An Improved Method for Cell Type-Selective Glycomic Analysis of Tissue Sections Assisted by Fluorescence Laser Microdissection
Abstract
:1. Introduction
2. Results
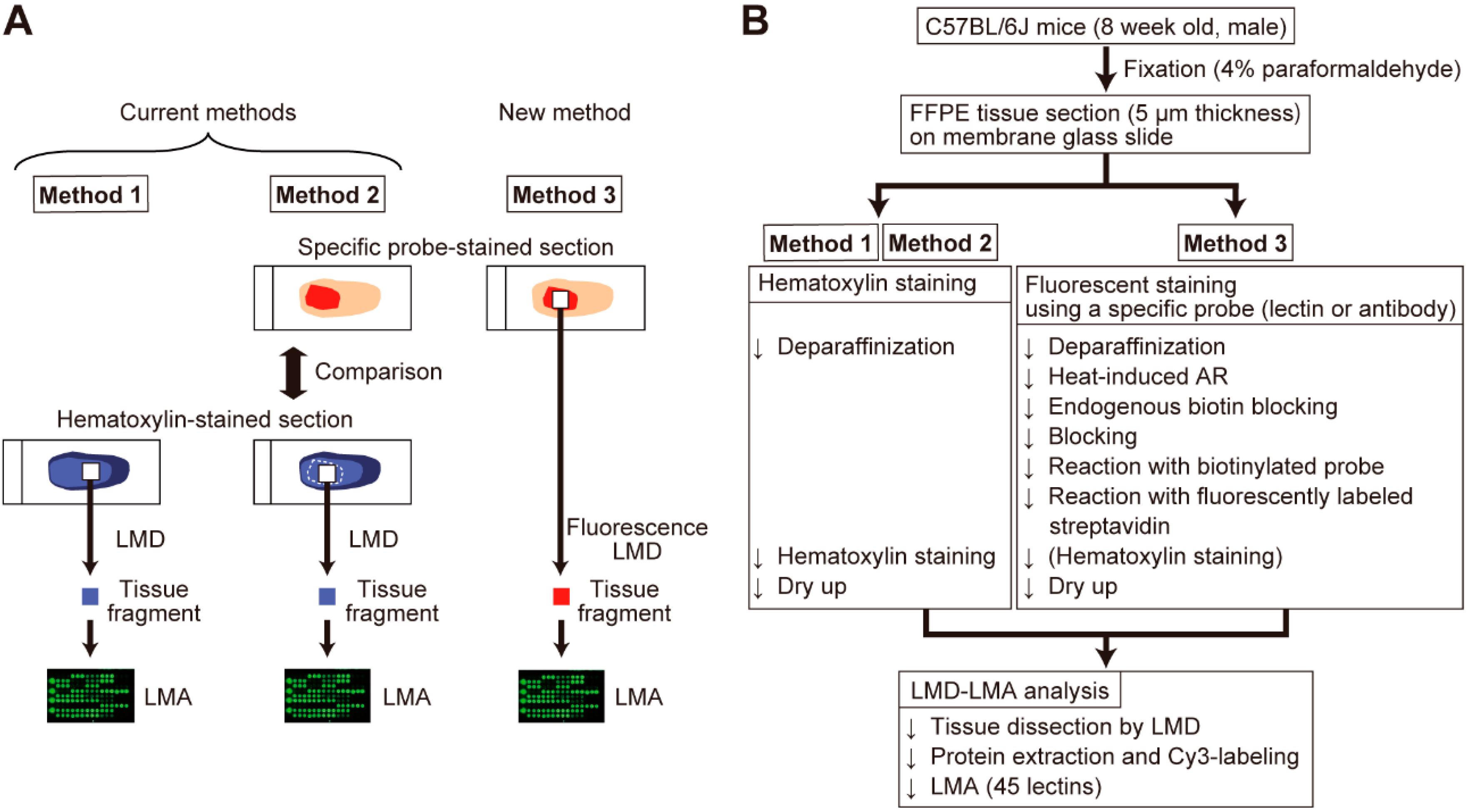
2.1. Optimization of Sample Preparation for Specific Probe-Stained Sections
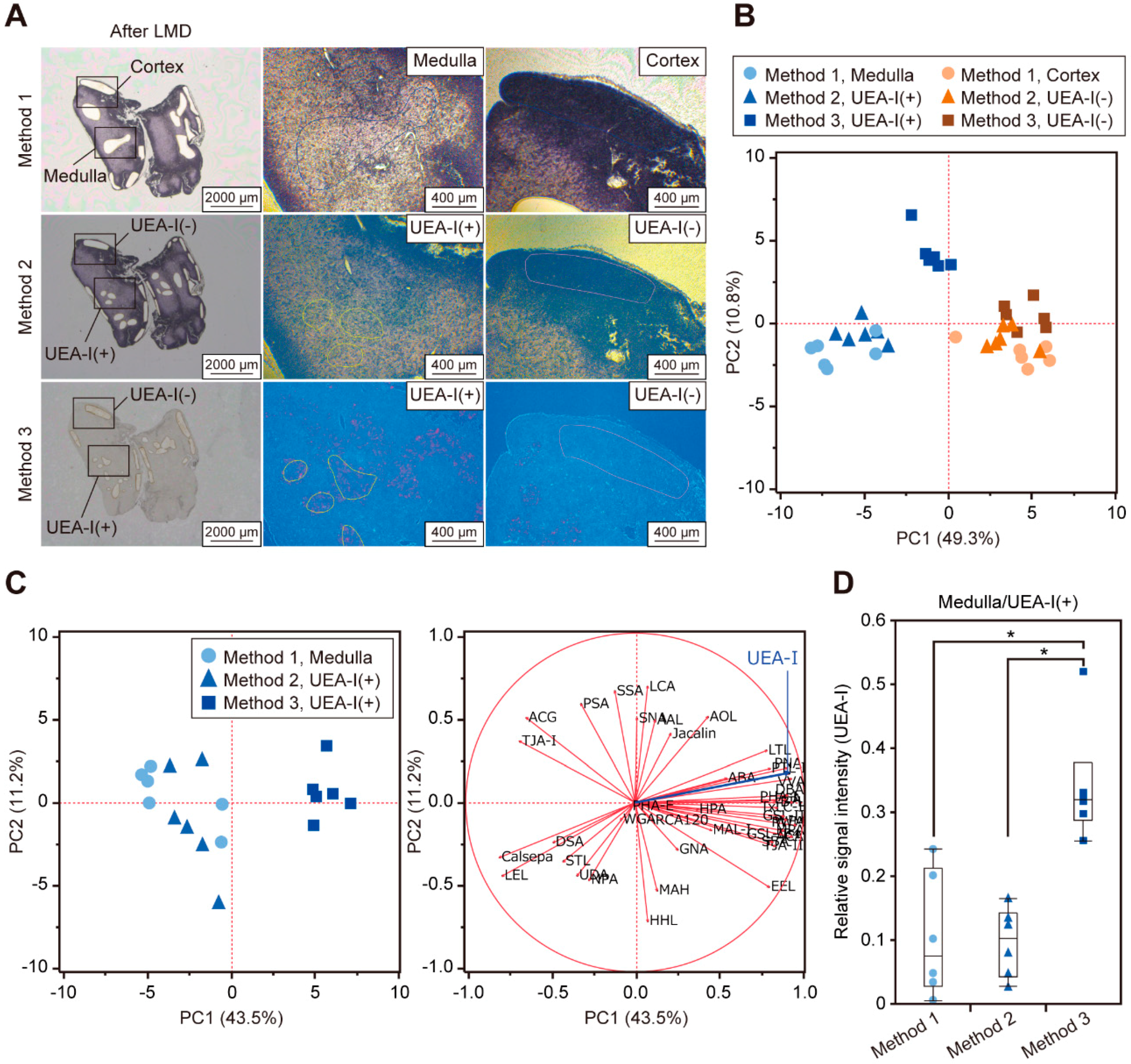
2.2. Effects of Fluorescent Staining Procedures on Glycomic Profiling
2.3. Comparison of Differential Glycomic Profiles Obtained by the Current and New Methods
2.4. Differential Glycomic Profiling Using Histochemical Sections Stained with Different Probes
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Tissue Section Preparation
4.3. Tissue Section Staining
4.4. Tissue Dissection and Protein Extraction
4.5. LMA Analysis
4.6. Western Blot Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AAL | Aleuria aurantia lectin |
| AOL | Aspergillus oryzae lectin |
| AR | antigen retrieval |
| CK5 | cytokeratin 5 |
| ConA | Concanavalin A |
| CSC | cancer stem cell |
| DBA | Dolichos biflorus agglutinin |
| DSA | Datura stramonium agglutinin |
| FFPE | formalin-fixed paraffin-embedded |
| GNA | Galanthus nivalis agglutinin |
| HPA | Helix pomatia agglutinin |
| LEL | Lycopersicon esculentum lectin |
| LMA | lectin microarray |
| LMD | laser microdissection |
| MAL-I | Maackia amurensis lectin-I |
| mTEC | medullary thymic epithelial cell |
| NPA | Narcissus pseudonarcissus agglutinin |
| PBS | phosphate-buffered saline |
| PCA | principle component analysis |
| PEN | polyethylene naphthalate |
| PNA | peanut agglutinin |
| PPS | polyphenylene sulfide |
| PSA | Pisum sativum agglutinin |
| RT | room temperature |
| SBA | soybean agglutinin |
| SNA | Sambucus nigra agglutinin |
| SSA | Sambucus sieboldiana agglutinin |
| TJA-I | Trichosanthes japonica agglutinin-I |
| UEA-I | Ulex europaeus agglutinin-I |
| VVA | Vicia villosa agglutinin |
| WFA | Wisteria floribunda agglutinin |
References
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462. [Google Scholar] [CrossRef] [PubMed]
- Bousseau, S.; Vergori, L.; Soleti, R.; Lenaers, G.; Martinez, M.C.; Andriantsitohaina, R. Glycosylation as new pharmacological strategies for diseases associated with excessive angiogenesis. Pharmacol. Ther. 2018, 191, 92–122. [Google Scholar] [CrossRef] [PubMed]
- Narimatsu, H.; Sawaki, H.; Kuno, A.; Kaji, H.; Ito, H.; Ikehara, Y. A strategy for discovery of cancer glyco-biomarkers in serum using newly developed technologies for glycoproteomics. FEBS J. 2010, 277, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, J.; Yamada, M.; Kuno, A.; Tateno, H. Lectin microarrays: Concept, principle and applications. Chem. Soc. Rev. 2013, 42, 4443–4458. [Google Scholar] [CrossRef] [PubMed]
- Kuno, A.; Uchiyama, N.; Koseki-Kuno, S.; Ebe, Y.; Takashima, S.; Yamada, M.; Hirabayashi, J. Evanescent-field fluorescence-assisted lectin microarray: A new strategy for glycan profiling. Nat. Methods 2005, 2, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Kuno, A.; Matsuda, A.; Ikehara, Y.; Narimatsu, H.; Hirabayashi, J. Chapter 7—Differential Glycan Profiling by Lectin Microarray Targeting Tissue Specimens. Methods Enzymol. 2010, 478, 165–179. [Google Scholar] [PubMed]
- Narimatsu, H.; Kaji, H.; Vakhrushev, S.Y.; Clausen, H.; Zhang, H.; Noro, E.; Togayachi, A.; Nagai-Okatani, C.; Kuno, A.; Zou, X.; et al. Current technologies for complex glycoproteomics and their applications to biology/disease-driven glycoproteomics. J. Proteome Res. 2018, 17, 4097–4112. [Google Scholar] [CrossRef]
- Kaji, H.; Ocho, M.; Togayachi, A.; Kuno, A.; Sogabe, M.; Ohkura, T.; Nozaki, H.; Angata, T.; Chiba, Y.; Ozaki, H.; et al. Glycoproteomic discovery of serological biomarker candidates for HCV/HBV infection-associated liver fibrosis and hepatocellular carcinoma. J. Proteome Res. 2013, 12, 2630–2640. [Google Scholar] [CrossRef]
- Matsuda, A.; Kuno, A.; Ishida, H.; Kawamoto, T.; Shoda, J.-I.; Hirabayashi, J. Development of an all-in-one technology for glycan profiling targeting formalin-embedded tissue sections. Biochem. Biophys. Res. Commun. 2008, 370, 259–263. [Google Scholar] [CrossRef]
- Matsuda, A.; Kuno, A.; Kawamoto, T.; Matsuzaki, H.; Irimura, T.; Ikehara, Y.; Zen, Y.; Nakanuma, Y.; Yamamoto, M.; Ohkohchi, N.; et al. Wisteria floribunda agglutinin-positive mucin 1 is a sensitive biliary marker for human cholangiocarcinoma. Hepatology 2010, 52, 174–182. [Google Scholar] [CrossRef] [Green Version]
- Tan, B.; Matsuda, A.; Zhang, Y.; Kuno, A.; Narimatsu, H. Multilectin-assisted fractionation for improved single-dot tissue glycome profiling in clinical glycoproteomics. Mol. Biosyst. 2014, 10, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Hirao, Y.; Matsuzaki, H.; Iwaki, J.; Kuno, A.; Kaji, H.; Ohkura, T.; Togayachi, A.; Abe, M.; Nomura, M.; Noguchi, M.; et al. Glycoproteomics approach for identifying Glycobiomarker candidate molecules for tissue type classification of non-small cell lung carcinoma. J. Proteome Res. 2014, 13, 4705–4716. [Google Scholar] [CrossRef]
- Espina, V.; Wulfkuhle, J.D.; Calvert, V.S.; VanMeter, A.; Zhou, W.; Coukos, G.; Geho, D.H.; Petricoin, E.F.; Liotta, L.A. Laser-capture microdissection. Nat. Protoc. 2006, 1, 586–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, X.; Yoshida, M.; Nagai-Okatani, C.; Iwaki, J.; Matsuda, A.; Tan, B.; Hagiwara, K.; Sato, T.; Itakura, Y.; Noro, E.; et al. A standardized method for lectin microarray-based tissue glycome mapping. Sci. Rep. 2017, 7, 43560. [Google Scholar] [CrossRef] [Green Version]
- Pearse, G. Normal Structure, Function and Histology of the Thymus. Toxicol. Pathol. 2006, 34, 504–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, L.; Kyewski, B.; Allen, P.M.; Hogquist, K.A. Positive and negative selection of the T cell repertoire: What thymocytes see (and don’t see). Nat. Rev. Immunol. 2014, 14, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Farr, A.G.; Anderson, S.K. Epithelial heterogeneity in the murine thymus: Fucose-specific lectins bind medullary epithelial cells. J. Immunol. 1985, 134, 2971–2977. [Google Scholar]
- Reisner, Y.; Linker-Israeli, M.; Sharon, N. Separation of mouse thymocytes into two subpopulations by the use of peanut agglutinin. Cell. Immunol. 1976, 25, 129–134. [Google Scholar] [CrossRef]
- Klug, D.B.; Carter, C.; Crouch, E.; Roop, D.; Conti, C.J.; Richie, E.R. Interdependence of cortical thymic epithelial cell differentiation and T-lineage commitment. Proc. Natl. Acad. Sci. USA 1998, 95, 11822–11827. [Google Scholar] [CrossRef] [Green Version]
- Eberle, F.C.; Hanson, J.C.; Killian, J.K.; Wei, L.; Ylaya, K.; Hewitt, S.M.; Jaffe, E.S.; Emmert-Buck, M.R.; Rodriguez-Canales, J. Immunoguided Laser Assisted Microdissection Techniques for DNA Methylation Analysis of Archival Tissue Specimens. J. Mol. Diagn. 2010, 12, 394–401. [Google Scholar] [CrossRef] [Green Version]
- Yoshioka, W.; Endo, N.; Kurashige, A.; Haijima, A.; Endo, T.; Shibata, T.; Nishiyama, R.; Kakeyama, M.; Tohyama, C. Fluorescence laser microdissection reveals a distinct pattern of gene activation in the mouse hippocampal region. Sci. Rep. 2012, 2, 783. [Google Scholar] [CrossRef]
- Baum, L.G.; Derbin, K.; Perillo, N.L.; Wu, T.; Pang, M.; Uittenbogaart, C. Characterization of terminal sialic acid linkages on human thymocytes. Correlation between lectin-binding phenotype and sialyltransferase expression. J. Biol. Chem. 1996, 271, 10793–10799. [Google Scholar] [CrossRef]
- Paessens, L.C.; García-Vallejo, J.J.; Fernandes, R.J.; van Kooyk, Y. The glycosylation of thymic microenvironments. A microscopic study using plant lectins. Immunol. Lett. 2007, 110, 65–73. [Google Scholar] [CrossRef]
- Sawanobori, Y.; Ueta, H.; Dijkstra, C.D.; Park, C.G.; Satou, M.; Kitazawa, Y.; Matsuno, K. Three distinct subsets of thymic epithelial cells in rats and mice defined by novel antibodies. PLoS ONE 2014, 9, e109995. [Google Scholar] [CrossRef] [PubMed]
- Farr, A.G.; Anderson, S.K.; Braddy, S.C.; Mejino, J.L. Selective binding of Dolichos biflorus agglutinin to L3T4-, Lyt-2-thymocytes. Expression of terminal alpha-linked N-acetyl-D-galactosamine residues defines a subpopulation of fetal and adult murine thymocytes. J. Immunol. 1988, 140, 1014–1021. [Google Scholar] [PubMed]
- Mallard, B.W.; Tiralongo, J. Cancer stem cell marker glycosylation: Nature, function and significance. Glycoconj. J. 2017, 34, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Barkeer, S.; Chugh, S.; Batra, S.K.; Ponnusamy, M.P. Glycosylation of Cancer Stem Cells: Function in Stemness, Tumorigenesis, and Metastasis. Neoplasia 2018, 20, 813–825. [Google Scholar] [CrossRef]
- Yamashita, T.; Honda, M.; Nakamoto, Y.; Baba, M.; Nio, K.; Hara, Y.; Zeng, S.S.; Hayashi, T.; Kondo, M.; Takatori, H.; et al. Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology 2013, 57, 1484–1497. [Google Scholar] [CrossRef]
- Higashi, M.; Yonezawa, S.; Ho, J.J.; Tanaka, S.; Irimura, T.; Kim, Y.S.; Sato, E. Expression of MUC1 and MUC2 mucin antigens in intrahepatic bile duct tumors: Its relationship with a new morphological classification of cholangiocarcinoma. Hepatology 1999, 30, 1347–1355. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, A.; Higashi, M.; Nakagawa, T.; Yokoyama, S.; Kuno, A.; Yonezawa, S.; Narimatsu, H. Assessment of tumor characteristics based on glycoform analysis of membrane-tethered MUC1. Lab. Investig. 2017, 97, 1103–1113. [Google Scholar] [CrossRef]
- Kuno, A.; Kato, Y.; Matsuda, A.; Kaneko, M.K.M.; Ito, H.; Amano, K.; Chiba, Y.; Narimatsu, H.; Hirabayashi, J. Focused differential glycan analysis with the platform antibody-assisted lectin profiling for glycan-related biomarker verification. Mol. Cell. Proteomics 2009, 8, 99–108. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagai-Okatani, C.; Nagai, M.; Sato, T.; Kuno, A. An Improved Method for Cell Type-Selective Glycomic Analysis of Tissue Sections Assisted by Fluorescence Laser Microdissection. Int. J. Mol. Sci. 2019, 20, 700. https://doi.org/10.3390/ijms20030700
Nagai-Okatani C, Nagai M, Sato T, Kuno A. An Improved Method for Cell Type-Selective Glycomic Analysis of Tissue Sections Assisted by Fluorescence Laser Microdissection. International Journal of Molecular Sciences. 2019; 20(3):700. https://doi.org/10.3390/ijms20030700
Chicago/Turabian StyleNagai-Okatani, Chiaki, Misugi Nagai, Takashi Sato, and Atsushi Kuno. 2019. "An Improved Method for Cell Type-Selective Glycomic Analysis of Tissue Sections Assisted by Fluorescence Laser Microdissection" International Journal of Molecular Sciences 20, no. 3: 700. https://doi.org/10.3390/ijms20030700





