Salivary Exosomes as Nanocarriers for Cancer Biomarker Delivery
Abstract
:1. Introduction
2. Salivary Diagnostics
3. Salivary Exosomes
3.1. Salivary Extracellular RNA
3.2. Salivary Circulating Tumor DNA
3.3. Salivary Protein Biomarkers
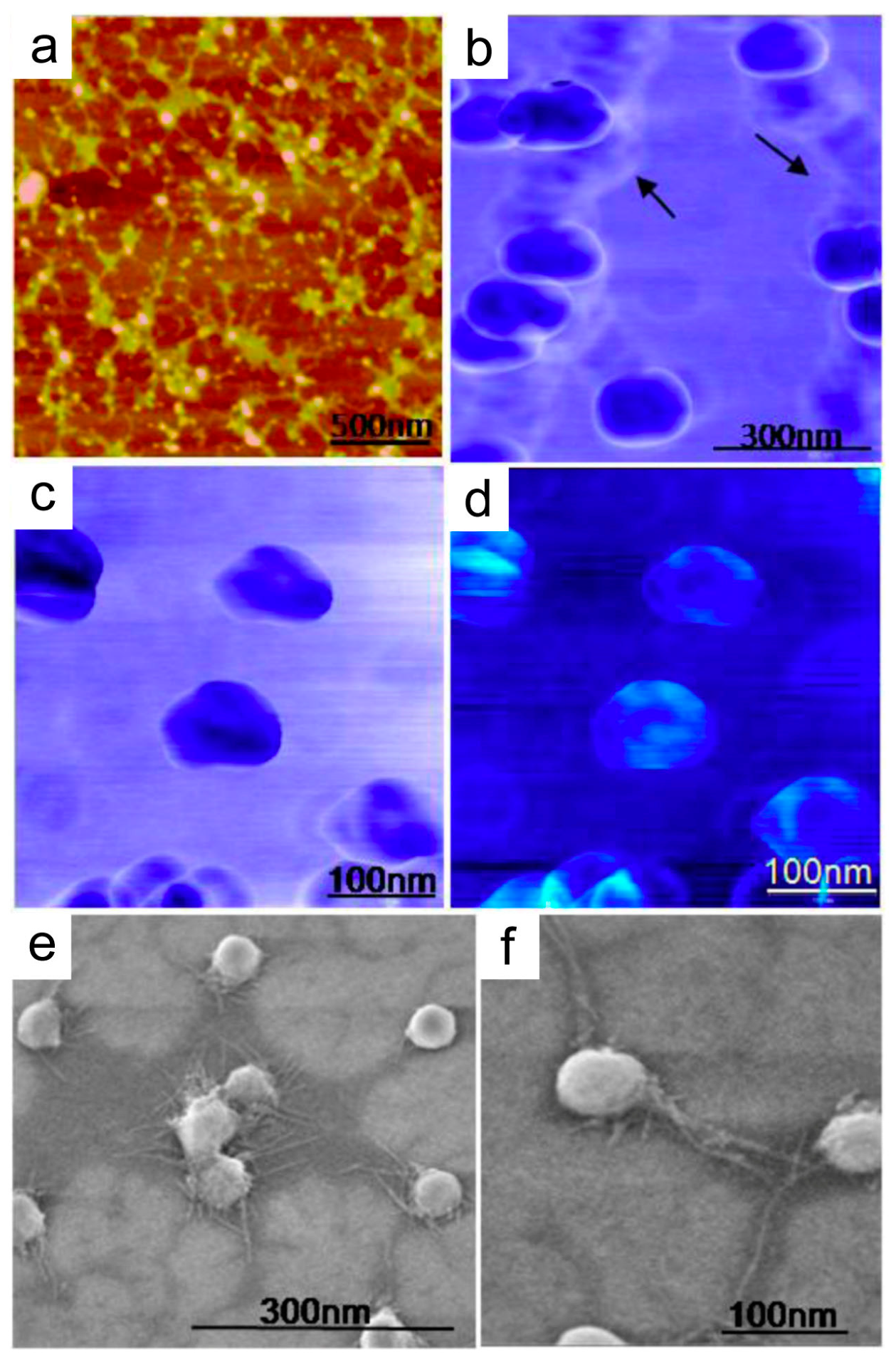
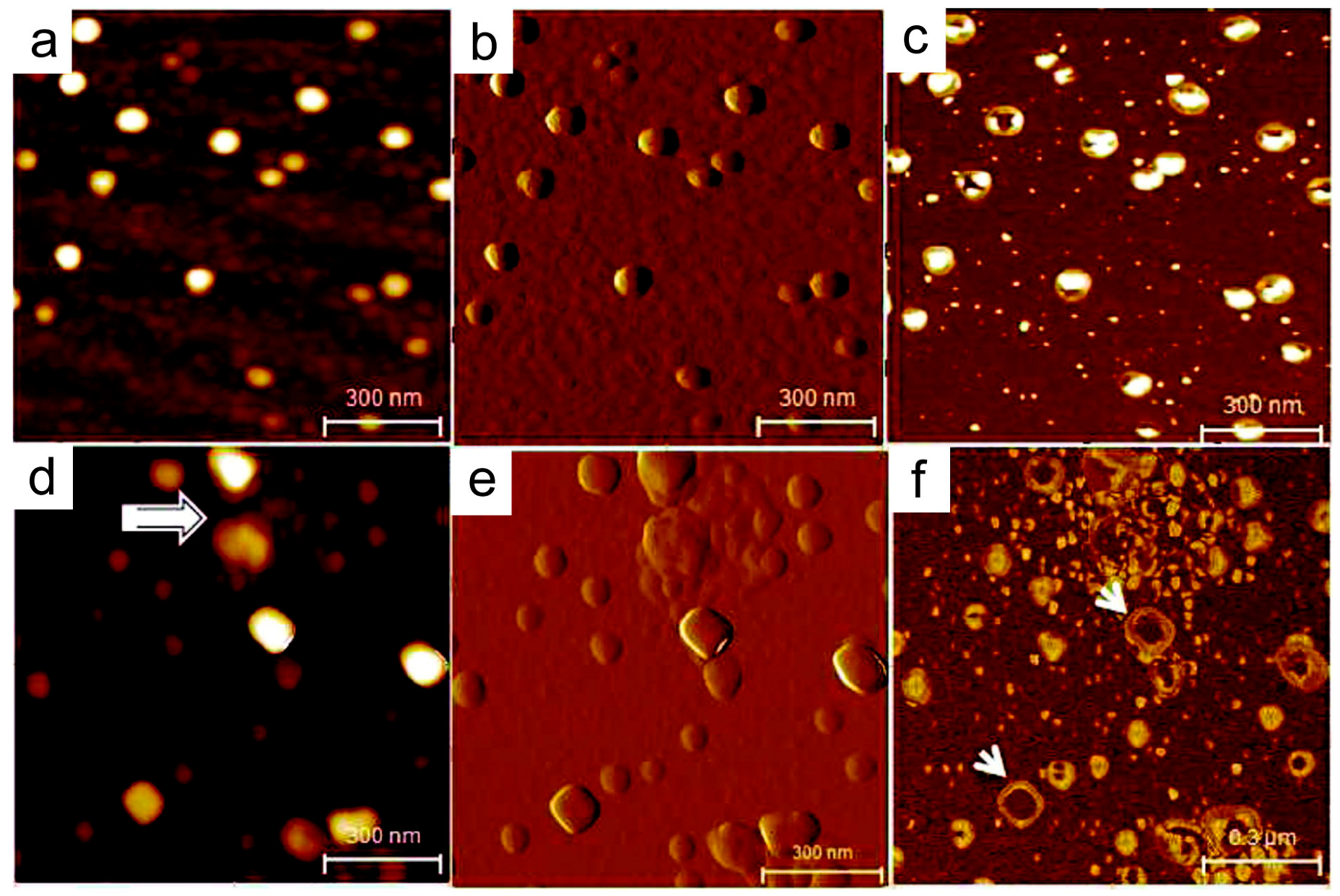
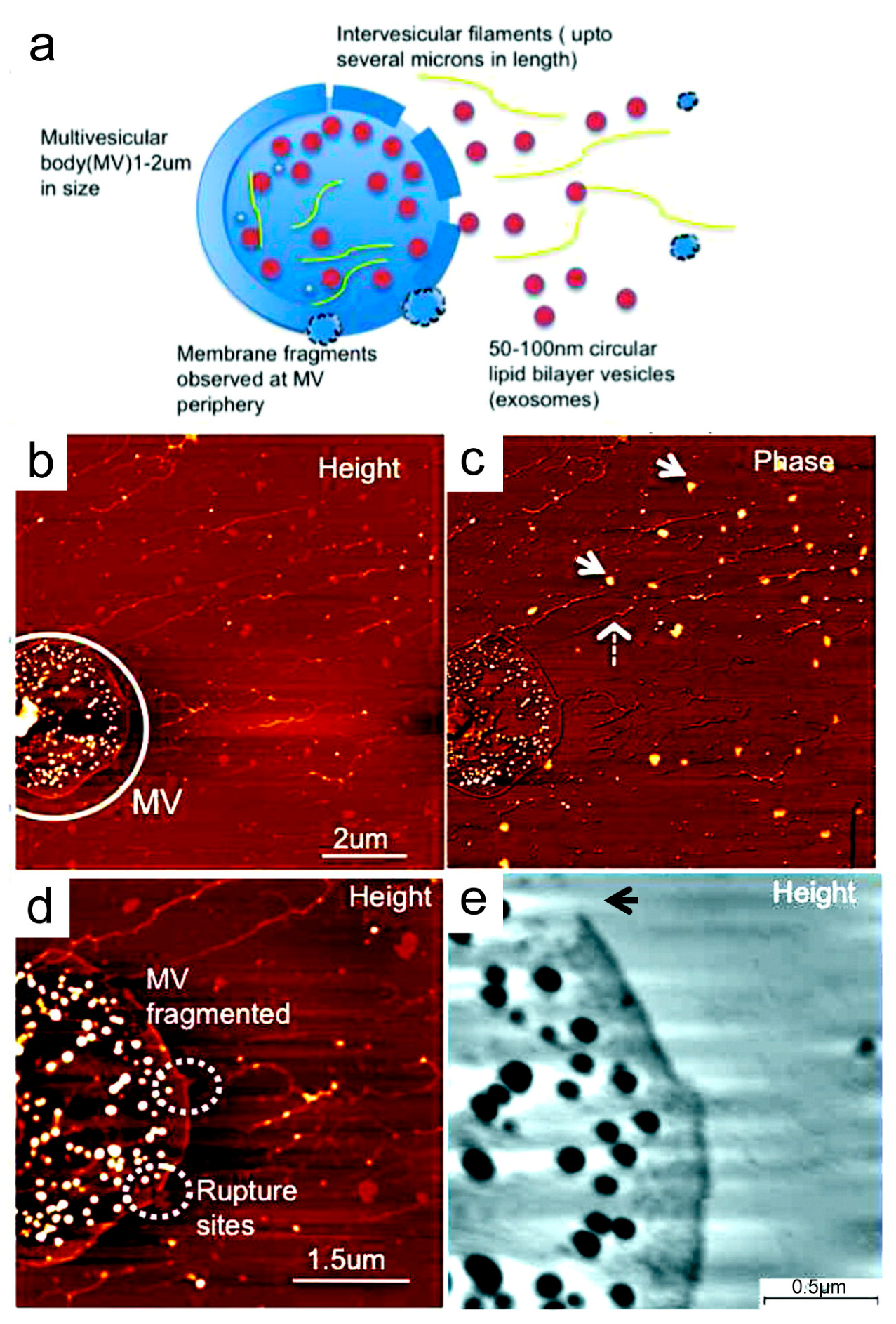
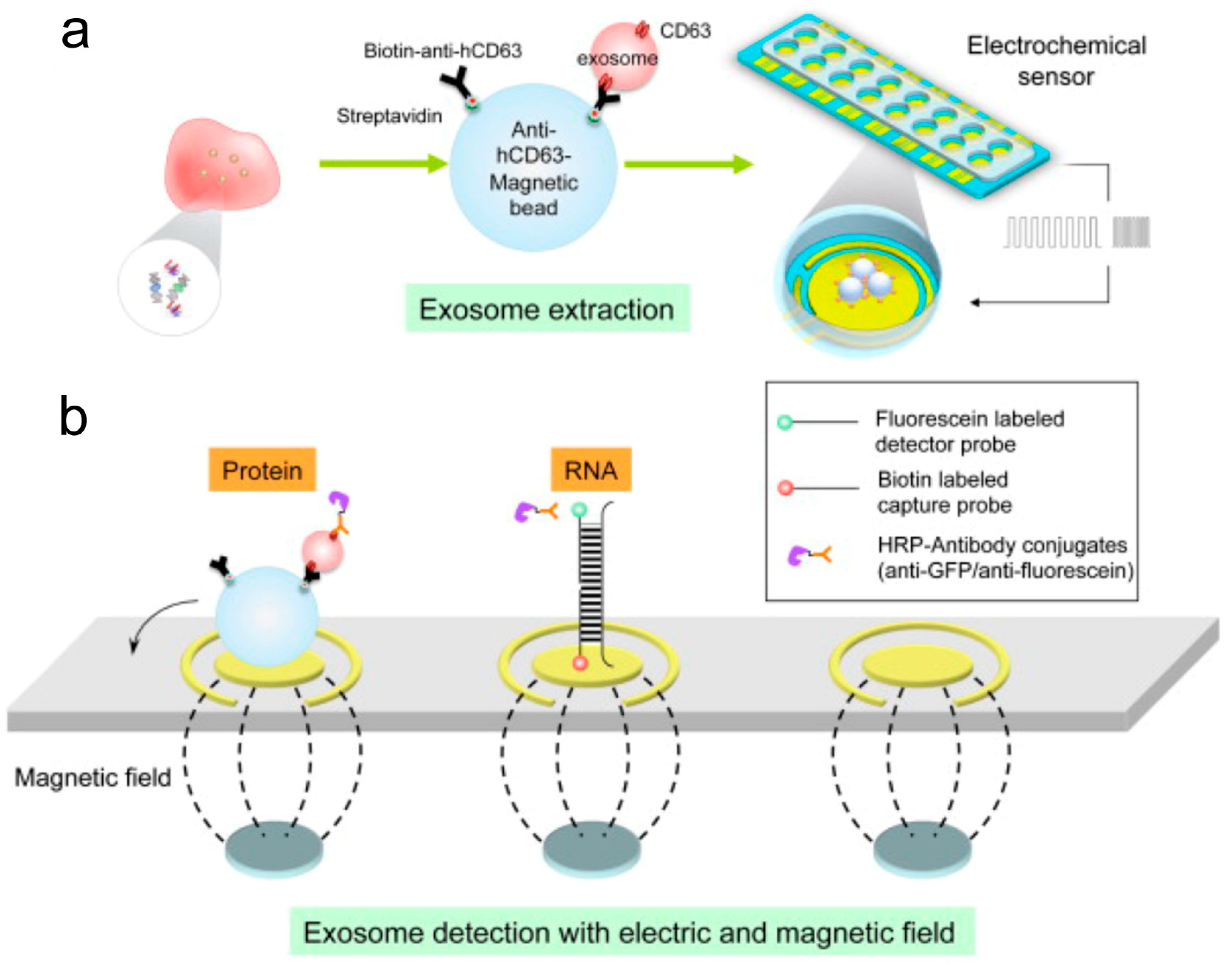
4. Nanostructural Properties of Salivary Exosomes
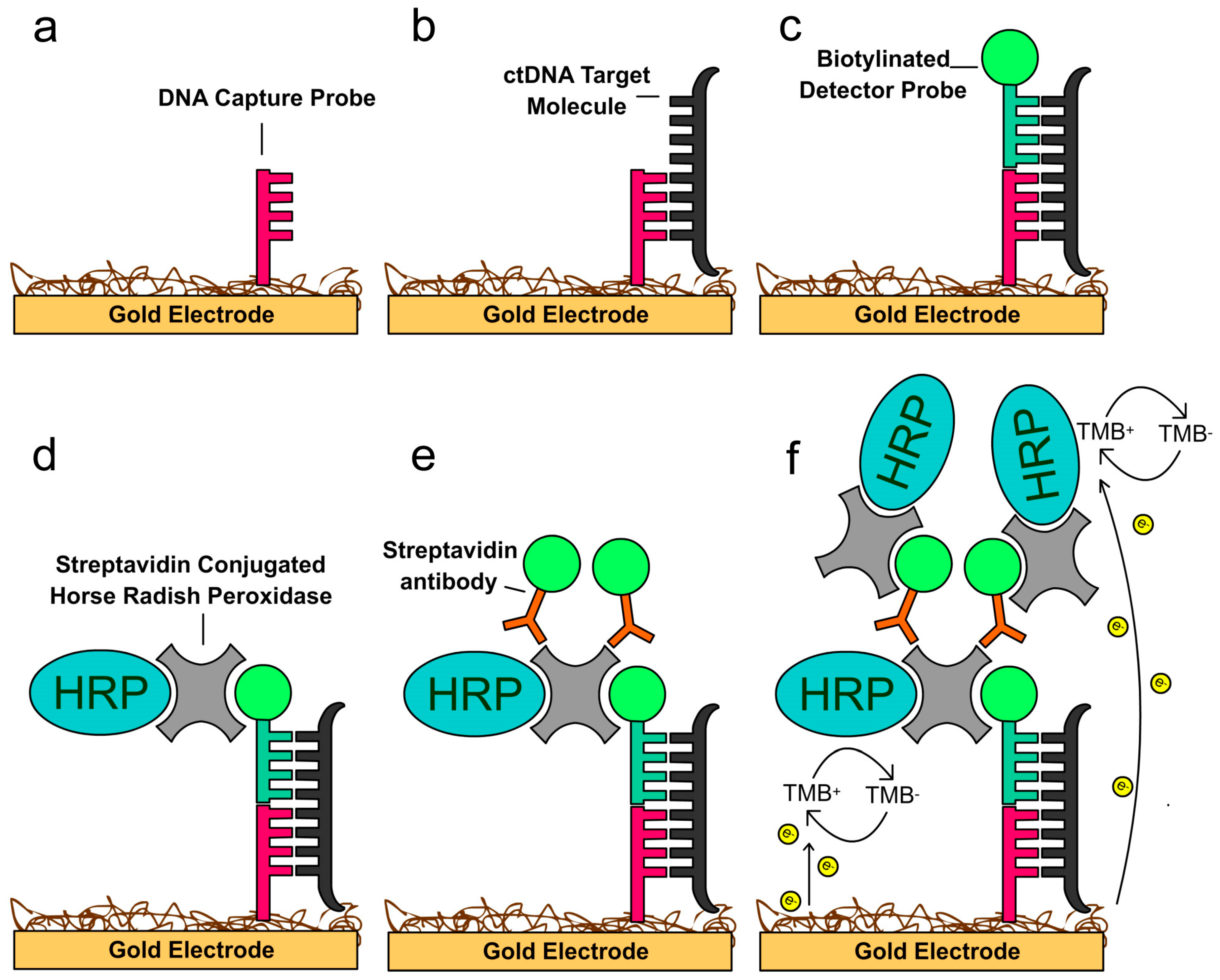
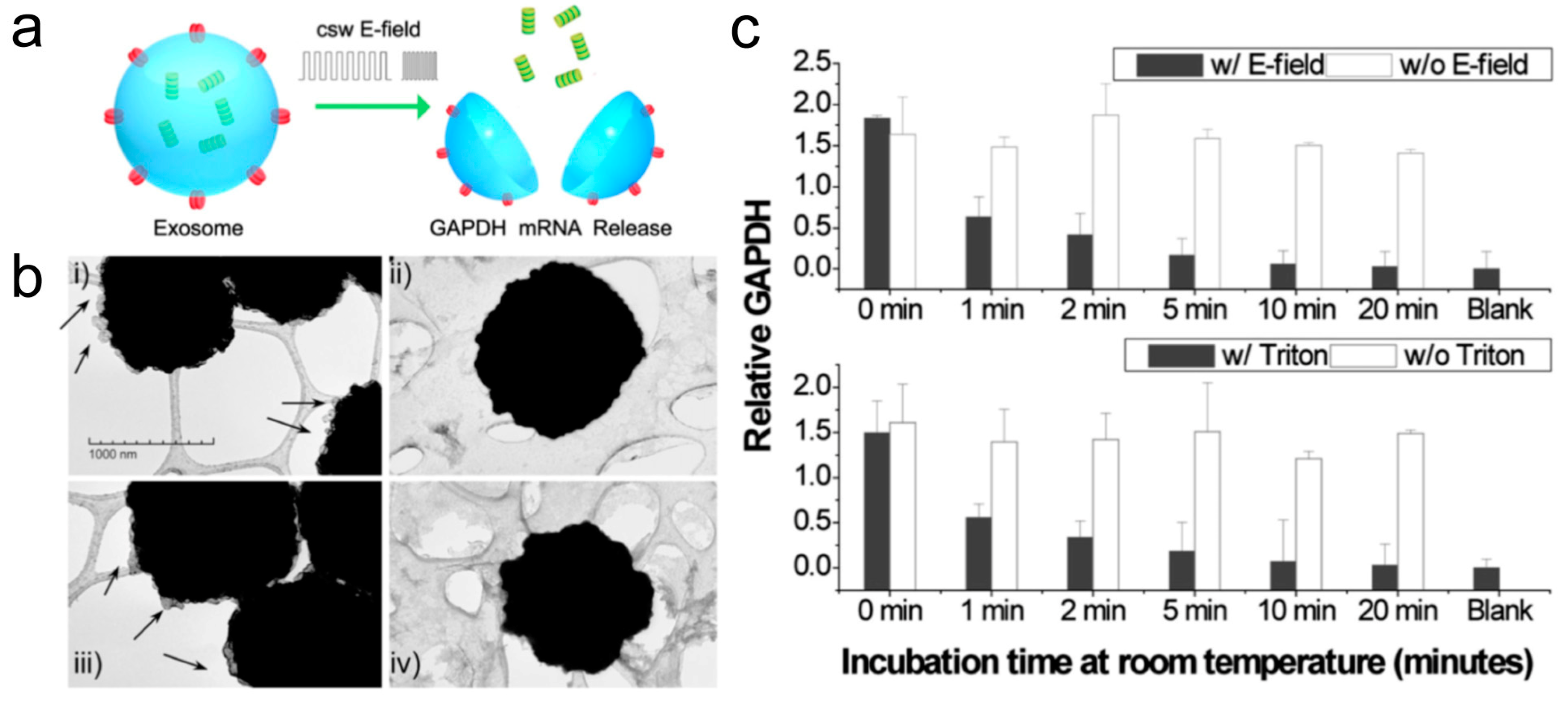
5. Electric Field-Induced Release and Measurement
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Brindle, K. New approaches for imaging tumour responses to treatment. Nat. Rev. Cancer 2008, 8, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Condeelis, J.; Weissleder, R. In vivo imaging in cancer. Cold Spring Harb. Perspect. Biol. 2010, 2, a003848. [Google Scholar] [CrossRef] [PubMed]
- Weissleder, R.; Pittet, M.J. Imaging in the era of molecular oncology. Nature 2008, 452, 580–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nonaka, T.; Wong, D.T.W. Liquid Biopsy in Head and Neck Cancer: Promises and Challenges. J. Dent. Res. 2018, 97, 701–708. [Google Scholar] [CrossRef]
- Lamy, E.; Mau, M. Saliva proteomics as an emerging, non-invasive tool to study livestock physiology, nutrition and diseases. J. Proteomics 2012, 75, 4251–4258. [Google Scholar] [CrossRef]
- Dawes, C.; Pedersen, A.M.; Villa, A.; Ekstrom, J.; Proctor, G.B.; Vissink, A.; Aframian, D.; McGowan, R.; Aliko, A.; Narayana, N.; et al. The functions of human saliva: A review sponsored by the World Workshop on Oral Medicine VI. Arch. Oral Biol. 2015, 60, 863–874. [Google Scholar] [CrossRef]
- Humphrey, S.P.; Williamson, R.T. A review of saliva: Normal composition, flow, and function. J. Prosthet. Dent. 2001, 85, 162–169. [Google Scholar] [CrossRef]
- Miller, C.S.; King, C.P.; Langub, M.C.; Kryscio, R.J.; Thomas, M.V. Salivary biomarkers of existing periodontal disease: A cross-sectional study. J. Am. Dent. Assoc. 2006, 137, 322–329. [Google Scholar] [CrossRef]
- Schipper, R.G.; Silletti, E.; Vingerhoeds, M.H. Saliva as research material: biochemical, physicochemical and practical aspects. Arch. Oral Biol. 2007, 52, 1114–1135. [Google Scholar] [CrossRef]
- Gupta, S.; Sandhu, S.V.; Bansal, H.; Sharma, D. Comparison of salivary and serum glucose levels in diabetic patients. J. Diabetes Sci. Technol. 2015, 9, 91–96. [Google Scholar] [CrossRef]
- Raff, H.; Findling, J.W. Biomarkers: Salivary cortisol or cortisone? Nat. Rev. Endocrinol. 2010, 6, 658–660. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Raposo, G. Exosomes-vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef]
- Niu, Z.; Pang, R.T.K.; Liu, W.; Li, Q.; Cheng, R.; Yeung, W.S.B. Polymer-based precipitation preserves biological activities of extracellular vesicles from an endometrial cell line. PLoS ONE 2017, 12, e0186534. [Google Scholar] [CrossRef]
- Sharma, P.; Ludwig, S.; Muller, L.; Hong, C.S.; Kirkwood, J.M.; Ferrone, S.; Whiteside, T.L. Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma. J. Extracell. Vesicles 2018, 7, 1435138. [Google Scholar] [CrossRef]
- He, M.; Zeng, Y. Microfluidic Exosome Analysis toward Liquid Biopsy for Cancer. J. Lab. Autom. 2016, 21, 599–608. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef] [Green Version]
- Morris, S.A.; Farrell, D.; Grodzinski, P. Nanotechnologies in cancer treatment and diagnosis. J. Natl. Compr. Cancer Netw. 2014, 12, 1727–1733. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.E.; Kwak, J.W.; Park, J.W. Nanotechnology for early cancer detection. Sensors 2010, 10, 428–455. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, T.; Wong, D.T.W. Saliva-Exosomics in Cancer: Molecular Characterization of Cancer-Derived Exosomes in Saliva. Enzymes 2017, 42, 125–151. [Google Scholar] [PubMed]
- Aro, K.; Wei, F.; Wong, D.T.; Tu, M. Saliva Liquid Biopsy for Point-of-Care Applications. Front Public Health 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, G.H. The secretion, components, and properties of saliva. Annu. Rev. Food Sci. Technol. 2013, 4, 267–276. [Google Scholar] [CrossRef]
- Groschl, M. Current status of salivary hormone analysis. Clin. Chem. 2008, 54, 1759–1769. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.K.; Christodoulides, N.J.; Floriano, P.N.; Miller, C.S.; Ebersole, J.L.; Weigum, S.E.; McDevitt, J.; Redding, S.W. Current development of saliva/oral fluid-based diagnostics. Tex Dent. J. 2010, 127, 651–661. [Google Scholar] [PubMed]
- Bandhakavi, S.; Stone, M.D.; Onsongo, G.; Van Riper, S.K.; Griffin, T.J. A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. J. Proteome Res. 2009, 8, 5590–5600. [Google Scholar] [CrossRef]
- Yan, W.; Apweiler, R.; Balgley, B.M.; Boontheung, P.; Bundy, J.L.; Cargile, B.J.; Cole, S.; Fang, X.; Gonzalez-Begne, M.; Griffin, T.J. Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin. Appl. 2009, 3, 116–134. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Zhang, W.; Wang, M.L. Sensing of Salivary Glucose Using Nano-Structured Biosensors. Biosensors 2016, 6, 10. [Google Scholar] [CrossRef]
- Zachary, D.; Mwenge, L.; Muyoyeta, M.; Shanaube, K.; Schaap, A.; Bond, V.; Kosloff, B.; de Haas, P.; Ayles, H. Field comparison of OraQuick ADVANCE Rapid HIV-1/2 antibody test and two blood-based rapid HIV antibody tests in Zambia. BMC Infect. Dis. 2012, 12, 183. [Google Scholar] [CrossRef] [PubMed]
- Floriano, P.N.; Christodoulides, N.; Miller, C.S.; Ebersole, J.L.; Spertus, J.; Rose, B.G.; Kinane, D.F.; Novak, M.J.; Steinhubl, S.; Acosta, S. Use of saliva-based nano-biochip tests for acute myocardial infarction at the point of care: a feasibility study. Clin. Chem. 2009, 55, 1530–1538. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Wong, D.T. Saliva: An emerging biofluid for early detection of diseases. Am. J. Dent. 2009, 22, 241–248. [Google Scholar] [PubMed]
- Cooke, F.; Bullen, C.; Whittaker, R.; McRobbie, H.; Chen, M.H.; Walker, N. Diagnostic accuracy of NicAlert cotinine test strips in saliva for verifying smoking status. Nicot. Tob. Res. 2008, 10, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Van Doornum, G.J.; Lodder, A.; Buimer, M.; van Ameijden, E.J.; Bruisten, S. Evaluation of hepatitis C antibody testing in saliva specimens collected by two different systems in comparison with HCV antibody and HCV RNA in serum. J. Med. Virol. 2001, 64, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Walt, D.R.; Blicharz, T.M.; Hayman, R.B.; Rissin, D.M.; Bowden, M.; Siqueira, W.L.; Helmerhorst, E.J.; Grand-Pierre, N.; Oppenheim, F.G.; Bhatia, J.S.; et al. Microsensor arrays for saliva diagnostics. Ann. N. Y. Acad. Sci. 2007, 1098, 389–400. [Google Scholar] [CrossRef]
- Christodoulides, N.; Floriano, P.N.; Miller, C.S.; Ebersole, J.L.; Mohanty, S.; Dharshan, P.; Griffin, M.; Lennart, A.; Ballard, K.L.; King, C.P. Lab-on-a-chip methods for point-of-care measurements of salivary biomarkers of periodontitis. Ann. N. Y. Acad. Sci. 2007, 1098, 411–428. [Google Scholar] [CrossRef]
- Ai, J.; Smith, B.; Wong, D.T. Saliva ontology: an ontology-based framework for a Salivaomics Knowledge Base. BMC Bioinf. 2010, 11, 302. [Google Scholar] [CrossRef]
- Wong, D.T. Salivaomics. J. Am. Dent. Assoc. 2012, 143, 19S–24S. [Google Scholar] [CrossRef]
- Herr, A.E.; Hatch, A.V.; Throckmorton, D.J.; Tran, H.M.; Brennan, J.S.; Giannobile, W.V.; Singh, A.K. Microfluidic immunoassays as rapid saliva-based clinical diagnostics. Proc. Natl. Acad. Sci. USA 2007, 104, 5268–5273. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, D.B.; Botchway, C.A. Circadian variations in the flow rate and composition of whole saliva stimulated by mastication. Arch. Oral Biol. 1979, 24, 877–881. [Google Scholar] [CrossRef]
- Mackie, D.A.; Pangborn, R.M. Mastication and its influence on human salivary flow and alpha-amylase secretion. Physiol. Behav. 1990, 47, 593–595. [Google Scholar] [CrossRef]
- Henson, B.S.; Wong, D.T. Collection, storage, and processing of saliva samples for downstream molecular applications. Methods Mol. Biol. 2010, 666, 21–30. [Google Scholar] [PubMed]
- Helmerhorst, E.J.; Oppenheim, F.G. Saliva: A dynamic proteome. J. Dent. Res. 2007, 86, 680–693. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.H.; Thomas, G.A.; Liao, W.; Grogan, T.; Buck, R.L.; Fuentes, L.; Yakob, M.; Laughlin, M.J.; Schafer, C.; Nazmul-Hossain, A.; et al. RNAPro*SAL: a device for rapid and standardized collection of saliva RNA and proteins. Biotechniques 2015, 58, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Wong, D.T. Method development for proteome stabilization in human saliva. Anal. Chim. Acta. 2012, 722, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borras, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [PubMed]
- Samos, J.; Garcia-Olmo, D.C.; Picazo, M.G.; Rubio-Vitaller, A.; Garcia-Olmo, D. Circulating nucleic acids in plasma/serum and tumor progression: Are apoptotic bodies involved? An experimental study in a rat cancer model. Ann. N. Y. Acad. Sci. 2006, 1075, 165–173. [Google Scholar] [CrossRef]
- Taylor, R.C.; Cullen, S.P.; Martin, S.J. Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008, 9, 231–241. [Google Scholar] [CrossRef]
- Andaloussi, S.E.; Mager, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Lasser, C.; Alikhani, V.S.; Ekstrom, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjostrand, M.; Gabrielsson, S.; Lotvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, M.T.; Obuse, C.; et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017, 8, 15287. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Kanai-Azuma, M.; Akimoto, Y.; Kawakami, H.; Yanoshita, R. Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Biol. Pharm. Bull. 2008, 31, 1059–1062. [Google Scholar] [CrossRef] [PubMed]
- Al-Tarawneh, S.K.; Border, M.B.; Dibble, C.F.; Bencharit, S. Defining salivary biomarkers using mass spectrometry-based proteomics: A systematic review. OMICS 2011, 15, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Park, N.J.; Li, Y.; Yu, T.; Brinkman, B.M.; Wong, D.T. Characterization of RNA in saliva. Clin. Chem. 2006, 52, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, X.; St John, M.A.; Wong, D.T. RNA profiling of cell-free saliva using microarray technology. J. Dent. Res. 2004, 83, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Park, N.J.; Zhou, X.; Yu, T.; Brinkman, B.M.; Zimmermann, B.G.; Palanisamy, V.; Wong, D.T. Characterization of salivary RNA by cDNA library analysis. Arch. Oral Biol. 2007, 52, 30–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spielmann, N.; Ilsley, D.; Gu, J.; Lea, K.; Brockman, J.; Heater, S.; Setterquist, R.; Wong, D.T. The human salivary RNA transcriptome revealed by massively parallel sequencing. Clin. Chem. 2012, 58, 1314–1321. [Google Scholar] [CrossRef]
- Bahn, J.H.; Zhang, Q.; Li, F.; Chan, T.M.; Lin, X.; Kim, Y.; Wong, D.T.; Xiao, X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015, 61, 221–230. [Google Scholar] [CrossRef]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef]
- Michael, A.; Bajracharya, S.D.; Yuen, P.S.; Zhou, H.; Star, R.A.; Illei, G.G.; Alevizos, I. Exosomes from human saliva as a source of microRNA biomarkers. Oral. Dis. 2010, 16, 34–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, Y.; Taketomi, Y.; Murakami, M.; Tsujimoto, M.; Yanoshita, R. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol. Pharm. Bull. 2013, 36, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Tsujimoto, M.; Yanoshita, R. Next-Generation sequencing of protein-coding and long non-protein-coding RNAs in two types of exosomes derived from Human whole saliva. Biol. Pharm. Bull. 2016, 39, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; St John, M.A.; Zhou, X.; Kim, Y.; Sinha, U.; Jordan, R.C.; Eisele, D.; Abemayor, E.; Elashoff, D.; Park, N.H. Salivary transcriptome diagnostics for oral cancer detection. Clin. Cancer Res. 2004, 10, 8442–8450. [Google Scholar] [CrossRef] [PubMed]
- Matse, J.H.; Yoshizawa, J.; Wang, X.; Elashoff, D.; Bolscher, J.G.; Veerman, E.C.; Bloemena, E.; Wong, D.T. Discovery and prevalidation of salivary extracellular microRNA biomarkers panel for the noninvasive detection of benign and malignant parotid gland tumors. Clin. Cancer Res. 2013, 19, 3032–3038. [Google Scholar] [CrossRef]
- Du, J.; Zhang, L. Analysis of salivary microRNA expression profiles and identification of novel biomarkers in esophageal cancer. Oncol. Lett. 2017, 14, 1387–1394. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Xiao, H.; Zhou, H.; Santiago, S.; Lee, J.M.; Garon, E.B.; Yang, J.; Brinkmann, O.; Yan, X.; Akin, D.; et al. Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol. Life Sci. 2012, 69, 3341–3350. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Farrell, J.J.; Zhou, H.; Elashoff, D.; Akin, D.; Park, N.H.; Chia, D.; Wong, D.T. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 2010, 138, 949–957. [Google Scholar] [CrossRef]
- Zhang, L.; Xiao, H.; Karlan, S.; Zhou, H.; Gross, J.; Elashoff, D.; Akin, D.; Yan, X.; Chia, D.; Karlan, B.; et al. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS ONE 2010, 5, e15573. [Google Scholar] [CrossRef]
- Lee, Y.H.; Kim, J.H.; Zhou, H.; Kim, B.W.; Wong, D.T. Salivary transcriptomic biomarkers for detection of ovarian cancer: For serous papillary adenocarcinoma. J. Mol. Med. (Berl) 2012, 90, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Rylander-Rudqvist, T.; Hakansson, N.; Tybring, G.; Wolk, A. Quality and quantity of saliva DNA obtained from the self-administrated oragene method-a pilot study on the cohort of Swedish men. Cancer Epidemiol. Biomarkers Prev. 2006, 15, 1742–1745. [Google Scholar] [CrossRef] [PubMed]
- Bonne, N.J.; Wong, D.T. Salivary biomarker development using genomic, proteomic and metabolomic approaches. Genome Med. 2012, 4, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, T.V.; Simonsen, M.K.; Nielsen, F.C.; Hundrup, Y.A. Collection of blood, saliva, and buccal cell samples in a pilot study on the Danish nurse cohort: comparison of the response rate and quality of genomic DNA. Cancer Epidemiol. Biomarkers Prev. 2007, 16, 2072–2076. [Google Scholar] [CrossRef] [PubMed]
- Looi, M.L.; Zakaria, H.; Osman, J.; Jamal, R. Quantity and quality assessment of DNA extracted from saliva and blood. Clin. Lab. 2012, 58, 307–312. [Google Scholar] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar] [PubMed]
- Diaz, L.A.; Bardelli, A. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef]
- Burnham, P.; Kim, M.S.; Agbor-Enoh, S.; Luikart, H.; Valantine, H.A.; Khush, K.K.; De Vlaminck, I. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci. Rep. 2016, 6, 27859. [Google Scholar] [CrossRef] [Green Version]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Garcia-Olmo, D.C.; Dominguez, C.; Garcia-Arranz, M.; Anker, P.; Stroun, M.; Garcia-Verdugo, J.M.; Garcia-Olmo, D. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 2010, 70, 560–567. [Google Scholar] [CrossRef]
- Aarthy, R.; Mani, S.; Velusami, S.; Sundarsingh, S.; Rajkumar, T. Role of Circulating Cell-Free DNA in Cancers. Mol. Diagn. Ther. 2015, 19, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.A.; Binkley, M.S.; Osmundson, E.C.; Alizadeh, A.A.; Diehn, M. Predicting Radiotherapy Responses and Treatment Outcomes Through Analysis of Circulating Tumor DNA. Semin. Radiat. Oncol. 2015, 25, 305–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichihara, E.; Lovly, C.M. Shades of T790M: Intratumor Heterogeneity in EGFR-Mutant Lung Cancer. Cancer Discov. 2015, 5, 694–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ignatiadis, M.; Lee, M.; Jeffrey, S.S. Circulating Tumor Cells and Circulating Tumor DNA: Challenges and Opportunities on the Path to Clinical Utility. Clin. Cancer Res. 2015, 21, 4786–4800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piotrowska, Z.; Niederst, M.J.; Karlovich, C.A.; Wakelee, H.A.; Neal, J.W.; Mino-Kenudson, M.; Fulton, L.; Hata, A.N.; Lockerman, E.L.; Kalsy, A.; et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov. 2015, 5, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Polivka, J.; Pesta, M.; Janku, F. Testing for oncogenic molecular aberrations in cell-free DNA-based liquid biopsies in the clinic: are we there yet? Expert Rev. Mol. Diagn. 2015, 15, 1631–1644. [Google Scholar] [CrossRef] [PubMed]
- Pepe, M.S.; Etzioni, R.; Feng, Z.; Potter, J.D.; Thompson, M.L.; Thornquist, M.; Winget, M.; Yasui, Y. Phases of biomarker development for early detection of cancer. J. Natl. Cancer Inst. 2001, 93, 1054–1061. [Google Scholar] [CrossRef]
- Navarro, M.A.; Mesia, R.; Diez-Gibert, O.; Rueda, A.; Ojeda, B.; Alonso, M.C. Epidermal growth factor in plasma and saliva of patients with active breast cancer and breast cancer patients in follow-up compared with healthy women. Breast Cancer Res. Treat. 1997, 42, 83–86. [Google Scholar] [CrossRef]
- Streckfus, C.; Bigler, L.; Dellinger, T.; Dai, X.; Kingman, A.; Thigpen, J.T. The presence of soluble c-erbB-2 in saliva and serum among women with breast carcinoma: a preliminary study. Clin. Cancer Res. 2000, 6, 2363–2370. [Google Scholar]
- Streckfus, C.; Bigler, L.; Tucci, M.; Thigpen, J.T. A preliminary study of CA15-3, c-erbB-2, epidermal growth factor receptor, cathepsin-D, and p53 in saliva among women with breast carcinoma. Cancer Invest. 2000, 18, 101–109. [Google Scholar] [CrossRef]
- Brooks, M.N.; Wang, J.; Li, Y.; Zhang, R.; Elashoff, D.; Wong, D.T. Salivary protein factors are elevated in breast cancer patients. Mol. Med. Rep. 2008, 1, 375–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, N.; Streckfus, C.F. The Expression of Lung Resistance Protein in Saliva: A Novel Prognostic Indicator Protein for Carcinoma of the Breast. Cancer Invest. 2015, 33, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Ohshiro, K.; Rosenthal, D.I.; Koomen, J.M.; Streckfus, C.F.; Chambers, M.; Kobayashi, R.; El-Naggar, A.K. Pre-analytic saliva processing affect proteomic results and biomarker screening of head and neck squamous carcinoma. Int. J. Oncol. 2007, 30, 743–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Arellano, M.; Boontheung, P.; Wang, J.; Zhou, H.; Jiang, J.; Elashoff, D.; Wei, R.; Loo, J.A.; Wong, D.T. Salivary proteomics for oral cancer biomarker discovery. Clin. Cancer Res. 2008, 14, 6246–6452. [Google Scholar] [CrossRef] [PubMed]
- Dowling, P.; Wormald, R.; Meleady, P.; Henry, M.; Curran, A.; Clynes, M. Analysis of the saliva proteome from patients with head and neck squamous cell carcinoma reveals differences in abundance levels of proteins associated with tumour progression and metastasis. J. Proteomics 2008, 71, 168–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rai, B.; Kaur, J.; Jacobs, R.; Anand, S.C. Adenosine deaminase in saliva as a diagnostic marker of squamous cell carcinoma of tongue. Clin. Oral. Investig. 2011, 15, 347–349. [Google Scholar] [CrossRef]
- Elashoff, D.; Zhou, H.; Reiss, J.; Wang, J.; Xiao, H.; Henson, B.; Hu, S.; Arellano, M.; Sinha, U.; Le, A.; et al. Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol. Biomarkers Prev. 2012, 21, 664–672. [Google Scholar] [CrossRef]
- Winck, F.V.; Prado Ribeiro, A.C.; Ramos Domingues, R.; Ling, L.Y.; Riano-Pachon, D.M.; Rivera, C.; Brandao, T.B.; Gouvea, A.F.; Santos-Silva, A.R.; Coletta, R.D.; et al. Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. Sci. Rep. 2015, 5, 16305. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.Z.; Wang, J.G.; Zhang, X.L. Diagnostic model of saliva protein finger print analysis of patients with gastric cancer. World J. Gastroenterol. 2009, 15, 865–870. [Google Scholar] [CrossRef]
- Xiao, H.; Zhang, Y.; Kim, Y.; Kim, S.; Kim, J.J.; Kim, K.M.; Yoshizawa, J.; Fan, L.Y.; Cao, C.X.; Wong, D.T. Differential proteomic analysis of Human saliva using tandem mass tags quantification for gastric cancer detection. Sci. Rep. 2016, 6, 22165. [Google Scholar] [CrossRef]
- Xiao, H.; Zhang, L.; Zhou, H.; Lee, J.M.; Garon, E.B.; Wong, D.T. Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol. Cell Proteomics 2012, 11, M111.012112. [Google Scholar] [CrossRef]
- Sun, Y.; Xia, Z.; Shang, Z.; Sun, K.; Niu, X.; Qian, L.; Fan, L.Y.; Cao, C.X.; Xiao, H. Facile preparation of salivary extracellular vesicles for cancer proteomics. Sci. Rep. 2016, 6, 24669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.X.; Schwartz, P.E.; Li, F.Q. Saliva and serum CA 125 assays for detecting malignant ovarian tumors. Obstet. Gynecol. 1990, 75, 701–704. [Google Scholar] [PubMed]
- Taylor, D.D.; Lyons, K.S.; Gercel-Taylor, C. Shed membrane fragment-associated markers for endometrial and ovarian cancers. Gynecol. Oncol. 2002, 84, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, V.; Sharma, S.; Deshpande, A.; Zhou, H.; Gimzewski, J.; Wong, D.T. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS ONE 2010, 5, e8577. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Rasool, H.I.; Palanisamy, V.; Mathisen, C.; Schmidt, M.; Wong, D.T.; Gimzewski, J.K. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano 2010, 4, 1921–1926. [Google Scholar] [CrossRef]
- Sharma, S.; Gillespie, B.M.; Palanisamy, V.; Gimzewski, J.K. Quantitative nanostructural and single-molecule force spectroscopy biomolecular analysis of human-saliva-derived exosomes. Langmuir 2011, 27, 14394–14400. [Google Scholar] [CrossRef]
- Han, X.; Wang, J.; Sun, Y. Circulating tumor dna as biomarkers for cancer detection. Genomics Proteomics Bioinformatics 2017, 15, 59–72. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Mulvey, C.L.; Silliman, N.; Schaefer, J.; Sausen, M.; James, N.; Rettig, E.M.; Guo, T.; Pickering, C.R.; et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med. 2015, 7, 293ra104. [Google Scholar] [CrossRef]
- Basik, M.; Aguilar-Mahecha, A.; Rousseau, C.; Diaz, Z.; Tejpar, S.; Spatz, A.; Greenwood, C.M.; Batist, G. Biopsies: next-generation biospecimens for tailoring therapy. Nat. Rev. Clin. Oncol. 2013, 10, 437–450. [Google Scholar] [CrossRef]
- Bidard, F.C.; Weigelt, B.; Reis-Filho, J.S. Going with the flow: from circulating tumor cells to DNA. Sci. Transl. Med. 2013, 5, 207ps14. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabieres, C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res. 2013, 73, 6384–6388. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Song, P.; Zhou, G.; Zuo, X.; Aldalbahi, A.; Lou, X.; Shi, J.; Fan, C. Electrochemical detection of nucleic acids, proteins, small molecules and cells using a DNA-nanostructure-based universal biosensing platform. Nat. Protoc. 2016, 11, 1244–1263. [Google Scholar] [CrossRef] [PubMed]
- Odenthal, K.J.; Gooding, J.J. An introduction to electrochemical DNA biosensors. Analyst 2007, 132, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Soleymani, L.; Fang, Z.; Sargent, E.H.; Kelley, S.O. Programming the detection limits of biosensors through controlled nanostructuring. Nat. Nanotechnol. 2009, 4, 844–848. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Lin, C.C.; Joon, A.; Feng, Z.; Troche, G.; Lira, M.E.; Chia, D.; Mao, M.; Ho, C.L.; Su, W.C.; et al. Noninvasive saliva-based EGFR gene mutation detection in patients with lung cancer. Am. J. Respir. Crit. Care Med. 2014, 190, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Pu, D.; Liang, H.; Wei, F.; Akin, D.; Feng, Z.; Yan, Q.; Li, Y.; Zhen, Y.; Xu, L.; Dong, G.; et al. Evaluation of a novel saliva-based epidermal growth factor receptor mutation detection for lung cancer: A pilot study. Thorac Cancer 2016, 7, 428–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, F.; Yang, J.; Wong, D.T. Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM). Biosens. Bioelectron. 2013, 44, 115–121. [Google Scholar] [CrossRef] [Green Version]
- Inal, C.; Yilmaz, E.; Piperdi, B.; Perez-Soler, R.; Cheng, H. Emerging treatment for advanced lung cancer with EGFR mutation. Expert Opin. Emerg. Drugs 2015, 20, 597–612. [Google Scholar] [CrossRef]
- Liga, A.; Vliegenthart, A.D.; Oosthuyzen, W.; Dear, J.W.; Kersaudy-Kerhoas, M. Exosome isolation: A microfluidic road-map. Lab. Chip. 2015, 15, 2388–2394. [Google Scholar] [CrossRef]
| Disease/Condition | Platform | Salivary Biomarker | Reference |
|---|---|---|---|
| Diabetes | Screen-printed electro chemical sensor | Glucose | [30] |
| HIV | OraQuick | HIV-1/2 antibody | [31] |
| Hepatitis C | Mono-Lisa anti-HCV Plus | HCV antibody | [35] |
| Acute myocardial infarction | Luminex lab-on-a-chip | C-reactive protein, myoglobin, MMP-9, IL-1B, slCAM-1, myeloperoxidase | [32] |
| Asthma and chronic obstructive pulmonary disease (COPD) | Multiplexed fiber optic microsphere-based cytokine array | IFNg, IP-10, RANTES, eotaxin-3, VEGF | [36] |
| Periodontitis | Lab-on-a-chip | C-reactive protein, MMP-8, IL-1B | [37] |
| Tobacco use | NicAlert test strips | Cotinine | [34] |
| Cancer | RNA Type | Salivary RNA Biomarker | Reference |
|---|---|---|---|
| Oral cancer | messenger RNA | DUSP1, H3F3A, IL1B, IL8, OAZ1, S100P, SAT | [65] |
| microRNA | miR-125a, miR-200a | [66] | |
| Esophageal cancer | microRNA | miR-144, miR-451, miR-98, miR-10b, miR-363 | [67] |
| Lung cancer | messenger RNA | CCNI, FGF19, GREB1, FRS2, EGFR | [68] |
| Pancreatic cancer | messenger RNA | KRAS, MBD3L2, ACRV1, DPM1 | [69] |
| Breast cancer | messenger RNA | CSTA, TPT1, IGF2BP1, GRM1, GRIK1, H6PD, MDM4, S100A8 | [70] |
| Ovarian cancer | messenger RNA | AGPAT1, B2M, IER3, IL1B, BASP1 | [71] |
| Cancer | Sample | Salivary Protein Biomarker | Reference |
|---|---|---|---|
| Breast cancer | Whole saliva | EGF | [88] |
| c-erbB-2 | [89] | ||
| CA15-3, c-erbB-2 | [90] | ||
| VEGF, EGF, CEA | [91] | ||
| CA6 | [70] | ||
| LRP | [92] | ||
| Oral cancer | Whole saliva | A1BG, CFB | [93] |
| M2BP, MRP14, CD59, CAT, PFN | [94] | ||
| FGB, S100, TF, IGHG, CFL1 | [95] | ||
| ADA | [96] | ||
| IL-8, M2BP, IL-1B | [97] | ||
| Salivary EVs | A2M, HPa, MUC5B, LGALS3BP, IGHA1, PIP, PKM1/M2, GAPDH | [98] | |
| Gastric cancer | Whole saliva | 1472.78Da, 2936.49Da, 6556.81Da, 7081.17Da | [99] |
| CSTB, TPI1, DMBT1, CALML3, IGH, IL1RA | [100] | ||
| Lung cancer | Whole saliva | HP, AZGP1, CALPR | [101] |
| Salivary EVs | Annexin A1, A2, A3, A5, A6, A11, NPRL2, CEACAM1, HIST1H4A, MUC1, PROM1, TNFAIP3 | [102] | |
| Ovarian cancer | Whole saliva | CA125 | [103] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, J.; Nonaka, T.; Wong, D.T.W. Salivary Exosomes as Nanocarriers for Cancer Biomarker Delivery. Materials 2019, 12, 654. https://doi.org/10.3390/ma12040654
Cheng J, Nonaka T, Wong DTW. Salivary Exosomes as Nanocarriers for Cancer Biomarker Delivery. Materials. 2019; 12(4):654. https://doi.org/10.3390/ma12040654
Chicago/Turabian StyleCheng, Jordan, Taichiro Nonaka, and David T.W. Wong. 2019. "Salivary Exosomes as Nanocarriers for Cancer Biomarker Delivery" Materials 12, no. 4: 654. https://doi.org/10.3390/ma12040654
APA StyleCheng, J., Nonaka, T., & Wong, D. T. W. (2019). Salivary Exosomes as Nanocarriers for Cancer Biomarker Delivery. Materials, 12(4), 654. https://doi.org/10.3390/ma12040654






