Application of Hydration Thermodynamics to the Evaluation of Protein Structures and Protein-Ligand Binding
Abstract
:1. Introduction
2. Theoretical Background and Molecular Models of Protein and Water
2.1. Major Driving Force of Protein Folding: Increase in Translational Entropy of Water
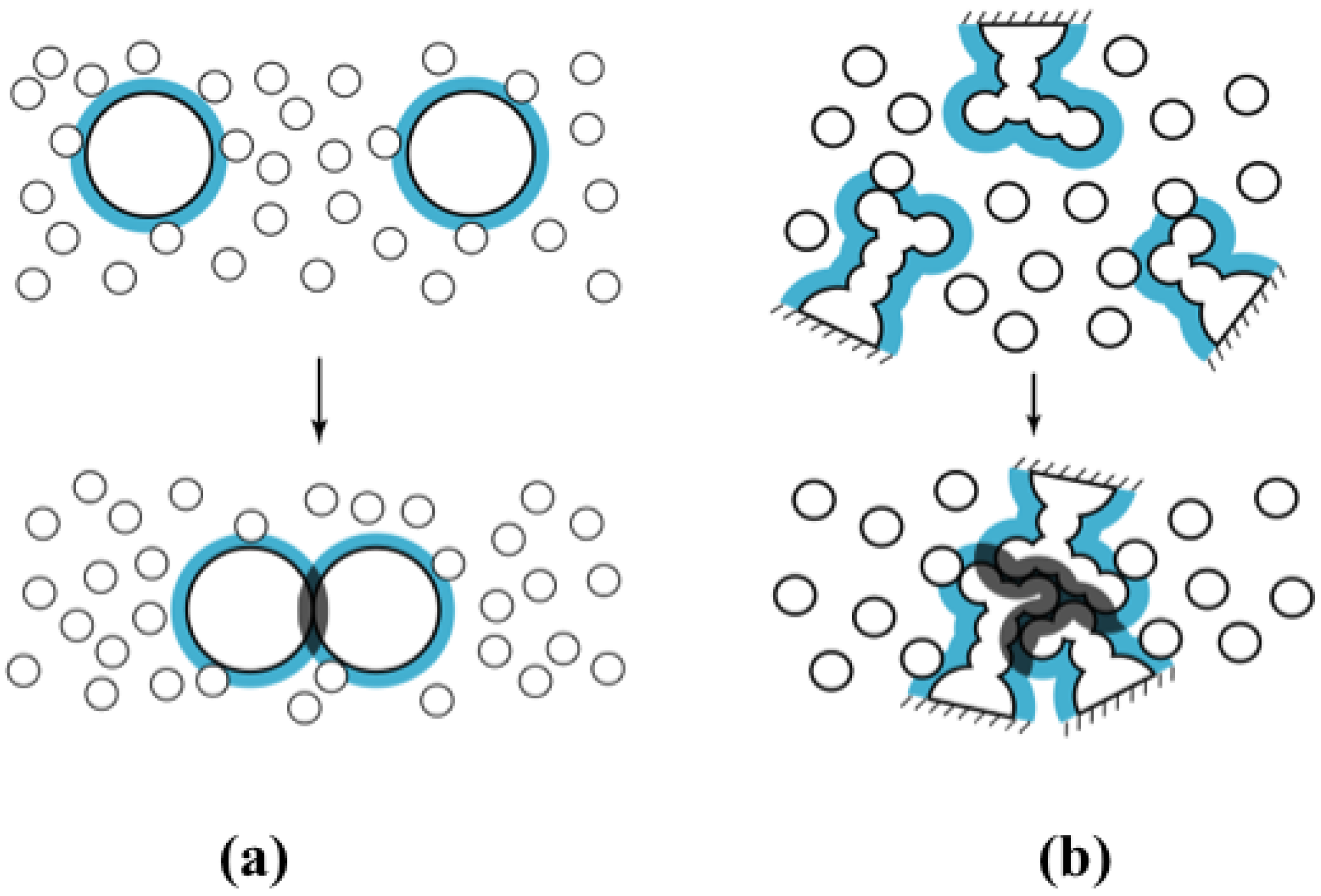


2.2. Free Energy Function Extracting the Physical Essence of Protein Stability
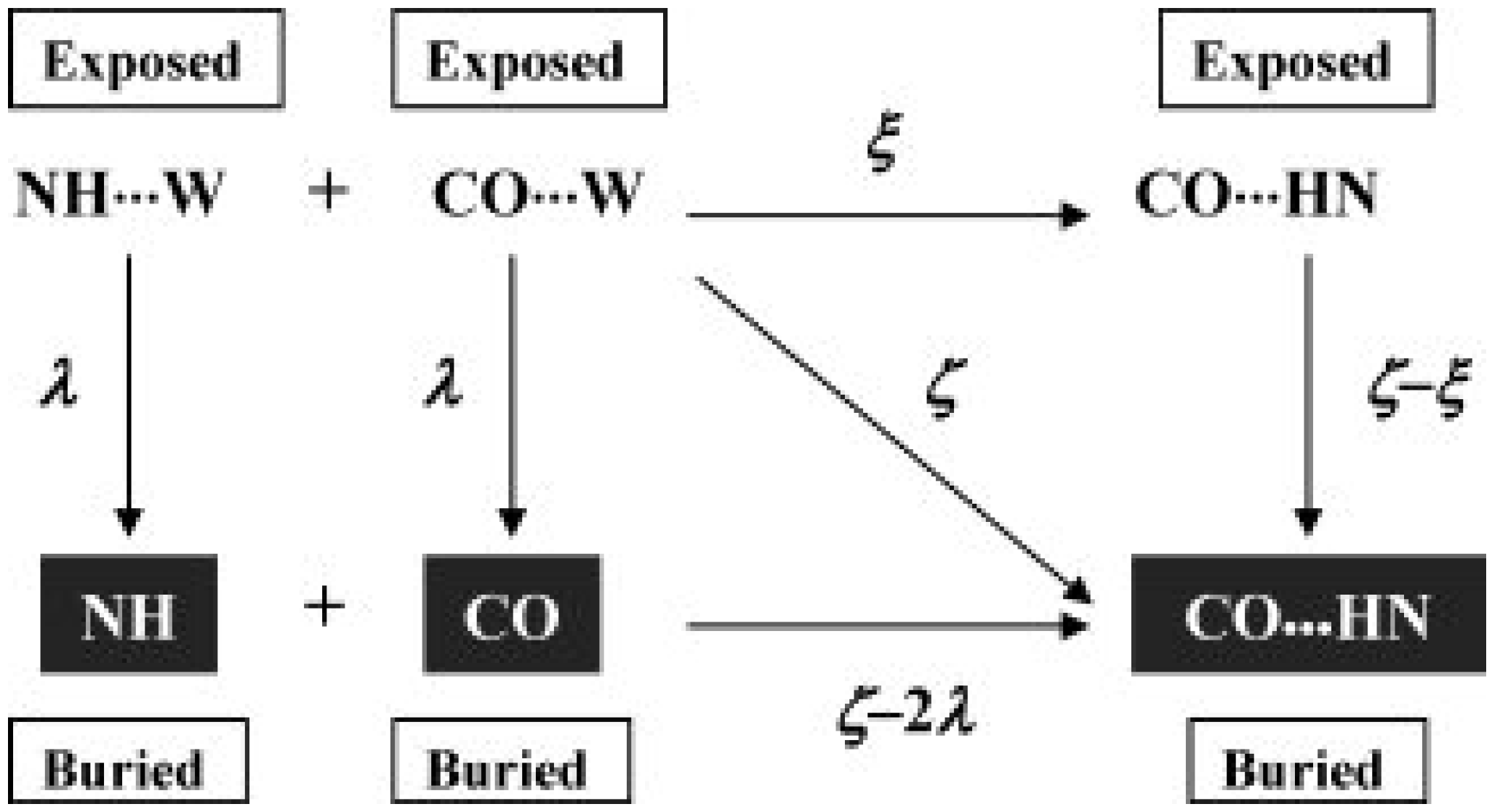
2.3. Computational Methods & Molecular Models
3. Validation of the Free Energy Function
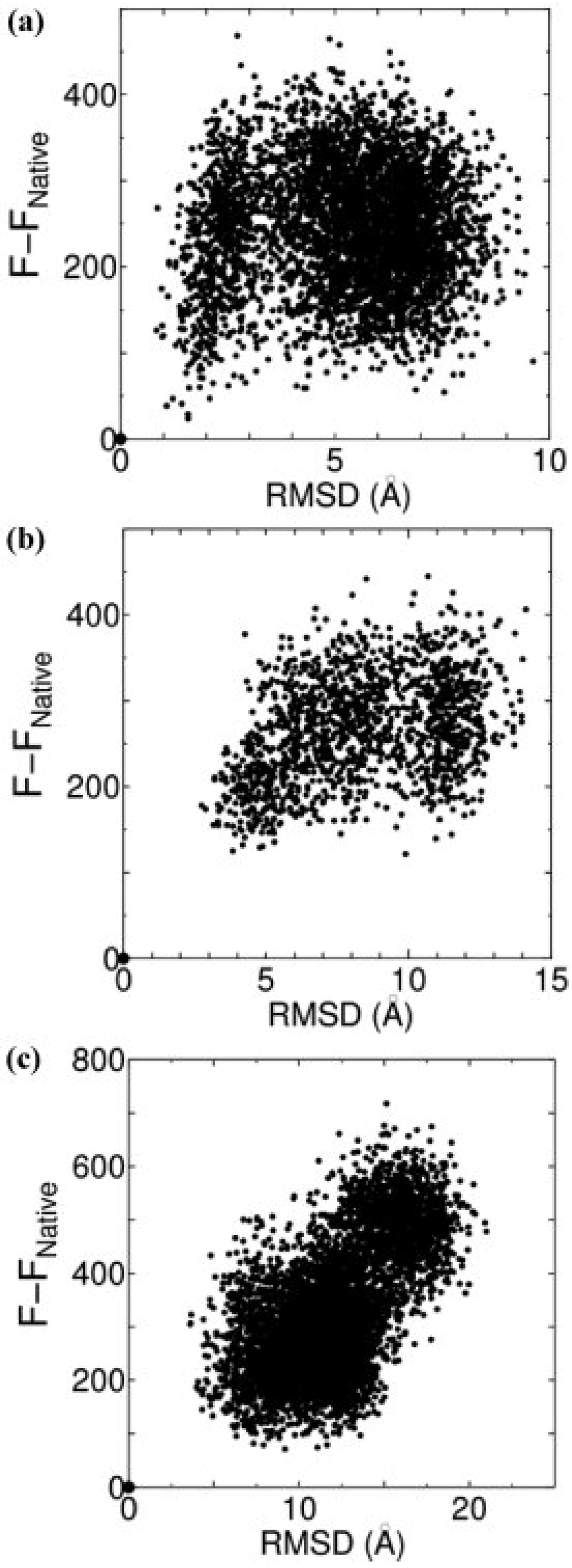
3.1. Discrimination of the Native Structures from Misfolded Decoys
| Scoring Function | Top1/Total No. | Mean Z |
|---|---|---|
| 4state_reduced | ||
| F | 7/7 | −5.19 |
| Lu et al., 2008 [60] | 7/7 | −4.41 |
| Miyazawa and Jernigan, 2005 [61] | 7/7 | −4.44 |
| Lee and Duan, 2004 [62] | 7/7 | −4.95 |
| Zhang et al., 2004 [63] | 6/7 | −3.94 |
| Onizuka et al., 2002 [64] | 7/7 | −3.41 |
| Zhou and Zhou, 2002 [65] | 6/7 | −3.49 |
| Toby and Elber, 2000 [66] | 3/6 | −3.14 |
| Fisa | ||
| F | 3/3 | −6.97 |
| Lu et al., 2008 [60] | 3/3 | −5.52 |
| Miyazawa and Jernigan, 2005 [61] | 2/4 | −2.55 |
| Lee and Duan, 2004 [62] | 2/3 | −3.73 |
| Zhang et al., 2004 [63] | 3/4 | −4.70 |
| Onizuka et al., 2002 [64] | 1/3 | −1.38 |
| Zhou and Zhou, 2002 [65] | 3/3 | −5.91 |
| Toby and Elber, 2000 [66] | 2/3 | −3.34 |
| Fisa_casp3 | ||
| F | 5/5 | −5.51 |
| Lu et al., 2008 [60] | 5/5 | −5.35 |
| Miyazawa and Jernigan, 2005 [61] | 2/5 | −3.61 |
| Lee and Duan, 2004 [62] | 2/5 | −3.19 |
| Zhang et al., 2004 [63] | 3/3 | −6.05 |
| Onizuka et al., 2002 [64] | 1/3 | −2.01 |
| Zhou and Zhou, 2002 [65] | 3/3 | −5.37 |
| Toby and Elber, 2000 [66] | 1/3 | −2.01 |
| Rosetta (X-ray) | ||
| F | 41/41 | −7.12 |
| Lu et al., 2008 [60] | 37/41 | −6.56 |
| lattice_ssfit | ||
| F | 8/8 | −7.54 |
| Miyazawa and Jernigan, 2005 [61] | 8/8 | −11.12 |
| Lu et al., 2008 [60] | 8/8 | −6.52 |
| Lmds | ||
| F | 9/10(10/10) | −6.29 (−6.79) |
| Miyazawa and Jernigan, 2005 [61] | 8/10 | −5.34 |
| Lu et al., 2008 [60] | 8/10 | −5.20 |
| Semfold | ||
| F | 8/8 | −6.76 |
| Miyazawa and Jernigan, 2005 [61] | 4/6 | −3.86 |


| Structure | α-helical Content (%) | MM+GBMV/SA/(kBT0) | −S | Λ | F |
|---|---|---|---|---|---|
| Native | 74 | 0.0 | 0.0 | 0.0 | 0.0 |
| Decoy | 79 | −5.7 | 97.5 | 35 | 132.5 |
| All α | 100 | −29.1 | 204.5 | −21 | 183.5 |
3.2. Characteristics of Native Proteins
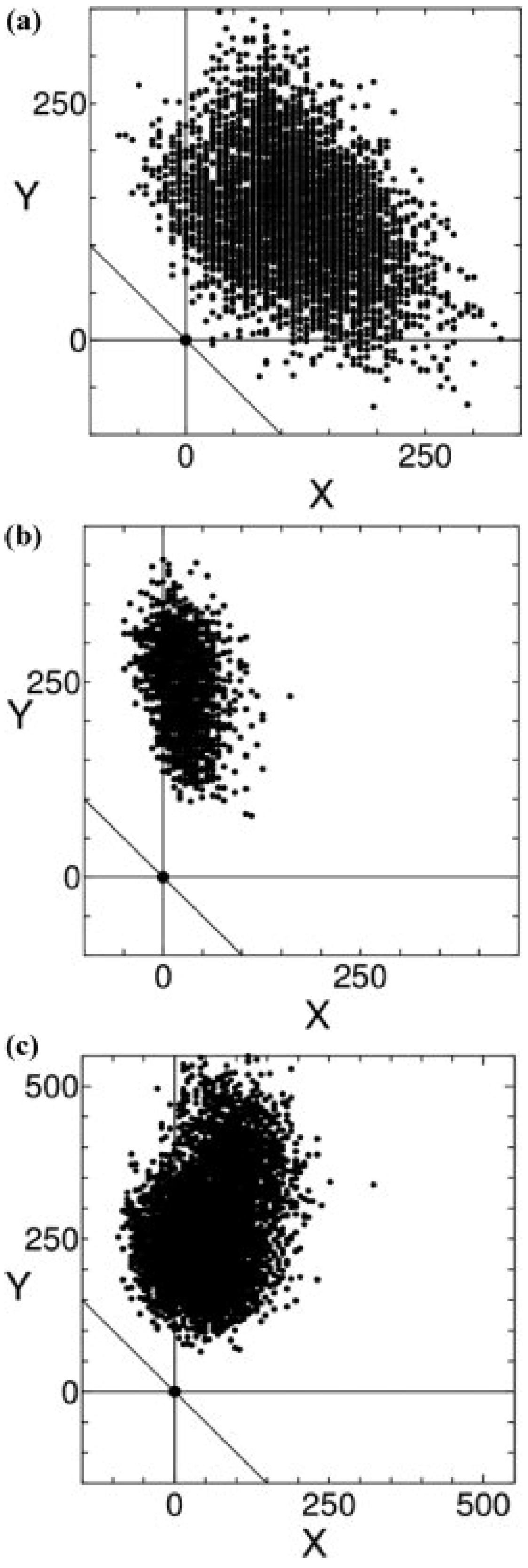
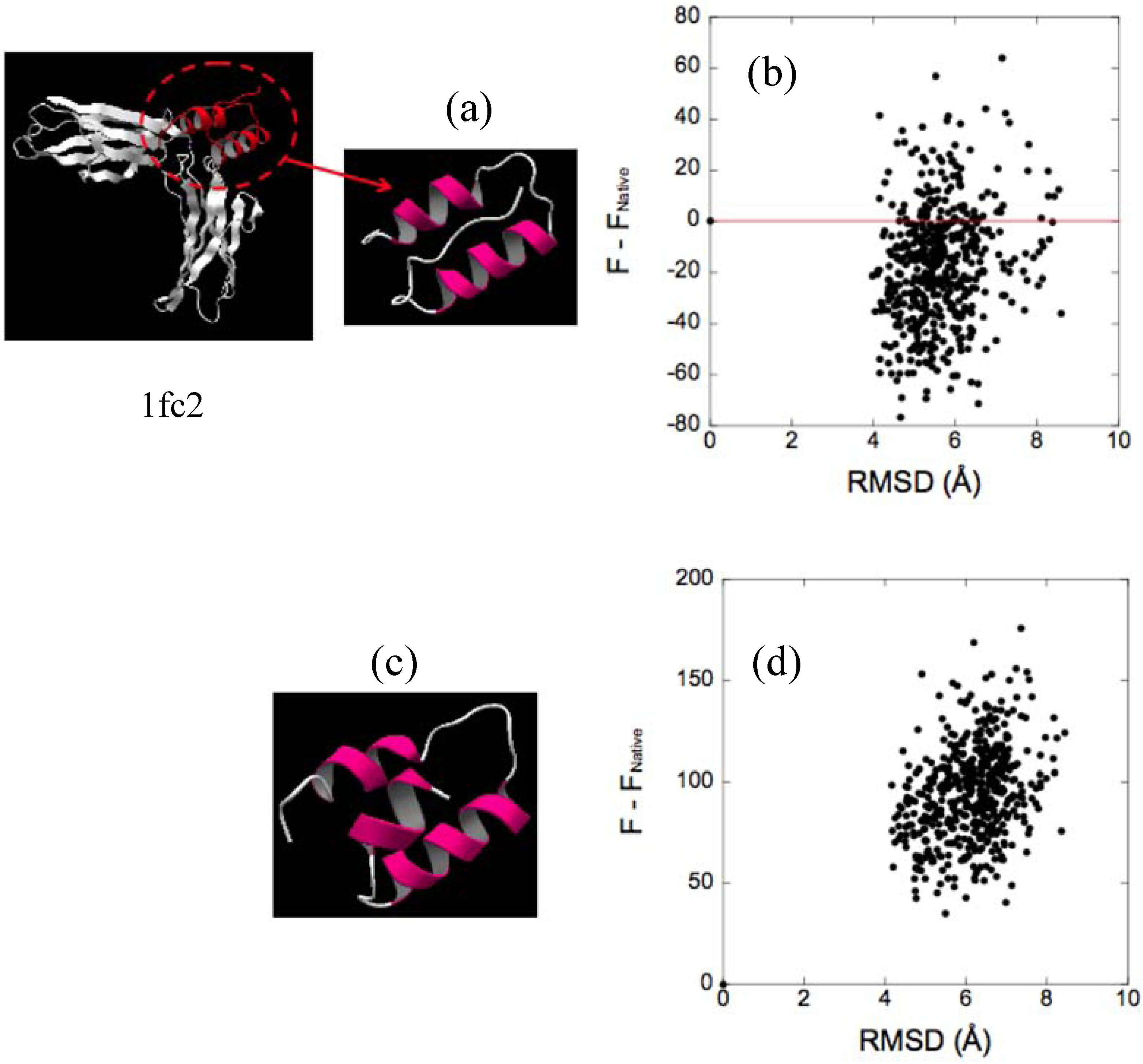
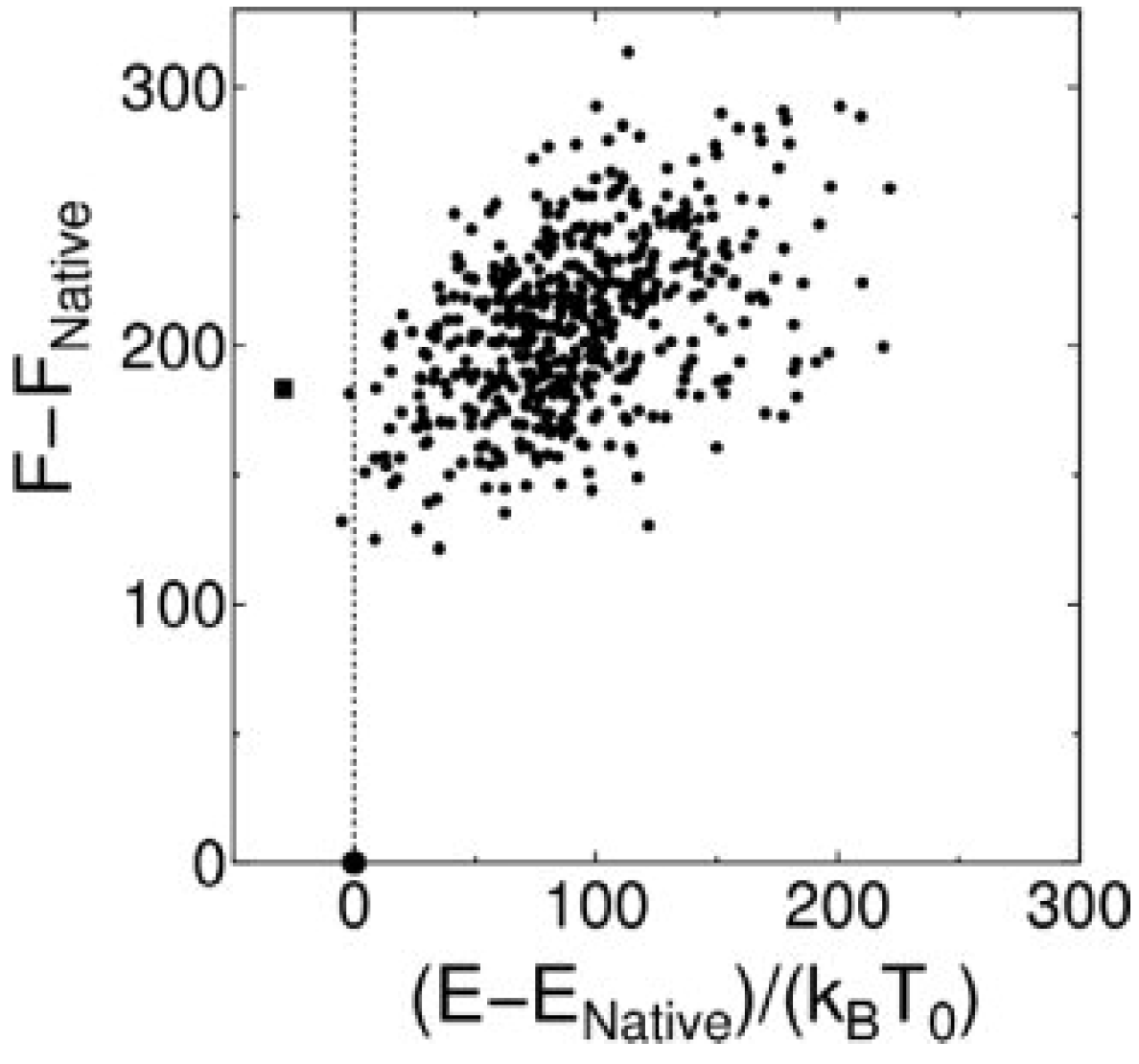
3.3. Evaluation of Protein-Ligand Binding Free Energy
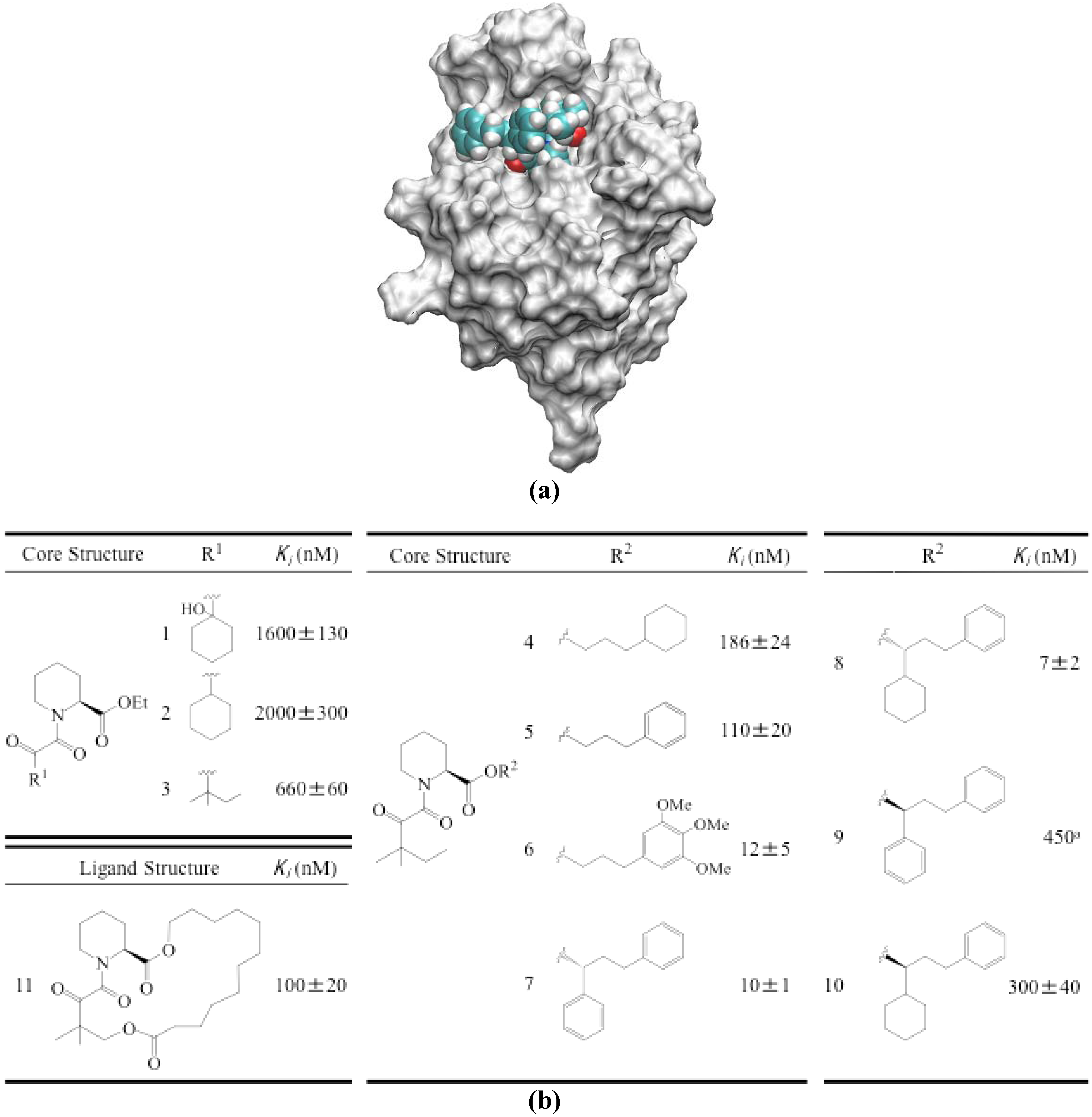
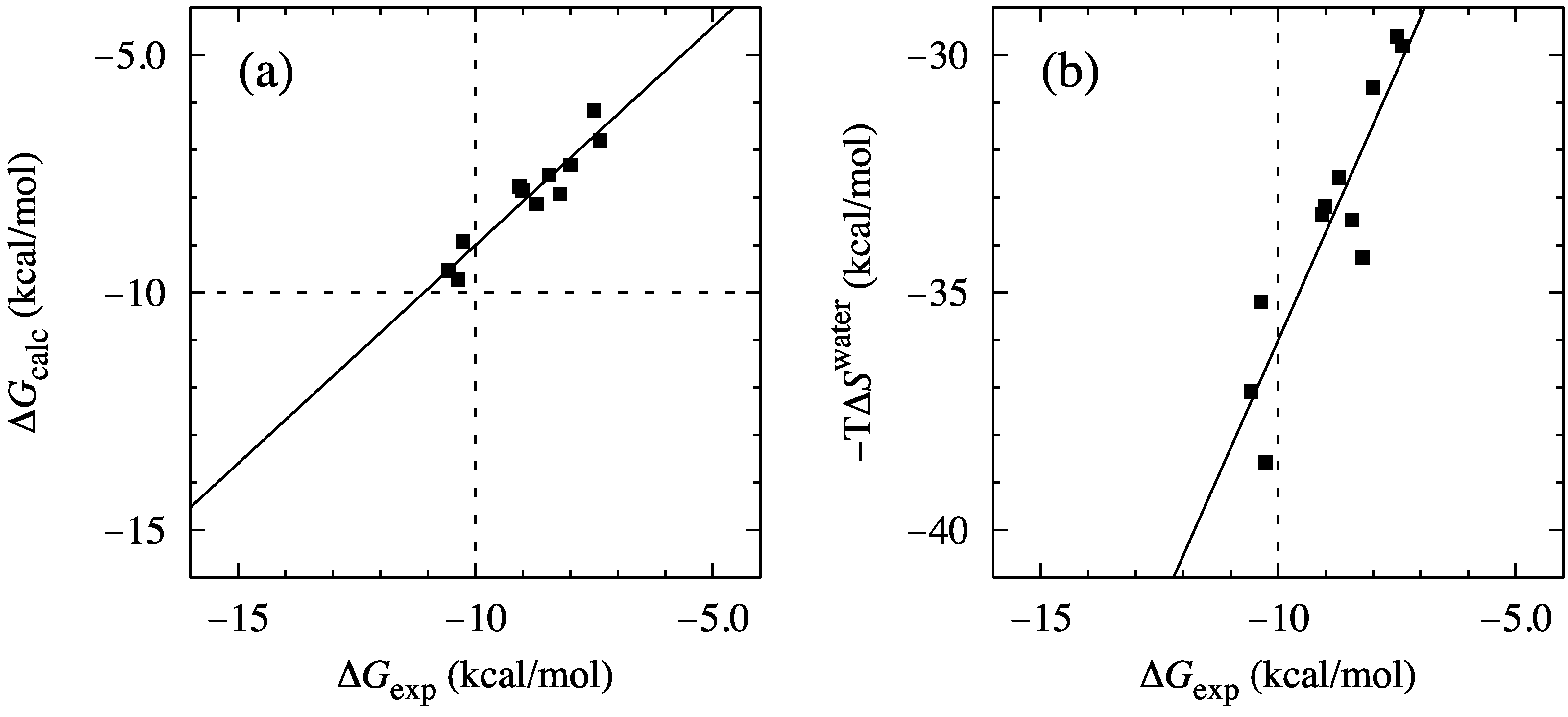
| ΔSconfig | |||||||
|---|---|---|---|---|---|---|---|
| Ligand # | trans | rot | vib | total | ΔSwater | ΔGcalc | ΔGexp |
| 1 | 12.1 | 9.7 | −2.5 | 19.2 | −29.6 | −10.4 | −7.5 |
| 2 | 12.1 | 9.6 | −2.8 | 18.8 | −29.8 | −11.0 | −7.4 |
| 3 | 12.0 | 9.5 | −2.4 | 19.2 | −30.7 | −11.5 | −8.0 |
| 4 | 12.3 | 10.1 | −2.2 | 20.2 | −32.6 | −12.3 | −8.7 |
| 5 | 12.2 | 10.2 | −1.3 | 21.1 | −33.2 | −12.0 | −9.0 |
| 6 | 12.4 | 10.6 | 2.4 | 25.4 | −38.6 | −13.1 | −10.3 |
| 7 | 12.4 | 10.4 | −1.5 | 21.3 | −35.2 | −13.9 | −10.4 |
| 8 | 12.4 | 10.4 | 0.5 | 23.3 | −37.1 | −13.7 | −10.6 |
| 9 | 12.4 | 10.4 | −0.6 | 22.1 | −34.3 | −12.1 | −8.2 |
| 10 | 12.4 | 10.3 | −1.0 | 21.8 | −33.5 | −11.7 | −8.5 |
| 11 | 12.4 | 10.3 | −1.3 | 21.4 | −33.4 | −12.0 | −9.1 |
4. Conclusions
Acknowledgements
References
- Anfinsen, C.B. Principles that govern folding of protein chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A. Dominant forces in protein folding. Biochemistry 1990, 29, 7133–7155. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Lin, S.L.; Nussiov, R. Protein binding versus protein folding: The role of hydrophilic bridges in protein associations. J. Mol. Biol. 1997, 265, 68–84. [Google Scholar] [CrossRef] [PubMed]
- Hendsch, Z.S.; Tidor, B. Do salt bridges stabilize proteins: A continuum electrostatic analysis. Protein Sci. 1994, 3, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Takano, K.; Yamagata, Y.; Fujii, S.; Yutani, K. Contribution of the hydrophobic effect to the stability of human lysozyme: Calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants. Biochemistry 1997, 36, 688–689. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N. Polar group burial contribute more to protein stability than nonpolar group burial. Biochemistry 2001, 40, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.L.; Davidson, A.R. Mutagenesis of a buried polar interaction in an SH3 domain: Sequence conservation provides the best prediction of stability effects. Biochemistry 1998, 37, 16172–16182. [Google Scholar] [CrossRef] [PubMed]
- Isom, D.G.; Cataneda, C.A.; Cannon, B.R.; Velu, P.D.; Garcia-Moreno, E.B. Charges in the hydrophobic interior of proteins. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 16096–16100. [Google Scholar] [CrossRef] [PubMed]
- Weeks, J.D.; Pratt, L.R. Introduction to special issue on water and associated liquids. J. Stat. Phys. 2011, 145, 207–208. [Google Scholar] [CrossRef]
- Klapper, M.H. On the nature of protein interior. Biochem. Biophys. Acta 1971, 229, 557–566. [Google Scholar] [CrossRef]
- Tsai, J.; Taylor, R.; Chothia, C.; Gerstein, M. The packing density in proteins: Standard radii and volumes. J. Mol. Biol. 1999, 290, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Vitkup, D.; Karplus, M. Native proteins are surface-molten solids: Application of the Lindemann criterion for the solid versus liquid state. J. Mol. Biol. 1999, 285, 1371–1375. [Google Scholar] [CrossRef] [PubMed]
- Fleming, P.J.; Richards, F.M. Protein packing: Dependence on protein size, secondary structure and amino acid composition. J. Mol. Biol. 2000, 299, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.B.O.; Price, S.L. The nature of the N-H O=C hydrogen-bond: An intermolecular perturbation-theory study of the formamide formaldehyde complex. J. Comput. Chem. 1990, 11, 1217–1233. [Google Scholar] [CrossRef]
- Mitchell, J.B.O.; Price, S.L. On the relative strengths of amide…amide and amide…water hydrogen bonds. Chem. Phys. Lett. 1991, 180, 517–523. [Google Scholar] [CrossRef]
- Honig, B.; Cohen, F.E. Adding backbone to protein folding: Why proteins are polypeptides. Fold. Des. 1996, 1, R17–R20. [Google Scholar] [CrossRef]
- Baldwin, R.L. In search of the energetic role of peptide hydrogen bonds. J. Biol. Chem. 2003, 278, 17581–17588. [Google Scholar] [CrossRef] [PubMed]
- Kauzmann, W. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 1959, 14, 1–63. [Google Scholar] [PubMed]
- Lesser, G.J.; Rose, G.D. Hydrophobicity of amino acid subgroup in proteins. Proteins 1990, 8, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Sugai, Y. Methodology of predicting approximate shapes and size distribution of micelles: Illustration for simple models. J. Comput. Chem. 2002, 23, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Asakura, S.; Oosawa, F. Interaction between particles suspended in solutions of macromolecules. J. Polym. Sci. 1958, 33, 183–192. [Google Scholar] [CrossRef]
- Dinsmore, A.D.; Yodh, A.G.; Pine, D.J. Entropic control of particle motion using passive surface microstructure. Nature 1996, 383, 239–242. [Google Scholar] [CrossRef]
- Anderson, V.J.; Lekkerkerker, H.N.W. Insights into phase transition kinetics from colloid science. Nature 2002, 416, 811–815. [Google Scholar] [CrossRef] [PubMed]
- Harano, Y.; Kinoshita, M. Large gain in translational entropy of water is a major driving force in protein folding. Chem. Phys. Lett. 2004, 399, 342–348. [Google Scholar] [CrossRef]
- Harano, Y.; Kinoshita, M. Translational-entropy gain of solvent upon protein folding. Biophys. J. 2005, 89, 2701–2710. [Google Scholar] [CrossRef] [PubMed]
- Doig, A.J.; Sternberg, M.J.E. Side-chain conformational entropy in protein folding. Protein Sci. 1995, 4, 2247–2251. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.; Harano, Y.; Kinoshita, M. Morphometric approach to the solvation free energy of complex molecules. Phys. Rev. Lett. 2006, 97, 78101. [Google Scholar] [CrossRef]
- Harano, Y.; Roth, R.; Sugita, Y.; Ikeguchi, M.; Kinoshita, M. Physical basis for characterizing native structures of proteins. Chem. Phys. Lett. 2007, 437, 112–116. [Google Scholar] [CrossRef]
- Cann, N.M.; Patey, G.N. An investigation of the influence of solute size and insertion conditions on solvation thermodynamics. J. Chem. Phys. 1997, 106, 8165–8195. [Google Scholar] [CrossRef]
- Kinoshita, M.; Harano, Y.; Akiyama, R. Changes in thermodynamic quantities upon contact of two solutes in solvent under isochoric and isobaric conditions. J. Chem. Phys. 2006, 125, 244504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, T.; Harano, Y.; Kinoshita, M.; Kovalenko, A.; Hirata, F. Theoretical analysis on hydration thermodynamics of proteins. J. Chem. Phys. 2007, 125, 024911. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Matubayasi, N.; Harano, Y.; Nakahara, M. Pair-correlation entropy of hydrophobic hydration: Decomposition into translational and orientational contributions and analysis of solute-size effects. J. Chem. Phys. 2006, 124, 244512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, T.; Harano, Y.; Kinoshita, M.; Kovalenko, A.; Hirata, F. Theoretical analysis on changes in thermodynamic quantities upon protein folding: Essential role of hydration. J. Chem. Phys. 2007, 126, 225102. [Google Scholar] [CrossRef] [PubMed]
- Kusalik, P.G.; Patey, G.N. On the molecular theory of aqueous electrolyte solutions. I. The solution of the RHNC approximation for models in finite concentration. J. Chem. Phys. 1988, 88, 7715–7738. [Google Scholar] [CrossRef]
- Kusalik, P.G.; Patey, G.N. The solution of the reference hypernetted-chain approximation for water-like models. Mol. Phys. 1988, 65, 1105–1119. [Google Scholar] [CrossRef]
- Kinoshita, M.; Bérard, D.R. Analysis of the bulk and surface-induced structure of electrolyte solutions using integral equation theories. J. Comput. Phys. 1996, 124, 230–241. [Google Scholar] [CrossRef]
- Kinoshita, M. Molecular origin of the hydrophobic effect: Analysis using the angle-dependent integral equation theory. J. Chem. Phys. 2008, 128, 024507. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M. Water structure and phase transition near a surface. J. Sol. Chem. 2004, 33, 661–687. [Google Scholar] [CrossRef]
- Ren, P.; Ponder, J.W. Polarizable atomic multipole water model for molecular mechanics simulation. J. Phys. Chem. B 2003, 107, 5933–5947. [Google Scholar] [CrossRef]
- Morita, T. Theory of classical fluids: Hyper-netted chain approximation. III: A new integral equation for the pair distribution function. Prog. Theor. Phys. 1960, 23, 829–845. [Google Scholar] [CrossRef]
- Morita, T.; Hiroike, K. A new approach of the theory of classical fluids. III: General treatment of classical systems. Prog. Theor. Phys. 1961, 25, 537–578. [Google Scholar] [CrossRef]
- Connolly, M.L. Solvent-accessible surfaces of proteins and nucleic acids. Science 1983, 221, 709–713. [Google Scholar] [CrossRef] [PubMed]
- McDonald, I.K.; Thornton, J.M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994, 1994, 132, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Snedon, S.F.; Tobias, D.J.; Brooks, C.L., III. Thermodynamics of amide hydrogen bond formation in polar and apolar solvents. J. Mol. Biol. 1989, 209, 817–820. [Google Scholar] [CrossRef]
- Fleming, P.J.; Rose, G.D. Do all backbone polar groups in proteins form hydrogen bonds? Protein Sci. 2005, 14, 1911–1917. [Google Scholar] [CrossRef] [PubMed]
- Yoshidome, T.; Oda, K.; Harano, Y.; Roth, R.; Sugita, Y.; Ikeguchi, M.; Kinoshita, M. Free-energy function based on an all-atom model for proteins. Proteins 2009, 77, 950–961. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, S.; Yoshidome, T.; Harano, Y.; Roth, R.; Oshima, H.; Oda, K.; Sugita, Y.; Ikeguchi, M.; Kinoshita, M. Free-energy function for discriminating the native fold of a protein from misfolded decoys. Proteins 2011, 79, 2161–2171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, B.; Levitt, M. Energy functions that discriminates X-ray and near native folds from well-constructed decoys. J. Mol. Biol. 1996, 258, 367–392. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.T.; Kooperberg, C.; Huang, E.; Baker, D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and bayesian scoring functions. J. Mol. Biol. 1997, 268, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.T.; Bonneau, R.; Ruczinski, I.; Baker, D. Ab initio protein structure prediction of CASP III targets using ROSETTA. Proteins 1999, 37, 171–176. [Google Scholar] [CrossRef]
- Samudrala, R.; Xia, Y.; Levitt, M.; Huang, E. S. A combined approach for ab initio construction of low resolution protein tertiary structures from sequence. In Proceedings of the Pacific Symposium on Biocomputing, Mauni Lani, Big Island, HI, USA, 4–9 January 1999; Volume 4, pp. 505–516.
- Xia, Y.; Huang, E.S.; Levitt, M.; Samudrala, R. Ab initio construction of protein tertiary structures using a hierarchical approach. J. Mol. Biol. 2000, 300, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Keasar, C.; Levitt, M. A novel approach to decoy set generation: Designing a physical energy function having local minima with native structure characteristics. J. Mol. Biol. 2003, 329, 159–174. [Google Scholar] [CrossRef]
- Samudrala, R.; Levitt, M. A comprehensive analysis of 40 blind protein structure predictions. BMC Struct. Biol. 2002, 2, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Samudrala, R.; Levitt, M. Decoys ‘R’ Us: A database of incorrect protein conformations to improve protein structure prediction. Protein Sci. 2000, 9, 1399–1401. [Google Scholar] [CrossRef] [PubMed]
- Güntert, P.; Braun, W.; Wüthrich, K. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J. Mol. Biol. 1991, 217, 517–530. [Google Scholar] [CrossRef]
- Bowie, J.U.; Lüthy, R.; Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Sippl, M.J. Boltzmann's principle, knowledge-based mean fields and protein folding. An approach to the computational determination of protein structures. J. Comput. Aided Mol. Des. 1993, 7, 473–501. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Dousis, A.D.; Ma, J. OPUS-PSP: An orientation-dependent statistical all-atom potential derived from side-chain packing. J. Mol. Biol. 2008, 376, 288–301. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, S.; Jernigan, R.L. How effective for fold recognition is a potential of mean force that includes relative orientations between contacting residues in proteins? J. Chem. Phys. 2005, 122, 024901:1–024901:18. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.C.; Duan, Y. Distinguish protein decoys by using a scoring function based on a new AMBER force field, short molecular dynamics simulations, and the generalized born solvent model. Proteins 2004, 55, 620–634. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, S.; Zhou, H.; Zhou, Y. An accurate, residue-level, pair potential of mean force for folding and binding based on the distance-scaled, ideal-gas reference state. Protein Sci. 2004, 13, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Onizuka, K.; Noguchi, T.; Akiyama, Y.; Matsuda, H. Using data compression for multidimensional distribution analysis. Control Intell. Syst. 2002, 17, 48–54. [Google Scholar]
- Zhou, H.; Zhou, Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002, 11, 2714–2726. [Google Scholar] [CrossRef] [PubMed]
- Toby, D.; Elber, R. Distance-dependent, pair potential for protein folding: Results from linear optimization. Proteins 2000, 41, 40–46. [Google Scholar]
- MacKerell, A.D., Jr.; Bashford, D.; Bellott, M.; Dunbrack, R.L., Jr.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef] [PubMed]
- MacKerell, A.D., Jr.; Feig, M.; Brooks, C.L., III. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004, 25, 1400–1415. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Salsbury, F.R., Jr.; Brooks, C.L., III. Novel generalized Born methods. J. Chem. Phys. 2002, 116, 10606–10614. [Google Scholar] [CrossRef]
- Lee, M.S.; Salsbury, F.R., Jr.; Brooks, C.L., III. New analytic approximation to the standard molecular volume definition and its application to generalized Born calculations. J. Comput. Chem. 2003, 24, 1348–1356. [Google Scholar] [CrossRef] [PubMed]
- Chocholousšová, J.; Feig, M. Balancing an accurate representation of the molecular surface in generalized born formalisms with integrator stability in molecular dynamics simulations. J. Comput. Chem. 2006, 27, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.E. The alanine dipeptide free energy surface in solution. J. Chem. Phys. 1999, 111, 5568–5579. [Google Scholar] [CrossRef]
- Mu, Y.; Kosov, D.S.; Stock, G. Conformational dynamics of trialanine in water. 2. Comparison of AMBER, CHARMM, GROMOS, and OPLS force fields to NMR and infrared experiments. J. Phys. Chem. B 2003, 107, 5064–5073. [Google Scholar] [CrossRef]
- Chiba, S.; Harano, Y.; Roth, R.; Kinoshita, M.; Sakurai, M. Evaluation of protein-ligand binding free energy focused on its entropic components. J. Comput. Chem. 2012, 33, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Adcock, S.A.; McCammon, J.A. Molecular dynamics: Survey of methods for simulating the activity of proteins. Chem. Rev. 2006, 106, 1589–1615. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.A.; Chen, W.; Gilson, M.K. Ligand configurational entropy and protein binding. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 1534. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, G.D.; Cramer, C.J.; Truhlar, D.G. Pairwise solute descreening of solute charges from a dielectric medium. Chem. Phys. Lett. 1995, 246, 122–l29. [Google Scholar] [CrossRef]
- Lamb, M.L.; Tirado-Rives, J.; Jorgensen, W.L. Estimation of the binding affinities of FKBP12 inhibitors using a linear response method. Bioorg. Med. Chem. 1999, 7, 851. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, R. A computational analysis of the binding affinities of FKBP12 inhibitors using the MM-PB/SA method. Proteins 2006, 64, 1058. [Google Scholar] [CrossRef] [PubMed]
- Jayachandran, G.; Shirts, M.R.; Park, S.; Pande, V.S. Parallelized-over-parts computation of absolute binding free energy with docking and molecular dynamics. J. Chem. Phys. 2006, 125, 084901. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Deng, Y.; Roux, B. Absolute binding free energy calculations using molecular dynamics simulations with restraining potentials. Biophys. J. 2006, 91, 2798. [Google Scholar] [CrossRef] [PubMed]
- Fujitani, H.; Tanida, Y.; Ito, M.; Jayachandran, G.; Snow, C.D.; Shirts, M.R.; Sorin, E.J.; Pande, V.S. Direct calculation of the binding free energies of FKBP ligands. J. Chem. Phys. 2005, 123, 084108. [Google Scholar] [CrossRef] [PubMed]
- Fujitani, H.; Tanida, Y.; Matsuura, A. Massively parallel computation of absolute binding free energy with well-equilibrated states. Phys. Rev. E 2009, 79, 021914. [Google Scholar] [CrossRef]
- Szep, S.; Park, S.; Boder, E.T.; Van Duyne, G.D.; Saven, J.G. Structural coupling between FKBP12 and buried water. Proteins 2009, 74, 603. [Google Scholar] [CrossRef] [PubMed]
- Connelly, P.R.; Aldape, R.A.; Bruzzese, F.J.; Chambers, S.P.; Fitzgibbon, M.J.; Fleming, M.A.; Itoh, S.; Livingston, D.J.; Navia, M.A.; Thomson, J.A.; et al. Enthalpy of hydrogen bond formation in a protein-ligand binding reaction. Proc. Natl. Acad. Sci. U. S. A. 1994, 91, 1964–1968. [Google Scholar] [CrossRef] [PubMed]
- Holt, D.A.; Luengo, J.I.; Yamashita, D.S.; Oh, H.-J.; Konialian, A.L.; Yen, H.-K.; Rozamus, L.W.; Brandt, M.; Bossard, M.J.; Levy, M.A.; et al. Design, synthesis, and kinetic evaluation of high-affinity FKBP ligands and the X-ray crystal structures of their complexes with FKBP12. J. Am. Chem. Soc. 1993, 115, 9925. [Google Scholar] [CrossRef]
- Marlow, M.S.; Dogan, J.; Frederick, K.K.; Valentine, K.G.; Wand, A.J. The role of conformational entropy in molecular recognition by calmodulin. Nat. Chem. Biol. 2010, 6, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Irudayam, S.J.; Henchman, R.H. Entropic cost of protein-ligand binding and its dependence on the entropy in solution. J. Phys. Chem. 2009, 113, 5871–5884. [Google Scholar] [CrossRef] [PubMed]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Harano, Y. Application of Hydration Thermodynamics to the Evaluation of Protein Structures and Protein-Ligand Binding. Entropy 2012, 14, 1443-1468. https://doi.org/10.3390/e14081443
Harano Y. Application of Hydration Thermodynamics to the Evaluation of Protein Structures and Protein-Ligand Binding. Entropy. 2012; 14(8):1443-1468. https://doi.org/10.3390/e14081443
Chicago/Turabian StyleHarano, Yuichi. 2012. "Application of Hydration Thermodynamics to the Evaluation of Protein Structures and Protein-Ligand Binding" Entropy 14, no. 8: 1443-1468. https://doi.org/10.3390/e14081443




