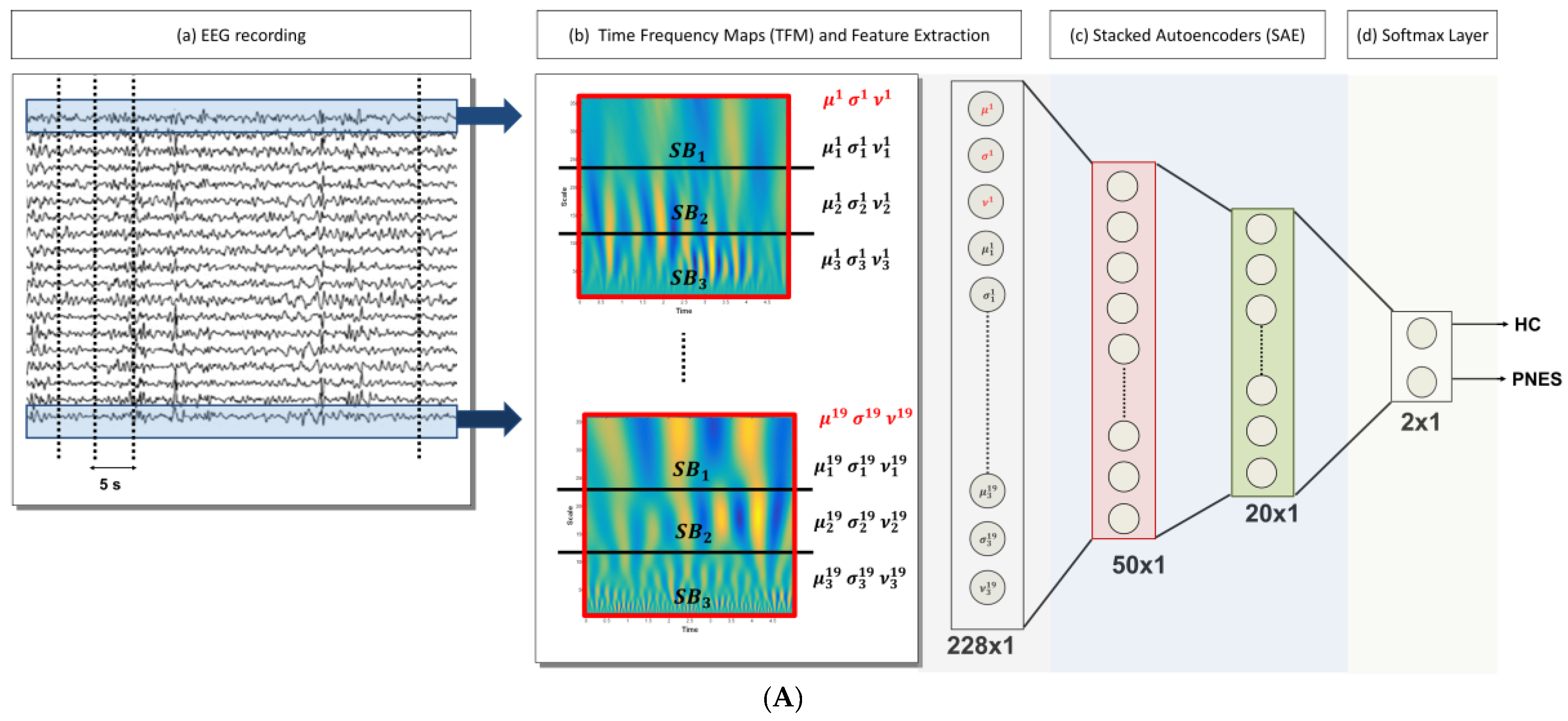
2.4. DL-Based Processing System for EEG Classification
- (1)
- Artifact rejection: rejection of the artifacts through the visual inspection of each EEG recording; the EEG segments clearly affected by artefactual components are discarded, Figure 1A (a);
- (2)
- EEG signal decomposition: the cleaned EEG recording is subdivided in non-overlapping T = 5 s epochs, Figure 1A (a);
- (3)
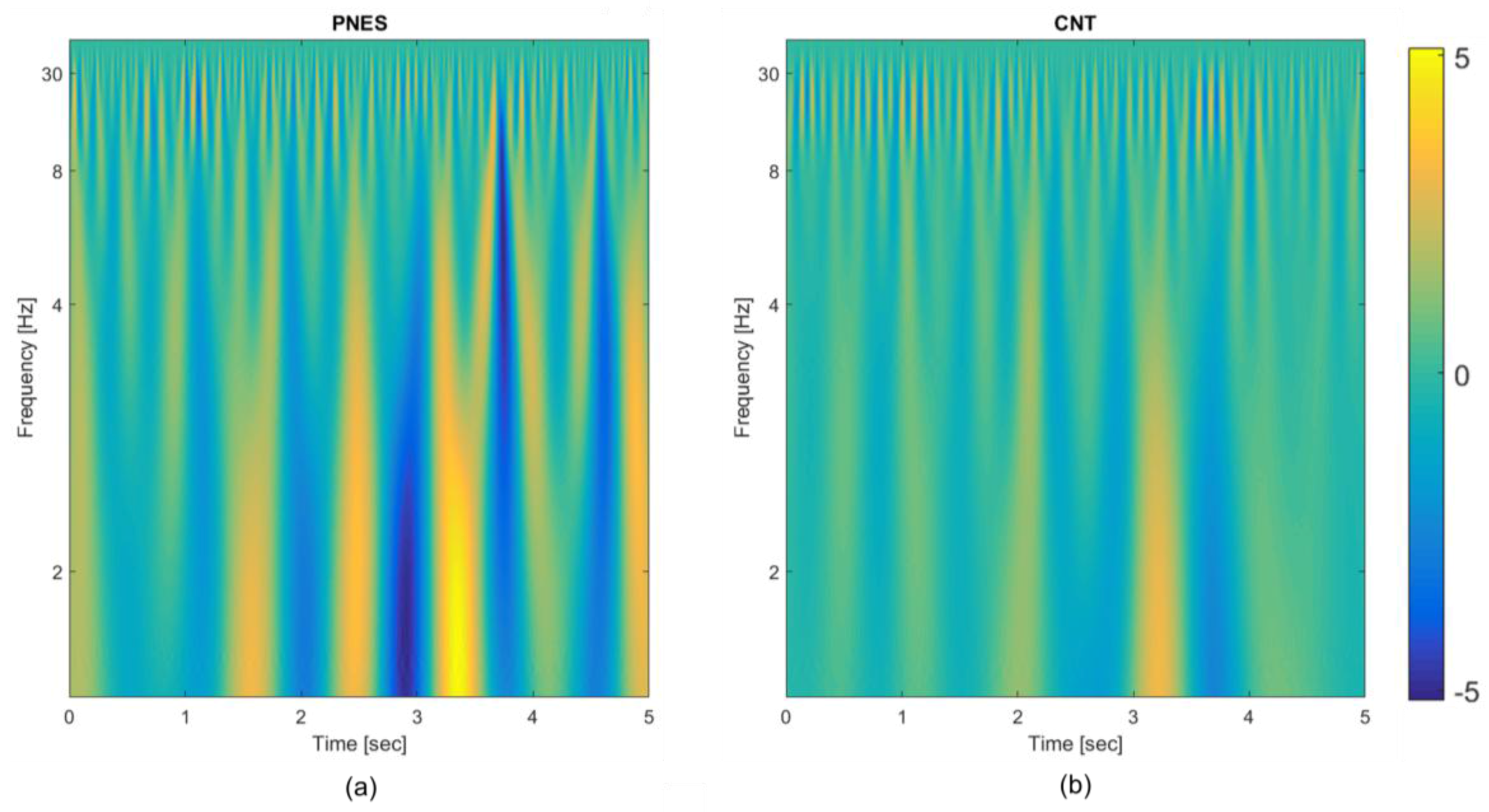
- TF transformation: each EEG epoch is time-frequency transformed by using CWT, as in (1), by using a Mexican hat function as mother wavelet (2), Figure 1A (b); the use of CWT showed significant advantages on simple spectrograms probably because of the choice of the mother wavelet function, which is particularly suitable for EEG signals;
- (4)
- Engineered feature extraction: partitioning of the CWT map into three parts (sub-bands maps) and estimation of the mean value (µ), the standard deviation (σ) and the skewness (υ) either of the three sub-bands maps and of the whole CWT map, Figure 1A (b); the widths of the three non-overlapping sub-bands have been selected by an optimization algorithm and do not exactly correspond to the brain rhythms [12]; the two higher bands in Figure 1A (b) roughly include delta and theta rhythms;
- (5)
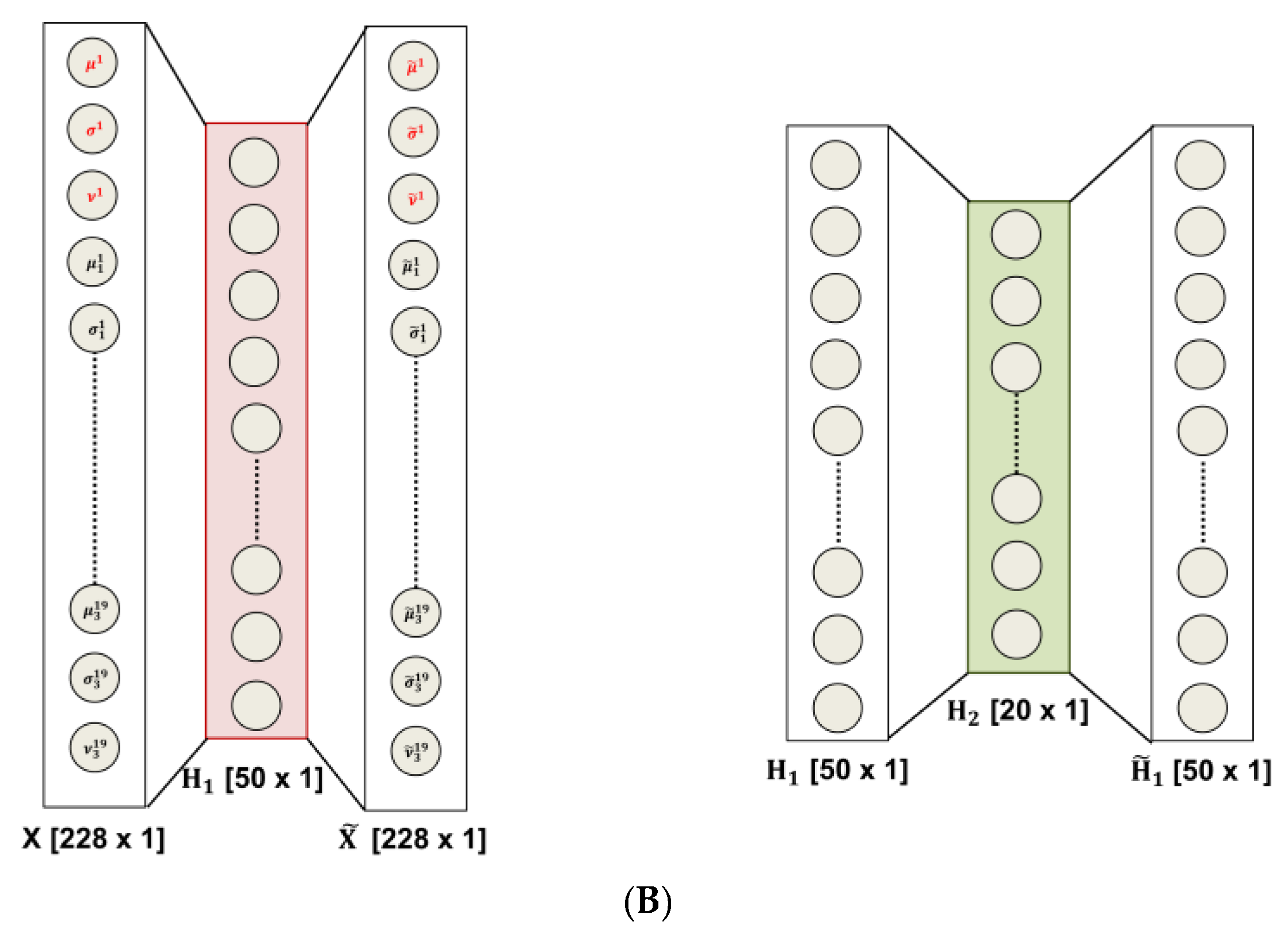
- Preparation of the feature vector: the resulting feature vector includes three features per electrode (µ, σ, and υ for each of the three sub-bands maps and the µ, σ, and υ of the whole CWT map); thus, the input vector of the autoencoders chain has a length of 12 (features) × 19 (electrodes) = 228 elements, Figure 1A (c);
- (6)
- Data-driven feature compression: two stages of autoencoding are used as compressors giving 50 and 20 successively extracted data-driven features; at this level the features extracted from each channel are combined outputting an unsupervised learned vector that mixes the characteristics of the channels, Figure 1A (c); the size of the second hidden layers has been related to the number of the electrodes; the first hidden layer is only approximatively sized, as the sparsification induced by the cost function automatically find a sub-optimal size;
- (7)
- Classification step: a softmax layer is trained by supervised learning (backprop) giving the relative probabilities of the two classes, Figure 1A (d).
2.5. Entropy-Based Interpretation of Hidden Layers
3. Results
3.1. Electroencephalography (EEG) Data Preprocessing
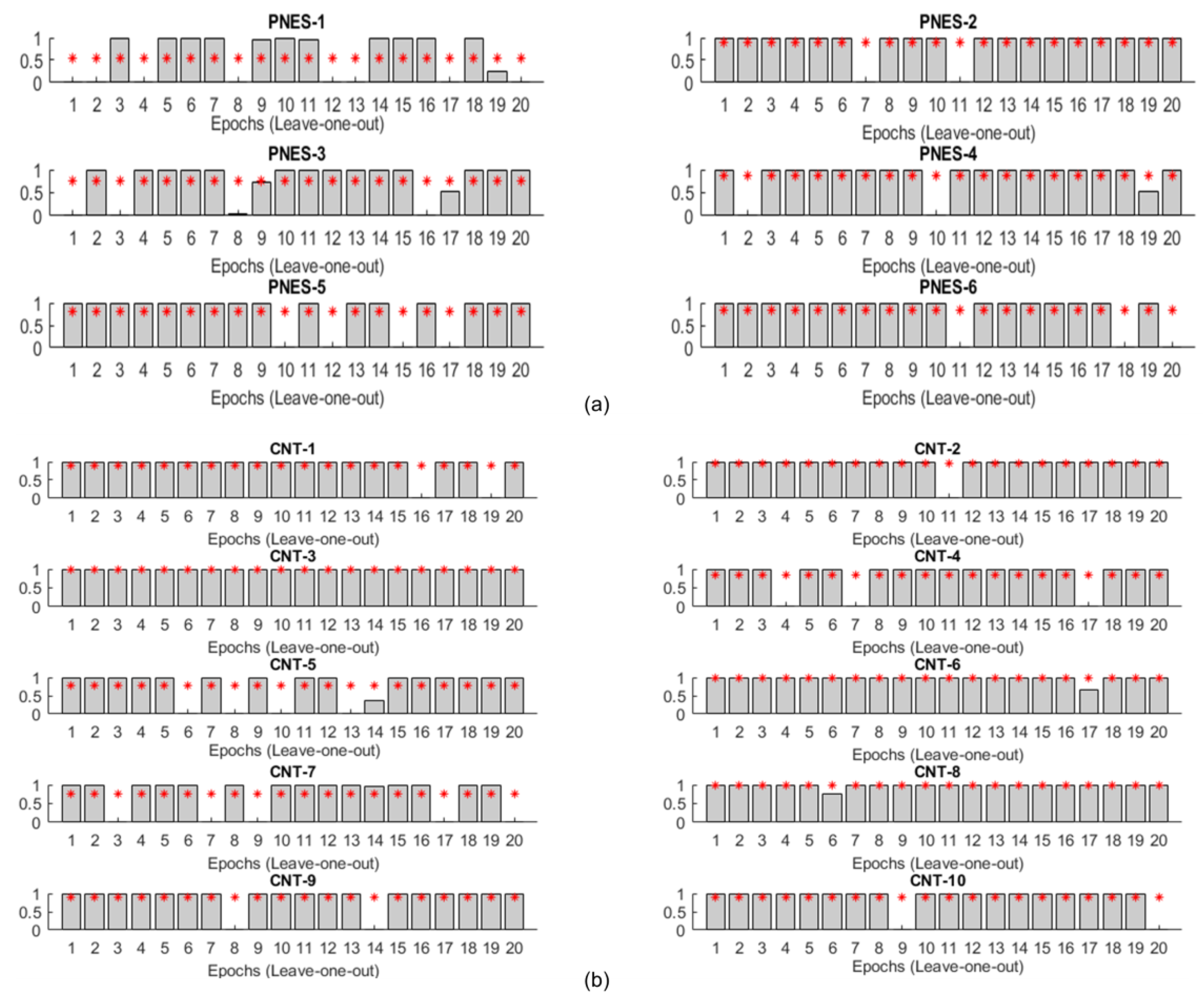
3.2. Performance of the Deep Learning (DL) Classification System
3.3. Entropic Interpretation of DL Classification
- (1)
- As recently noted in the literature [21], most of the information encoded in the input epochs is exploited in compression to generate an efficient representation regardless of the training labels, as the compression phase ignores the labels (considered just in the final classification stage);
- (2)
- The mean entropy indeed decreased as the layers deepened, which is intuitively rather expected as the successive representations gradually build the final vectors’ representation [26];
- (3)
- In contrast to the first stage of compression, the hidden layer of the second encoder seems clearly extracting the class information, i.e., the latent differences between the classes, even though in absence of any label information. This is an original result not previously reported in the literature, at our best knowledge. This is the first study where the behavior of the compressing stages has been discussed from an information-theoretical perspective in classification networks. In our opinion, the noted behavior can justify the use of a deep structure to extract high-level features that can widely facilitate the classification procedure [27].
4. Discussion and Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ban, G.-Y.; Karoui, N.E.; Lim, A.E.B. Machine learning and portfolio optimization. Manag. Sci. 2016. [Google Scholar] [CrossRef]
- Sirignano, J.; Sadhwani, A.; Giesecke, K. Deep Learning for Mortgage Risk. Available online: https://ssrn.com/abstract=2799443 (accessed on 19 January 2018).
- Socher, R.; Huval, B.; Bhat, B.; Manning, C.D.; Ng, A.Y. Convolutional recursive deep learning for 3D object classification. Adv. Neural Inf. Process. Syst. 2012, 1, 656–664. Available online: http://papers.nips.cc/paper/4773-convolutional-recursive-deep-learning-for-3d-object-classification.pdf (accessed on 19 January 2018).
- Sofman, B.; Lin, E.; Bagnell, J.A.; Cole, J.; Vandapel, N.; Stentz, A. Improving robot navigation through self-supervised online learning. J. Field Robot. 2006, 23, 1059–1075. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [PubMed]
- Wulsin, D.F.; Gupta, J.R.; Mani, R.; Blanco, J.; Litt, A. Modeling electroencephalography waveforms with semi-supervised deep belief nets: Fast classification and anomaly measurement. J. Neural Eng. 2011, 8, 036015. [Google Scholar] [CrossRef] [PubMed]
- Mirowski, P.; Madhavan, D.; LeCun, Y.; Kuzniecky, R. Classification of patterns of EEG synchronization for seizure prediction. Clin. Neurophysiol. 2009, 120, 1927–1940. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; He, L. Deep learning in the EEG diagnosis of Alzheimer’s disease. In Asian Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Morabito, F.C.; Campolo, M.; Mammone, N.; Versaci, M.; Franceschetti, S.; Tagliavini, F.; Sofia, V.; Fatuzzo, D.; Gambardella, A.; Labate, A.; et al. Deep learning representation from electroencephalography of Early-Stage Creutzfeldt-Jakob disease and features for differentiation from rapidly progressive dementia. Int. J. Neural Syst. 2017, 27, 1650039. [Google Scholar] [CrossRef] [PubMed]
- Morabito, F.C.; Campolo, M.; Ieracitano, C.; Ebadi, J.M.; Bonanno, L.; Bramanti, A.; Desalvo, S.; Mammone, N.; Bramanti, P. Deep convolutional neural networks for classification of mild cognitive impaired and Alzheimer’s disease patients from scalp EEG recordings. In Proceedings of the 2016 IEEE 2nd International Forum on Research and Technologies for Society and Industry Leveraging a Better Tomorrow (RTSI), Bologna, Italy, 7–9 September 2016. [Google Scholar]
- Bodde, N.M.G.; Brooks, J.L.; Baker, G.A.; Boon, P.A.J.M.; Hendriksen, J.G.M.; Mulder, O.G.; Aldenkamp, A.P. Psychogenic non-epileptic seizures—Definition, etiology, treatment and prognostic issues: A critical review. Seizure 2009, 18, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Reuber, M.; Baker, G.A.; Gill, R.; Smith, D.F.; Chadwick, D.W. Failure to recognize psychogenic nonepileptic seizures may cause death. Neurology 2004, 62, 834–835. [Google Scholar] [CrossRef] [PubMed]
- LaFrance, W.C., Jr.; Benbadis, S.R. Avoiding the costs of unrecognized psychological nonepileptic seizures. Neurology 2006, 66, 1620–1621. [Google Scholar] [CrossRef] [PubMed]
- LaFrance, W.C.; Baker, G.A.; Duncan, R.; Goldstein, L.H.; Reuber, M. Minimum requirements for the diagnosis of psychogenic nonepileptic seizures: A staged approach. Epilepsia 2013, 54, 2005–2018. [Google Scholar] [CrossRef] [PubMed]
- Devinsky, O.; Gazzola, D.; LaFrance, W.C. Differentiating between nonepileptic and epileptic seizures. Nat. Rev. Neurol. 2011, 7, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Bengio, Y.; Goodfellow, I.J.; Courville, A. Deep Learning. MIT Press, 2016. Available online: https://icdm2016.eurecat.org/wp-content/uploads/2016/05/ICDM-Barcelona-13Dec2016-YoshuaBengio.pdf (accessed on 19 January 2018).
- Bengio, Y.; Lamblin, P.; Popovici, D.; Larochelle, H. Greedy layer-wise training of deep networks. In Proceedings of the 19th International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 4–7 December 2006. [Google Scholar]
- Erhan, D.; Bengio, Y.; Courville, A.; Manzagol, P.A.; Vincent, P.; Bengio, S. Why does unsupervised pre-training help deep learning? J. Mach. Learn. Res. 2010, 11, 625–660. [Google Scholar]
- Larochelle, H.; Bengio, Y.; Louradour, J.; Lamblin, P. Exploring strategies for training deep neural networks. J. Mach. Learn. Res. 2009, 10, 1–40. [Google Scholar]
- Hinton, G.E.; Srivastava, N.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R.R. Improving Neural Networks by Preventing Co-Adaptation of Feature Detectors. arXiv 2012, arXiv:1207.0580. Available online: https://arxiv.org/abs/1207.0580 (accessed on 19 January 2018).
- Parikh, R.; Mathai, A.; Parikh, S.; Sekhar, G.C.; Thomas, R. Understanding and using sensitivity, specificity and predictive values. Indian J. Ophthalmol. 2008, 56, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Burges, C.J. A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- McLachlan, G. Discriminant Analysis and Statistical Pattern Recognition; John Wiley & Sons: Hoboken, NJ, USA, 2004. [Google Scholar]
- Van der Kruijs, S.J.; Bodde, N.M.; Vaessen, M.J.; Lazeron, R.H.; Vonck, K.; Boon, P.; Hofman, P.A.; Backes, W.H.; Aldenkamp, A.P.; Jansen, J.F. Functional connectivity of dissociation in patients with psychogenic non-epileptic seizures. J. Neurol Neurosurg. Psychiatr. 2012, 83, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Vincent, P.; Larochelle, H.; Lajoie, I.; Bengio, Y.; Manzagol, P.A. Stacked denoising autoencoders: Learning useful representations in a deep network with a local denoising criterion. J. Mach. Learn. Res. 2010, 11, 3371–3408. [Google Scholar]
- Shwartz-Ziv, R.; Tishby, N. Opening the Black Box of Deep Neural Networks via Information. arXiv 2017, arXiv:1703.00810. Available online: https://arxiv.org/abs/1703.00810 (accessed on 19 January 2018).
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]

| Classifier | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|---|---|---|---|---|
| SAE | 88.8 | 90.7 | 86.2 | 88.6 | 86.5 |
| LDA | 84.1 | 72.1 | 83.6 | 73.5 | 79.7 |
| QDA | 88.0 | 54.2 | 76.2 | 73.3 | 75.3 |
| L-SVM | 88.0 | 82.5 | 88.7 | 86.5 | 84.4 |
| Q-SVM | 92.3 | 57.5 | 78.6 | 85.2 | 80.3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gasparini, S.; Campolo, M.; Ieracitano, C.; Mammone, N.; Ferlazzo, E.; Sueri, C.; Tripodi, G.G.; Aguglia, U.; Morabito, F.C. Information Theoretic-Based Interpretation of a Deep Neural Network Approach in Diagnosing Psychogenic Non-Epileptic Seizures. Entropy 2018, 20, 43. https://doi.org/10.3390/e20020043
Gasparini S, Campolo M, Ieracitano C, Mammone N, Ferlazzo E, Sueri C, Tripodi GG, Aguglia U, Morabito FC. Information Theoretic-Based Interpretation of a Deep Neural Network Approach in Diagnosing Psychogenic Non-Epileptic Seizures. Entropy. 2018; 20(2):43. https://doi.org/10.3390/e20020043
Chicago/Turabian StyleGasparini, Sara, Maurizio Campolo, Cosimo Ieracitano, Nadia Mammone, Edoardo Ferlazzo, Chiara Sueri, Giovanbattista Gaspare Tripodi, Umberto Aguglia, and Francesco Carlo Morabito. 2018. "Information Theoretic-Based Interpretation of a Deep Neural Network Approach in Diagnosing Psychogenic Non-Epileptic Seizures" Entropy 20, no. 2: 43. https://doi.org/10.3390/e20020043
APA StyleGasparini, S., Campolo, M., Ieracitano, C., Mammone, N., Ferlazzo, E., Sueri, C., Tripodi, G. G., Aguglia, U., & Morabito, F. C. (2018). Information Theoretic-Based Interpretation of a Deep Neural Network Approach in Diagnosing Psychogenic Non-Epileptic Seizures. Entropy, 20(2), 43. https://doi.org/10.3390/e20020043







