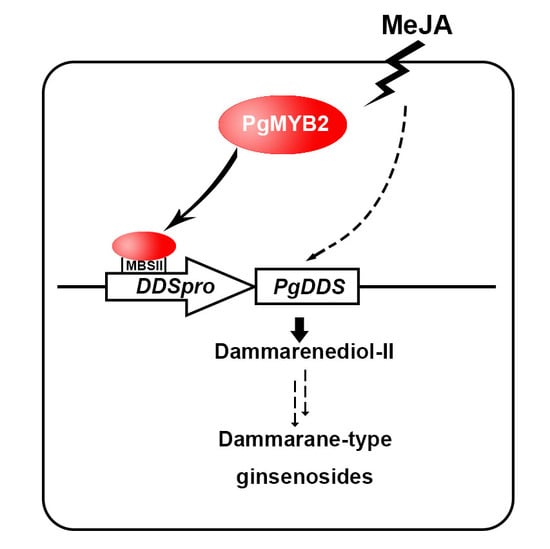
PgMYB2, a MeJA-Responsive Transcription Factor, Positively Regulates the Dammarenediol Synthase Gene Expression in Panax Ginseng
Abstract
:1. Introduction
2. Results
2.1. Characterization of PgMYB2 and Bioinformatics Analysis
2.2. Homology Analysis of PgMYB2 Protein
2.3. Subcellular Localization of PgMYB2
2.4. Expression Analysis of PgMYB2 in Different Tissue of P. ginseng
2.5. Expression Analysis of PgMYB2 and PgDDS under MeJA Treatments
2.6. DNA Binding Activity of PgMYB2
2.7. PgMYB2 Activates the Expression of PgDDS in A. thaliana Protoplasts
2.8. Transient Expression of PgMYB2 in Ginseng Leaves Promote the Expression of PgDDS
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Culture Environment
4.2. Total RNA Extraction and First Strand cDNA Synthesis
4.3. Bioinformatics Analysis and Prediction of PgMYB2
4.4. Subcellular Localization of PgMYB2
4.5. Hormone Treatments
4.6. The Expression Analysis of Related Genes by RT-PCR and qRT-PCR
4.7. Analysis of Transcriptional Activity in Yeast of PgMYB2
4.8. Expression of Fusion PgMYB2 Protein and Purification
4.9. Electrophoretic Mobility Shift Assay
4.10. Transient Expression Analysis of PgMYB2
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yun, T.K. Brief introduction of Panax ginseng C.A. Meyer. J. Korean Med. Sci. 2001, 16, S3–S5. [Google Scholar] [CrossRef]
- Chu, S.F.; Zhang, J.T. New achievements in ginseng research and its future prospects. Chin. J. Integr. Med. 2009, 15, 403–408. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Han, J.-Y.; Lim, S.; Choi, Y.-E. Ginseng metabolic engineering: Regulation of genes related to ginsenoside biosynthesis. J. Med. Plants Res. 2010, 3, 1270–1276. [Google Scholar]
- Augustin, J.M.; Kuzina, V.; Andersen, S.B.; Bak, S. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 2011, 72, 435–457. [Google Scholar] [CrossRef]
- Cao, H.; Nuruzzaman, M.; Xiu, H.; Huang, J.; Wu, K.; Chen, X.; Li, J.; Wang, L.; Jeong, J.H.; Park, S.J. Transcriptome Analysis of Methyl Jasmonate-Elicited Panax ginseng Adventitious Roots to Discover Putative Ginsenoside Biosynthesis and Transport Genes. Int. J. Mol. Sci. 2015, 16, 3035–3057. [Google Scholar] [CrossRef]
- Tansakul, P.; Shibuya, M.; Kushiro, T.; Ebizuka, Y. Dammarenediol-II synthase, the first dedicated enzyme for ginsenoside biosynthesis, in Panax ginseng. Febs Lett. 2006, 580, 5143–5149. [Google Scholar] [CrossRef]
- Kushiro, T.; Ohno, Y.; Shibuya, M.; Ebizuka, Y. In Vitro Conversion of 2,3-Oxidosqualene into Dammarenediol by Panax ginseng Microsomes. Biol. Pharm. Bull. 1997, 20, 292. [Google Scholar] [CrossRef]
- Kim, Y.J.; Zhang, D.; Yang, D.C. Biosynthesis and biotechnological production of ginsenosides. Biotechnol. Adv. 2015, 33, 717–735. [Google Scholar] [CrossRef]
- Han, J.Y.; Yong, S.K.; Yang, D.C.; Jung, Y.R.; Yong, E.C. Expression and RNA interference-induced silencing of the dammarenediol synthase gene in Panax ginseng. Plant Cell Physiol. 2006, 47, 1653. [Google Scholar] [CrossRef]
- Liang, Y.; Zhao, S.; Zhang, X. Antisense Suppression of Cycloartenol Synthase Results in Elevated Ginsenoside Levels in Panax ginseng Hairy Roots. Plant Mol. Biol. Report. 2009, 27, 298–304. [Google Scholar] [CrossRef]
- Zhou, M.; Memelink, J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016, 34, S0734975016300118. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Christian, D.; Ralf, S.; Erich, G.; Bernd, W.; Cathie, M.; Loïc, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Klempnauer, K.H.; Gonda, T.J.; Bishop, J.M. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: The architecture of a transduced oncogene. Cell 1982, 31, 453–463. [Google Scholar] [CrossRef]
- Pazares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. Embo J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef]
- Du, H.; Wang, Y.B.; Xie, Y.; Liang, Z.; Jiang, S.J.; Zhang, S.S.; Huang, Y.B.; Tang, Y.X. Genome-Wide Identification and Evolutionary and Expression Analyses of MYB-Related Genes in Land Plants. DNA Res. 2013, 20, 437–448. [Google Scholar] [CrossRef] [Green Version]
- Reyes, J.L.; Chua, N.H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2010, 49, 592–606. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, F.; Cheng, S.; Liao, Y. Characterization and functional analysis of a MYB gene (GbMYBFL) related to flavonoid accumulation in Ginkgo biloba. Genes Genom. 2018, 40, 49. [Google Scholar] [CrossRef]
- Gajjeraman, P.; Doddananjappa Theertha, P. Functional characterization of sugarcane MYB transcription factor gene promoter (PScMYBAS1) in response to abiotic stresses and hormones. Plant Cell Rep. 2012, 31, 661–669. [Google Scholar]
- Gális, I.; Simek, P.; Narisawa, T.; Sasaki, M.; Horiguchi, T.; Fukuda, H.; Matsuoka, K. A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonate-dependent regulator of phenylpropanoid-conjugate biosynthesis in tobacco. Plant J. 2010, 46, 573–592. [Google Scholar] [CrossRef]
- Sugimoto, K.; Takeda, S.; Hirochika, H. MYB-Related Transcription Factor NtMYB2 Induced by Wounding and Elicitors is a Regulator of the Tobacco Retrotransposon Tto1 and Defense-Related Genes. Plant Cell 2000, 12, 2511–2527. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, X.; He, K.; Liu, M.; Li, J.; Gao, Z.; Lin, Z.; Zhang, Y.; Wang, X.; Qiu, X. The MYB Transcription Factor Superfamily of Arabidopsis: Expression Analysis and Phylogenetic Comparison with the Rice MYB Family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar]
- Wei, S.Y.; Lou, Y.C.; Jia-Yin, T.; Meng-Ru, H.; Chun-Chi, C.; Rajasekaran, M.; Hong-Ming, H.; Tai, J.H.; Chwan-Deng, H.; Chen, C. Structure of theTrichomonas vaginalisMyb3 DNA-binding domain bound to a promoter sequence reveals a unique C-terminal β-hairpin conformation. Nucleic Acids Res. 2012, 40, 449–460. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef]
- Cheong, J.J.; Yang, D.C. Methyl jasmonate as a vital substance in plants. Trends Genet. 2003, 19, 409–413. [Google Scholar] [CrossRef]
- Yousaf, N.; Gould, D. Demonstrating Interactions of Transcription Factors with DNA by Electrophoretic Mobility Shift Assay. Methods Mol. Biol. 2017, 11–21. [Google Scholar]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.V.D.; Rouz, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [Green Version]
- Solano, R.; Nieto, C.; Avila, J.; Canas, L.; Diaz, I.; Paz-Ares, J. Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 1995, 14, 1773–1784. [Google Scholar] [CrossRef]
- Farmer, E.E. Plant biology: Jasmonate perception machines. Nature 2007, 448, 659–660. [Google Scholar] [CrossRef]
- Choi, D.W.; Jung, J.; Ha, Y.I.; Park, H.W.; In, D.S.; Chung, H.J.; Liu, J.R. Analysis of transcripts in methyl jasmonate-treated ginseng hairy roots to identify genes involved in the biosynthesis of ginsenosides and other secondary metabolites. Plant Cell Rep. 2005, 23, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Bedon, F.; Bomal, C.; Caron, S.; Levasseur, C.; Boyle, B.; Mansfield, S.D.; Schmidt, A.; Gershenzon, J.; Grimapettenati, J.; Séguin, A. Subgroup 4 R2R3-MYBs in conifer trees: Gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses. J. Exp. Bot. 2010, 61, 3847–3864. [Google Scholar] [CrossRef]
- Lee, M.H.; Jeong, J.H.; Seo, J.W.; Shin, C.G.; Kim, Y.S.; In, J.G.; Yang, D.C.; Yi, J.S.; Choi, Y.E. Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing squalene synthase gene. Plant Cell Physiol. 2004, 45, 976–984. [Google Scholar] [CrossRef]
- Oktae, K.; Bang, K.H.; Youngchang, K.; Dongyun, H.; Minyoung, K.; Seonwoo, C. Upregulation of ginsenoside and gene expression related to triterpene biosynthesis in ginseng hairy root cultures elicited by methyl jasmonate. Plant Cell Tissue Organ Cult. 2009, 98, 25–33. [Google Scholar]
- Kim, Y.S.; Hahn, E.J.; Murthy, H.N.; Paek, K.Y. Adventitious root growth and ginsenoside accumulation in Panax ginseng cultures as affected by methyl jasmonate. Biotechnol. Lett. 2004, 26, 1619–1622. [Google Scholar] [CrossRef]
- Zhao, J.; Davis, L.C.; Verpoorte, R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005, 23, 283–333. [Google Scholar] [CrossRef]
- Martin, C.; Paz-Ares, J. MYB transcription factors in plants. Trends Genet. 1997, 13, 67–73. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Li, C.; Lu, S. Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC Genom. 2014, 15, 277. [Google Scholar] [CrossRef]
- Gou, M.; Hou, G.; Yang, H.; Zhang, X.; Cai, Y.; Kai, G.; Liu, C.J. The MYB107 Transcription Factor Positively Regulates Suberin Biosynthesis. Plant Physiol. 2017, 173, 1045. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, Y.; Guo, M. Triterpenoid Saponins from the Seeds of Celosia argentea and Their Anti-inflammatory and Antitumor Activities. Chem. Pharm. Bull. 2011, 59, 666–671. [Google Scholar] [CrossRef] [Green Version]
- Osbourn, A.; Goss, R.J.M.; Field, R.A. The saponins—polar isoprenoids with important and diverse biological activities. Nat. Prod. Rep. 2011, 28, 1261–1268. [Google Scholar] [PubMed]
- Thang, N.V.; Thu, V.K.; Nhiem, N.X.; Dung, D.T.; Quang, T.H.; Tai, B.H.; Anh, H.L.T.; Yen, P.H.; Ngan, N.T.T.; Hoang, N.H. Oleanane-type Saponins from Glochidion hirsutum and Their Cytotoxic Activities. Chem. Biodivers. 2017, 14, e1600445. [Google Scholar] [CrossRef] [PubMed]
- Mahjoub, A.; Hernould, M.; Joubès, J.; Decendit, A.; Mars, M.; Barrieu, F.; Hamdi, S.; Delrot, S. Overexpression of a grapevine R2R3-MYB factor in tomato affects vegetative development, flower morphology and flavonoid and terpenoid metabolism. Plant Physiol. Biochem. 2009, 47, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.L.; Hou, H.L.; Zhu, X.M.; Shao, J.R.; Wu, Y.M.; Tang, Y.X. Molecular regulation of terpenoid indole alkaloids pathway in the medicinal plant, Catharanthus roseus. J. Med. Plant Res. 2011, 425, 2760–2772. [Google Scholar]
- Hu, W.; Liu, N.; Tian, Y.; Zhang, L. Molecular cloning, expression, purification, and functional characterization of dammarenediol synthase from Panax ginseng. Biomed Res. Int. 2012, 2013, 285740. [Google Scholar] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. Methods Mol. Biol. 1999, 112, 531. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von, H.G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Combet, C.; Blanchet, C.; Geourjon, C.; Deléage, G. NPS@: Network Protein Sequence Analysis. Trends Biochem. Sci. 2000, 25, 147–150. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Luo, T.; Guo, X.; Zou, X.; Zhou, D.; Afrin, S.; Li, G.; Zhang, Y.; Zhang, R.; Luo, Z. PgMYB2, a MeJA-Responsive Transcription Factor, Positively Regulates the Dammarenediol Synthase Gene Expression in Panax Ginseng. Int. J. Mol. Sci. 2019, 20, 2219. https://doi.org/10.3390/ijms20092219
Liu T, Luo T, Guo X, Zou X, Zhou D, Afrin S, Li G, Zhang Y, Zhang R, Luo Z. PgMYB2, a MeJA-Responsive Transcription Factor, Positively Regulates the Dammarenediol Synthase Gene Expression in Panax Ginseng. International Journal of Molecular Sciences. 2019; 20(9):2219. https://doi.org/10.3390/ijms20092219
Chicago/Turabian StyleLiu, Tuo, Tiao Luo, Xiangqian Guo, Xian Zou, Donghua Zhou, Sadia Afrin, Gui Li, Yue Zhang, Ru Zhang, and Zhiyong Luo. 2019. "PgMYB2, a MeJA-Responsive Transcription Factor, Positively Regulates the Dammarenediol Synthase Gene Expression in Panax Ginseng" International Journal of Molecular Sciences 20, no. 9: 2219. https://doi.org/10.3390/ijms20092219