Phytochemical Combination PB125 Activates the Nrf2 Pathway and Induces Cellular Protection against Oxidative Injury
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Cell Culture
2.3. Nrf2 Reporter Gene Assays
2.4. Gene Expression Assays
2.4.1. Cell Culture and RNA Isolation
2.4.2. Microarray Assays
2.4.3. Quantitative Reverse Transcription-PCR Assays
2.4.4. RNA-seq Assays
RNA-seq Library Preparation
Sequencing
mRNA-seq Profiling
2.5. Protein Assays
2.6. Assays to Measure Cytoprotective Effects
2.6.1. Cell Viability
2.6.2. LDH Release
2.7. Statistical Analysis
3. Results
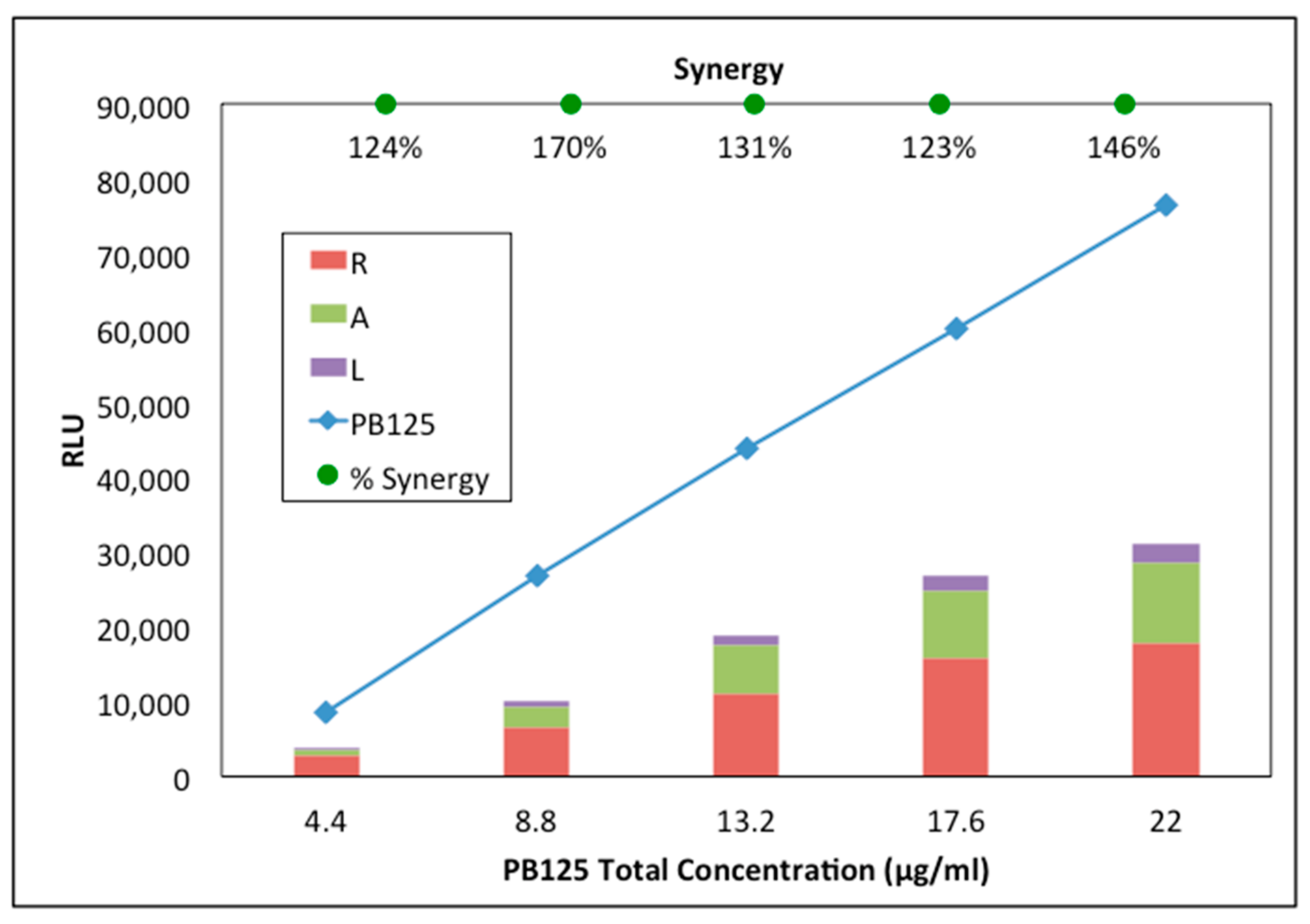
3.1. Synergy
3.2. Gene Expression
3.2.1. HepG2 Gene Expression by Microarray
3.2.2. HepG2 Gene Expression by qPCR
3.2.3. HepG2 RNA-seq Gene Expression Quantitation
3.2.4. Non-Canonical Nrf2 genes and Non-Nrf2 Genes
3.3. HMOX1 Protein ELISA
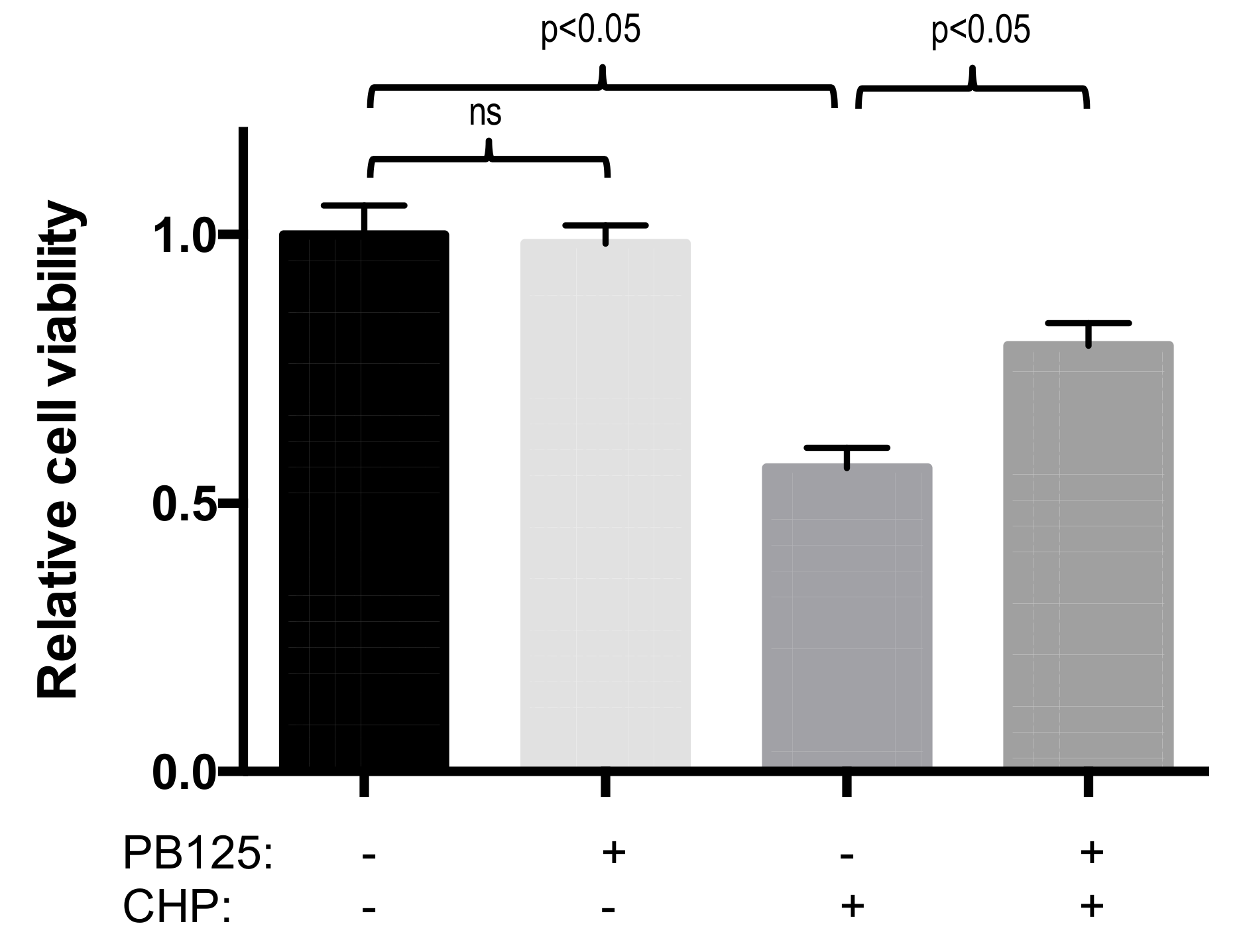
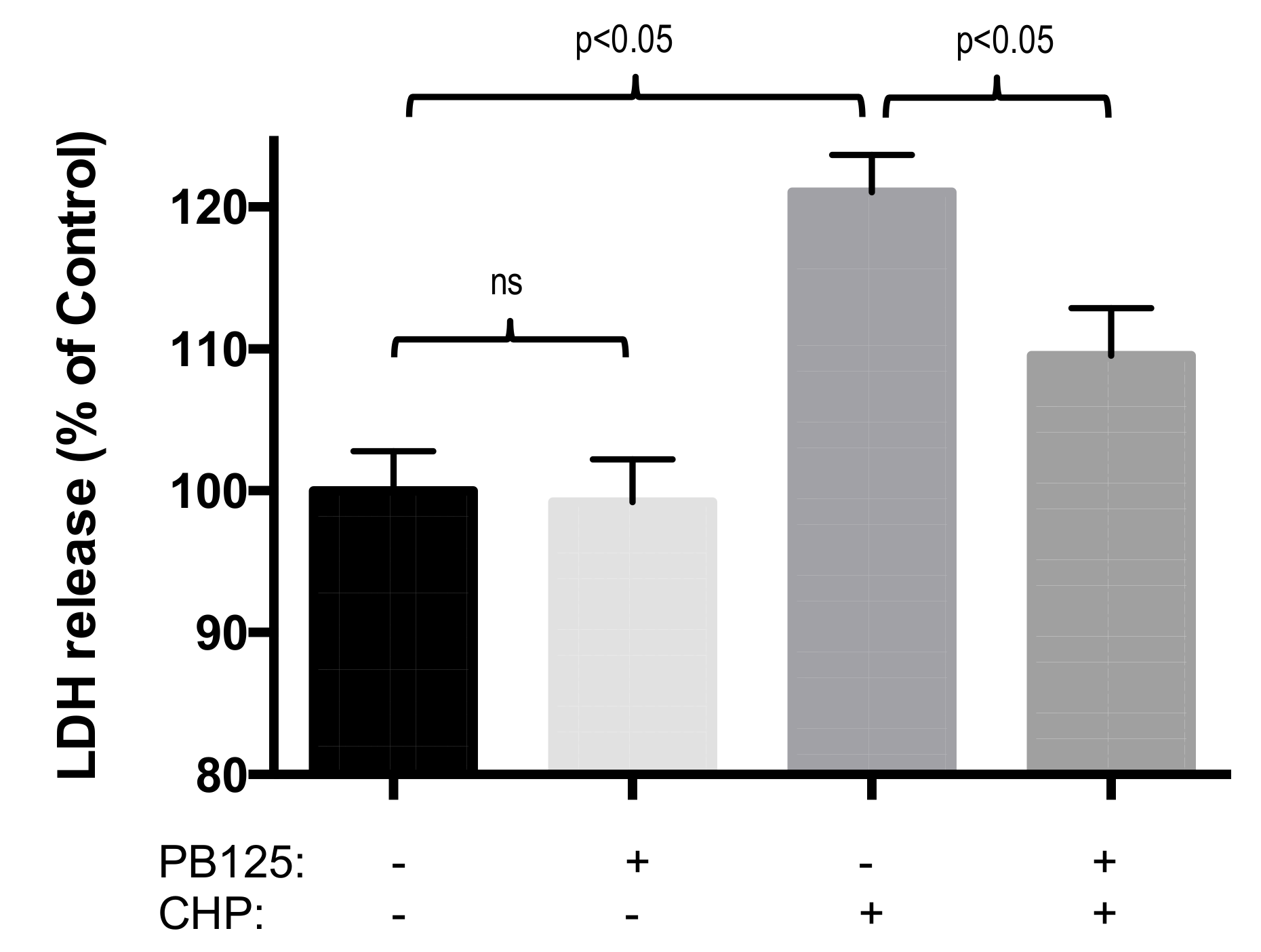
3.4. Functional Benefit
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harman, D. Aging: A theory based on free radical and radiation chemistry. J. Gerontol 1956, 11, 298–300. [Google Scholar] [CrossRef] [PubMed]
- Harman, D. The aging process. Proc. Natl. Acad. Sci. USA 1981, 78, 7124–7128. [Google Scholar] [CrossRef] [PubMed]
- Harman, D. Nutritional implications of the free-radical theory of aging. J. Am. Coll. Nutr. 1982, 1, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Beckman, K.B.; Ames, B.N. The Free Radical Theory of Aging Matures. Physiol. Rev. 1998, 78, 547–581. [Google Scholar] [CrossRef]
- Pomatto, L.C.D.; Davies, K.J.A. Adaptive homeostasis and the free radical theory of ageing. Free Radic. Biol. Med. 2018, 124, 420–430. [Google Scholar] [CrossRef]
- Kiefte-de Jong, J.C.; Mathers, J.C.; Franco, O.H. Nutrition and healthy ageing: The key ingredients. Proc. Nutr. Soc. 2014, 73, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Cannella, C.; Savina, C.; Donini, L.M. Nutrition, longevity and behavior. Arch. Gerontol. Geriatr. 2009, 49 Suppl 1, 19–27. [Google Scholar] [CrossRef]
- Harman, D. Free radical theory of aging: Dietary implications. Am. J. Clin. Nutr. 1972, 25, 839–843. [Google Scholar] [CrossRef]
- Prior, R.L.; Cao, G.; Prior, R.L.; Cao, G. Analysis of botanicals and dietary supplements for antioxidant capacity: A review. J. AOAC Int. 2000, 83, 950–956. [Google Scholar] [PubMed]
- Ninfali, P.; Mea, G.; Giorgini, S.; Rocchi, M.; Bacchiocca, M. Antioxidant capacity of vegetables, spices and dressings relevant to nutrition. Br. J. Nutr. 2005, 93, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.K.; Bose, S.K.; Grunwald, G.K.; Myhill, P.; McCord, J.M. The induction of human superoxide dismutase and catalase in vivo: A fundamentally new approach to antioxidant therapy. Free Radic. Biol. Med. 2006, 40, 341–347. [Google Scholar] [CrossRef]
- Lewis, K.N.; Mele, J.; Hayes, J.D.; Buffenstein, R. Nrf2, a guardian of healthspan and gatekeeper of species longevity. Integr. Comp. Biol. 2010, 50, 829–843. [Google Scholar] [CrossRef]
- Cardozo, L.F.; Pedruzzi, L.M.; Stenvinkel, P.; Stockler-Pinto, M.B.; Daleprane, J.B.; Leite, M., Jr.; Mafra, D. Nutritional strategies to modulate inflammation and oxidative stress pathways via activation of the master antioxidant switch Nrf2. Biochimie 2013, 95, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Kelsey, N.A.; Wilkins, H.M.; Linseman, D.A. Nutraceutical Antioxidants as Novel Neuroprotective Agents. Molecules 2010, 15, 7792–7814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.M.; Li, J.; Johnson, D.A.; Stein, T.D.; Kraft, A.D.; Calkins, M.J.; Jakel, R.J.; Johnson, J.A. Nrf2, a multi-organ protector? FASEB J. 2005, 19, 1061–1066. [Google Scholar] [CrossRef]
- Eggler, A.L.; Gay, K.A.; Mesecar, A.D. Molecular mechanisms of natural products in chemoprevention: Induction of cytoprotective enzymes by Nrf2. Mol. Nutr Food Res. 2008, 52, S84–S94. [Google Scholar] [CrossRef]
- Na, H.K.; Surh, Y.J. Modulation of Nrf2-mediated antioxidant and detoxifying enzyme induction by the green tea polyphenol EGCG. Food Chem. Toxicol. 2008, 46, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ichikawa, T.; Janicki, J.S.; Cui, T. Targeting the Nrf2 pathway against cardiovascular disease. Expert Opin. Ther. Targets 2009, 13, 785–794. [Google Scholar] [CrossRef]
- Cho, H.Y.; Kleeberger, S.R. Nrf2 protects against airway disorders. Toxicol. Appl. Pharmacol. 2010, 244, 43–56. [Google Scholar] [CrossRef]
- Wakabayashi, N.; Slocum, S.L.; Skoko, J.J.; Shin, S.; Kensler, T.W. When NRF2 talks, who’s listening? Antioxid. Redox Signal. 2010, 13, 1649–1663. [Google Scholar] [CrossRef] [PubMed]
- Klaassen, C.D.; Reisman, S.A. Nrf2 the rescue: Effects of the antioxidative/electrophilic response on the liver. Toxicol. Appl. Pharmacol. 2010, 244, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.K.; Kensler, T.W. Targeting NRF2 signaling for cancer chemoprevention. Toxicol. Appl. Pharmacol. 2010, 244, 66–76. [Google Scholar] [CrossRef]
- Sykiotis, G.P.; Bohmann, D. Stress-activated cap‘n’collar transcription factors in aging and human disease. Sci. Signal. 2010, 3, re3. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.H.; Shenvi, S.V.; Dixon, B.M.; Liu, H.; Jaiswal, A.K.; Liu, R.M.; Hagen, T.M. Decline in transcriptional activity of Nrf2 causes age-related loss of glutathione synthesis, which is reversible with lipoic acid. Proc. Natl. Acad. Sci. USA 2004, 101, 3381–3386. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.J.; Shay, K.P.; Thomas, N.O.; Butler, J.A.; Finlay, L.F.; Hagen, T.M. Age-related loss of hepatic Nrf2 protein homeostasis: Potential role for heightened expression of miR-146a. Free Radic. Biol. Med. 2015, 89, 1184–1191. [Google Scholar] [CrossRef] [Green Version]
- Shay, K.P.; Michels, A.J.; Li, W.; Kong, A.-N.T.; Hagen, T.M. Cap-independent Nrf2 translation is part of a lipoic acid-stimulated detoxification stress response. Biochim. Biophys. Acta 2012, 1823, 1102–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Zhang, H.; Davies, K.J.A.; Forman, H.J. Aging-related decline in the induction of Nrf2-regulated antioxidant genes in human bronchial epithelial cells. Redox Biol. 2018, 14, 35–40. [Google Scholar] [CrossRef]
- Kubo, E.; Chhunchha, B.; Singh, P.; Sasaki, H.; Singh, D.P. Sulforaphane reactivates cellular antioxidant defense by inducing Nrf2/ARE/Prdx6 activity during aging and oxidative stress. Sci. Rep. 2017, 7, 14130. [Google Scholar] [CrossRef] [Green Version]
- Petiwala, S.M.; Johnson, J.J. Diterpenes from rosemary (Rosmarinus officinalis): Defining their potential for anti-cancer activity. Cancer Lett. 2015, 367, 93–102. [Google Scholar] [CrossRef]
- Xiang, Q.; Liu, Z.; Wang, Y.; Xiao, H.; Wu, W.; Xiao, C.; Liu, X. Carnosic acid attenuates lipopolysaccharide-induced liver injury in rats via fortifying cellular antioxidant defense system. Food Chem. Toxicol. 2013, 53, 1–9. [Google Scholar] [CrossRef]
- Satoh, T.; McKercher, S.R.; Lipton, S.A. Nrf2/ARE-mediated antioxidant actions of pro-electrophilic drugs. Free Radic. Biol. Med. 2013, 65, 645–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh, T.; Kosaka, K.; Itoh, K.; Kobayashi, A.; Yamamoto, M.; Shimojo, Y.; Kitajima, C.; Cui, J.; Kamins, J.; Okamoto, S.; et al. Carnosic acid, a catechol-type electrophilic compound, protects neurons both in vitro and in vivo through activation of the Keap1/Nrf2 pathway via S-alkylation of targeted cysteines on Keap1. J. Neurochem. 2008, 104, 1116–1131. [Google Scholar] [CrossRef] [PubMed]
- Foresti, R.; Bains, S.K.; Pitchumony, T.S.; de Castro Bras, L.E.; Drago, F.; Dubois-Rande, J.L.; Bucolo, C.; Motterlini, R. Small molecule activators of the Nrf2-HO-1 antioxidant axis modulate heme metabolism and inflammation in BV2 microglia cells. Pharmacol. Res. 2013, 76C, 132–148. [Google Scholar] [CrossRef]
- Johnson, J.J. Carnosol: A promising anti-cancer and anti-inflammatory agent. Cancer Lett. 2011, 305, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, D.; Rojo, A.I.; Salinas, M.; Diaz, R.; Gallardo, G.; Alam, J.; de Galarreta, C.M.R.; Cuadrado, A. Regulation of Heme Oxygenase-1 Expression through the Phosphatidylinositol 3-Kinase/Akt Pathway and the Nrf2 Transcription Factor in Response to the Antioxidant Phytochemical Carnosol. J. Biol. Chem. 2004, 279, 8919–8929. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Wang, Z.; Wang, S.; Ravula, R.; Yang, L.; Xu, J.; Wang, C.; Zuo, Z.; Chow, M.S.; Shi, L.; et al. Discovery of molecular mechanisms of traditional Chinese medicinal formula Si-Wu-Tang using gene expression microarray and connectivity map. PLoS ONE 2011, 6, e18278. [Google Scholar] [CrossRef]
- Priyandoko, D.; Ishii, T.; Kaul, S.C.; Wadhwa, R. Ashwagandha leaf derived withanone protects normal human cells against the toxicity of methoxyacetic acid, a major industrial metabolite. PLoS ONE 2011, 6, e19552. [Google Scholar] [CrossRef]
- Velmurugan, K.; Alam, J.; McCord, J.M.; Pugazhenthi, S. Synergistic induction of heme oxygenase-1 by the components of the antioxidant supplement Protandim. Free Radic. Biol. Med. 2009, 46, 430–440. [Google Scholar] [CrossRef]
- Mishra, L.C.; Singh, B.B.; Dagenais, S. Scientific basis for the therapeutic use of Withania somnifera (ashwagandha): A review. Altern. Med. Rev. 2000, 5, 334–346. [Google Scholar] [PubMed]
- Paredes-Gonzalez, X.; Fuentes, F.; Jeffery, S.; Saw, C.L.; Shu, L.; Su, Z.Y.; Kong, A.T. Induction of NRF2-mediated gene expression by dietary phytochemical flavones apigenin and luteolin. Biopharm. Drug Dispos. 2015. [Google Scholar] [CrossRef]
- Xu, J.; Wang, H.; Ding, K.; Zhang, L.; Wang, C.; Li, T.; Wei, W.; Lu, X. Luteolin provides neuroprotection in models of traumatic brain injury via the Nrf2-ARE pathway. Free Radic. Biol. Med. 2014, 71, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.C.; Gan, F.F.; Shelar, S.B.; Ng, K.Y.; Chew, E.H. Antioxidant and Nrf2 inducing activities of luteolin, a flavonoid constituent in Ixeris sonchifolia Hance, provide neuroprotective effects against ischemia-induced cellular injury. Food Chem. Toxicol. 2013, 59, 272–280. [Google Scholar] [CrossRef]
- Sun, G.B.; Sun, X.; Wang, M.; Ye, J.X.; Si, J.Y.; Xu, H.B.; Meng, X.B.; Qin, M.; Sun, J.; Wang, H.W.; et al. Oxidative stress suppression by luteolin-induced heme oxygenase-1 expression. Toxicol. Appl Pharmacol. 2012, 265, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Wu, M.J.; Liu, I.Y.; Su, J.D.; Yen, J.H. Neurotrophic and cytoprotective action of luteolin in PC12 cells through ERK-dependent induction of Nrf2-driven HO-1 expression. J. Agric. Food Chem. 2010, 58, 4477–4486. [Google Scholar] [CrossRef]
- Raskovic, A.; Milanovic, I.; Pavlovic, N.; Cebovic, T.; Vukmirovic, S.; Mikov, M. Antioxidant activity of rosemary (Rosmarinus officinalis L.) essential oil and its hepatoprotective potential. BMC Complement. Altern. Med. 2014, 14, 225. [Google Scholar] [CrossRef] [PubMed]
- Ortuno, J.; Serrano, R.; Banon, S. Antioxidant and antimicrobial effects of dietary supplementation with rosemary diterpenes (carnosic acid and carnosol) vs vitamin E on lamb meat packed under protective atmosphere. Meat Sci. 2015, 110, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Klancnik, A.; Guzej, B.; Kolar, M.H.; Abramovic, H.; Mozina, S.S. In vitro antimicrobial and antioxidant activity of commercial rosemary extract formulations. J Food Prot. 2009, 72, 1744–1752. [Google Scholar] [CrossRef] [PubMed]
- Theoharides, T.C.; Asadi, S.; Panagiotidou, S. A case series of a luteolin formulation (NeuroProtek(R)) in children with autism spectrum disorders. Int. J. Immunopathol. Pharmacol. 2012, 25, 317–323. [Google Scholar] [CrossRef]
- Taliou, A.; Zintzaras, E.; Lykouras, L.; Francis, K. An open-label pilot study of a formulation containing the anti-inflammatory flavonoid luteolin and its effects on behavior in children with autism spectrum disorders. Clin. Ther. 2013, 35, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.F.; Braidy, N.; Gortzi, O.; Sobarzo-Sanchez, E.; Daglia, M.; Skalicka-Woźniak, K.; Nabavi, S.M. Luteolin as an anti-inflammatory and neuroprotective agent: A brief review. Brain Res. Bull. 2015, 119 Pt A, 1–11. [Google Scholar] [CrossRef]
- Gonzalez-Vallinas, M.; Reglero, G.; Ramirez de Molina, A. Rosemary (Rosmarinus officinalis L.) Extract as a Potential Complementary Agent in Anticancer Therapy. Nutr. Cancer 2015, 67, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Anadon, A.; Martinez-Larranaga, M.R.; Martinez, M.A.; Ares, I.; Garcia-Risco, M.R.; Senorans, F.J.; Reglero, G. Acute oral safety study of rosemary extracts in rats. J. Food Prot. 2008, 71, 790–795. [Google Scholar] [CrossRef]
- Kumar, G.; Srivastava, A.; Sharma, S.K.; Rao, T.D.; Gupta, Y.K. Efficacy & safety evaluation of Ayurvedic treatment (Ashwagandha powder & Sidh Makardhwaj) in rheumatoid arthritis patients: A pilot prospective study. Indian J. Med. Res. 2015, 141, 100–106. [Google Scholar] [PubMed]
- Chandrasekhar, K.; Kapoor, J.; Anishetty, S. A prospective, randomized double-blind, placebo-controlled study of safety and efficacy of a high-concentration full-spectrum extract of ashwagandha root in reducing stress and anxiety in adults. Indian J. Psychol. Med. 2012, 34, 255–262. [Google Scholar] [CrossRef]
- Emami, F.; Ali-Beig, H.; Farahbakhsh, S.; Mojabi, N.; Rastegar-Moghadam, B.; Arbabian, S.; Kazemi, M.; Tekieh, E.; Golmanesh, L.; Ranjbaran, M.; et al. Hydroalcoholic extract of Rosemary (Rosmarinus officinalis L.) and its constituent carnosol inhibit formalin-induced pain and inflammation in mice. Pak. J. Biol. Sci. 2013, 16, 309–316. [Google Scholar] [PubMed]
- Del Campo, J.; Amiot, M.J.; Nguyen-The, C. Antimicrobial effect of rosemary extracts. J. Food Prot. 2000, 63, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Bozin, B.; Mimica-Dukic, N.; Samojlik, I.; Jovin, E. Antimicrobial and antioxidant properties of rosemary and sage (Rosmarinus officinalis L. and Salvia officinalis L., Lamiaceae) essential oils. J. Agric. Food Chem. 2007, 55, 7879–7885. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Subramaneyaan, M.; Arora, V.K.; Banerjee, B.D.; Ahmed, R.S. Effect of Withania somnifera (Ashwagandha) root extract on amelioration of oxidative stress and autoantibodies production in collagen-induced arthritic rats. J. Complement. Integr. Med. 2015, 12, 117–125. [Google Scholar] [CrossRef]
- Rai, M.; Jogee, P.S.; Agarkar, G.; Santos, C.A. Anticancer activities of Withania somnifera: Current research, formulations, and future perspectives. Pharm. Biol. 2016, 54, 189–197. [Google Scholar] [CrossRef]
- Raghavan, A.; Shah, Z.A. Withania somnifera: A pre-clinical study on neuroregenerative therapy for stroke. Neural Regen. Res. 2015, 10, 183–185. [Google Scholar]
- Wankhede, S.; Langade, D.; Joshi, K.; Sinha, S.R.; Bhattacharyya, S. Examining the effect of Withania somnifera supplementation on muscle strength and recovery: A randomized controlled trial. J. Int. Soc. Sports Nutr. 2015, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Seelinger, G.; Merfort, I.; Schempp, C.M. Anti-oxidant, anti-inflammatory and anti-allergic activities of luteolin. Planta Med. 2008, 74, 1667–1677. [Google Scholar] [CrossRef]
- Kim, Y.J.; Park, M.Y.; Chang, N.; Kwon, O. Intake and major sources of dietary flavonoid in Korean adults: Korean National Health and Nutrition Examination Survey 2010–2012. Asia Pac. J. Clin. Nutr. 2015, 24, 456–463. [Google Scholar]
- Jun, S.; Shin, S.; Joung, H. Estimation of dietary flavonoid intake and major food sources of Korean adults. Br. J. Nutr. 2016, 115, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Chun, O.K.; Chung, S.J.; Song, W.O. Estimated dietary flavonoid intake and major food sources of U.S. adults. J. Nutr. 2007, 137, 1244–1252. [Google Scholar] [CrossRef]
- Zhang, D.; Lee, B.; Nutter, A.; Song, P.; Dolatabadi, N.; Parker, J.; Sanz-Blasco, S.; Newmeyer, T.; Ambasudhan, R.; McKercher, S.R.; et al. Protection from cyanide-induced brain injury by the Nrf2 transcriptional activator carnosic acid. J. Neurochem. 2015, 133, 898–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh, T.; Lipton, S.A. Redox regulation of neuronal survival mediated by electrophilic compounds. Trends Neurosci. 2007, 30, 37–45. [Google Scholar] [CrossRef]
- Satoh, T.; Lipton, S. Recent advances in understanding NRF2 as a druggable target: Development of pro-electrophilic and non-covalent NRF2 activators to overcome systemic side effects of electrophilic drugs like dimethyl fumarate. F1000Res 2017, 6, 2138. [Google Scholar] [CrossRef]
- Satoh, T.; Harada, N.; Hosoya, T.; Tohyama, K.; Yamamoto, M.; Itoh, K. Keap1/Nrf2 system regulates neuronal survival as revealed through study of keap1 gene-knockout mice. Biochem. Biophys. Res. Commun. 2009, 380, 298–302. [Google Scholar] [CrossRef]
- Palliyaguru, D.L.; Singh, S.V.; Kensler, T.W. Withania somnifera: From prevention to treatment of cancer. Mol. Nutr. Food Res. 2016, 60, 1342–1353. [Google Scholar] [CrossRef]
- Palliyaguru, D.L.; Chartoumpekis, D.V.; Wakabayashi, N.; Skoko, J.J.; Yagishita, Y.; Singh, S.V.; Kensler, T.W. Withaferin A induces Nrf2-dependent protection against liver injury: Role of Keap1-independent mechanisms. Free Radic. Biol. Med. 2016, 101, 116–128. [Google Scholar] [CrossRef]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef] [Green Version]
- Aliper, A.; Jellen, L.; Cortese, F.; Artemov, A.; Karpinsky-Semper, D.; Moskalev, A.; Swick, A.G.; Zhavoronkov, A. Towards natural mimetics of metformin and rapamycin. Aging (Albany NY) 2017, 9, 2245–2268. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Q.; Wu, R.; Xiao, X.; Yang, C.; Yang, Y.; Wang, C.; Lin, L.; Kong, A.N. The dietary flavone luteolin epigenetically activates the Nrf2 pathway and blocks cell transformation in human colorectal cancer HCT116 cells. J. Cell. Biochem. 2018, 119, 9573–9582. [Google Scholar] [CrossRef]
- Bustanji, Y.; Taha, M.O.; Almasri, I.M.; Al-Ghussein, M.A.; Mohammad, M.K.; Alkhatib, H.S. Inhibition of glycogen synthase kinase by curcumin: Investigation by simulated molecular docking and subsequent in vitro/in vivo evaluation. J. Enzym. Inhib. Med. Chem. 2009, 24, 771–778. [Google Scholar] [CrossRef]
- Kaspar, J.W.; Jaiswal, A.K. Tyrosine phosphorylation controls nuclear export of Fyn, allowing Nrf2 activation of cytoprotective gene expression. FASEB J. 2011, 25, 1076–1087. [Google Scholar] [CrossRef] [Green Version]
- Pauff, J.M.; Hille, R. Inhibition studies of bovine xanthine oxidase by luteolin, silibinin, quercetin, and curcumin. J. Nat. Prod. 2009, 72, 725–731. [Google Scholar] [CrossRef]
- McCord, J.M. Oxygen-derived free radicals in postischemic tissue injury. N. Engl. J. Med. 1985, 312, 159–163. [Google Scholar]
- Son, Y.O.; Pratheeshkumar, P.; Wang, Y.; Kim, D.; Zhang, Z.; Shi, X. Protection from Cr(VI)-induced malignant cell transformation and tumorigenesis of Cr(VI)-transformed cells by luteolin through Nrf2 signaling. Toxicol. Appl. Pharmacol. 2017, 331, 24–32. [Google Scholar] [CrossRef]
- Chian, S.; Thapa, R.; Chi, Z.; Wang, X.J.; Tang, X. Luteolin inhibits the Nrf2 signaling pathway and tumor growth in vivo. Biochem. Biophys. Res. Commun. 2014, 447, 602–608. [Google Scholar] [CrossRef]
- Kukoyi, A.T.; Fan, X.; Staitieh, B.S.; Hybertson, B.M.; Gao, B.; McCord, J.M.; Guidot, D.M. MiR-144 mediates Nrf2 inhibition and alveolar epithelial dysfunction in HIV-1 transgenic rats. Am. J. Physiol. Cell 2019, in press. [Google Scholar]
- Hybertson, B.M.; Gao, B.; Bose, S.K.; McCord, J.M. Oxidative stress in health and disease: The therapeutic potential of Nrf2 activation. Mol. Asp. Med. 2011, 32, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Krajka-Kuzniak, V.; Paluszczak, J.; Szaefer, H.; Baer-Dubowska, W. The activation of the Nrf2/ARE pathway in HepG2 hepatoma cells by phytochemicals and subsequent modulation of phase II and antioxidant enzyme expression. J. Physiol. Biochem. 2015, 71, 227–238. [Google Scholar] [CrossRef]
- Simmons, S.O.; Fan, C.Y.; Yeoman, K.; Wakefield, J.; Ramabhadran, R. NRF2 oxidative stress induced by heavy metals is cell type dependent. Curr. Chem. Genomics 2011, 5, 1–12. [Google Scholar] [CrossRef]
- Wu, T.D.; Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010, 26, 873–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511. [Google Scholar] [CrossRef]
- Baird, N.L.; Bowlin, J.L.; Cohrs, R.J.; Gilden, D.; Jones, K.L. Comparison of Varicella-Zoster virus RNA sequences in human neurons and fibroblasts. J. Virol. 2014, 88, 5877–5880. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]
- Reddy, N.M.; Kleeberger, S.R.; Yamamoto, M.; Kensler, T.W.; Scollick, C.; Biswal, S.; Reddy, S.P. Genetic dissection of the Nrf2-dependent redox signaling-regulated transcriptional programs of cell proliferation and cytoprotection. Physiol. Genom. 2007, 32, 74–81. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Rojo de la Vega, M.; Sammani, S.; Mascarenhas, J.B.; Kerins, M.; Dodson, M.; Sun, X.; Wang, T.; Ooi, A.; Garcia, J.G.N.; et al. RPA1 binding to NRF2 switches ARE-dependent transcriptional activation to ARE-NRE–dependent repression. Proc. Natl. Acad. Sci. USA 2018, 115, E10352–E10361. [Google Scholar] [CrossRef] [PubMed]
- Yates, M.S.; Tran, Q.T.; Dolan, P.M.; Osburn, W.O.; Shin, S.; McCulloch, C.C.; Silkworth, J.B.; Taguchi, K.; Yamamoto, M.; Williams, C.R.; et al. Genetic versus chemoprotective activation of Nrf2 signaling: Overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis 2009, 30, 1024–1031. [Google Scholar] [CrossRef]
- Thimmulappa, R.K.; Rangasamy, T.; Alam, J.; Biswal, S. Dibenzoylmethane activates Nrf2-dependent detoxification pathway and inhibits benzo(a)pyrene induced DNA adducts in lungs. Med. Chem. 2008, 4, 473–481. [Google Scholar] [CrossRef]
- Zhang, H.; Davies, K.J.A.; Forman, H.J. Oxidative stress response and Nrf2 signaling in aging. Free Radic. Biol. Med. 2015, 88, 314–336. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Chu, A.; Feng, Y.; Chen, L.; Shao, Y.; Luo, Q.; Deng, X.; Wu, M.; Shi, X.; Chen, Y. MicroRNA-146a: A Comprehensive Indicator of Inflammation and Oxidative Stress Status Induced in the Brain of Chronic T2DM Rats. Front. Pharmacol. 2018, 9, 478. [Google Scholar] [CrossRef]
- Chen, X.Y.; Dou, Y.X.; Luo, D.D.; Zhang, Z.B.; Li, C.L.; Zeng, H.F.; Su, Z.R.; Xie, J.H.; Lai, X.P.; Li, Y.C. beta-Patchoulene from patchouli oil protects against LPS-induced acute lung injury via suppressing NF-kappaB and activating Nrf2 pathways. Int. Immunopharmacol. 2017, 50, 270–278. [Google Scholar] [CrossRef]
- Li, N.; Muthusamy, S.; Liang, R.; Sarojini, H.; Wang, E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech. Ageing Dev. 2011, 132, 75–85. [Google Scholar] [CrossRef]
- Xue, W.L.; Bai, X.; Zhang, L. rhTNFR:Fc increases Nrf2 expression via miR-27a mediation to protect myocardium against sepsis injury. Biochem. Biophys. Res. Commun. 2015, 464, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Shan, W.; Gao, L.; Zeng, W.; Hu, Y.; Wang, G.; Li, M.; Zhou, J.; Ma, X.; Tian, X.; Yao, J. Activation of the SIRT1/p66shc antiapoptosis pathway via carnosic acid-induced inhibition of miR-34a protects rats against nonalcoholic fatty liver disease. Cell Death Dis. 2015, 6, e1833. [Google Scholar] [CrossRef]
- Sherratt, P.J.; Huang, H.C.; Nguyen, T.; Pickett, C.B. Role of protein phosphorylation in the regulation of NF-E2-related factor 2 activity. Methods Enzymol. 2004, 378, 286–301. [Google Scholar]
- Silva-Islas, C.A.; Maldonado, P.D. Canonical and non-canonical mechanisms of Nrf2 activation. Pharmacol. Res. 2018, 134, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Lall, D.; Baloh, R.H. Microglia and C9orf72 in neuroinflammation and ALS and frontotemporal dementia. J. Clin. Investig. 2017, 127, 3250–3258. [Google Scholar] [CrossRef] [Green Version]
- Brodeur, J.; Theriault, C.; Lessard-Beaudoin, M.; Marcil, A.; Dahan, S.; Lavoie, C. LDLR-related protein 10 (LRP10) regulates amyloid precursor protein (APP) trafficking and processing: Evidence for a role in Alzheimer’s disease. Mol. Neurodegener. 2012, 7, 31. [Google Scholar] [CrossRef]
- Mayne, J.; Dewpura, T.; Raymond, A.; Cousins, M.; Chaplin, A.; Lahey, K.A.; Lahaye, S.A.; Mbikay, M.; Ooi, T.C.; Chretien, M. Plasma PCSK9 levels are significantly modified by statins and fibrates in humans. Lipids Health Dis. 2008, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Genovese, G.; Friedman, D.J.; Ross, M.D.; Lecordier, L.; Uzureau, P.; Freedman, B.I.; Bowden, D.W.; Langefeld, C.D.; Oleksyk, T.K.; Uscinski Knob, A.L.; et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 2010, 329, 841–845. [Google Scholar] [CrossRef]
- Olabisi, O.A.; Zhang, J.Y.; VerPlank, L.; Zahler, N.; DiBartolo, S., 3rd; Heneghan, J.F.; Schlondorff, J.S.; Suh, J.H.; Yan, P.; Alper, S.L.; et al. APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc. Natl. Acad. Sci. USA 2016, 113, 830–837. [Google Scholar] [CrossRef]
- Vanzin, C.S.; Mescka, C.P.; Donida, B.; Hammerschimidt, T.G.; Ribas, G.S.; Kolling, J.; Scherer, E.B.; Vilarinho, L.; Nogueira, C.; Coitinho, A.S.; et al. Lipid, Oxidative and Inflammatory Profile and Alterations in the Enzymes Paraoxonase and Butyrylcholinesterase in Plasma of Patients with Homocystinuria Due CBS Deficiency: The Vitamin B12 and Folic Acid Importance. Cell. Mol. Neurobiol. 2015, 35, 899–911. [Google Scholar] [CrossRef]
- Kumar, M.; Sandhir, R. Neuroprotective Effect of Hydrogen Sulfide in Hyperhomocysteinemia Is Mediated Through Antioxidant Action Involving Nrf2. Neuromol. Med. 2018, 20, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Mescher, M.; Haarmann-Stemmann, T. Modulation of CYP1A1 metabolism: From adverse health effects to chemoprevention and therapeutic options. Pharmacol. Ther. 2018, 187, 71–87. [Google Scholar] [CrossRef]
- Uno, S.; Dalton, T.P.; Derkenne, S.; Curran, C.P.; Miller, M.L.; Shertzer, H.G.; Nebert, D.W. Oral exposure to benzo[a]pyrene in the mouse: Detoxication by inducible cytochrome P450 is more important than metabolic activation. Mol. Pharmacol. 2004, 65, 1225–1237. [Google Scholar] [CrossRef]
- Nebert, D.W.; Shi, Z.; Galvez-Peralta, M.; Uno, S.; Dragin, N. Oral benzo[a]pyrene: Understanding pharmacokinetics, detoxication, and consequences--Cyp1 knockout mouse lines as a paradigm. Mol. Pharmacol. 2013, 84, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Sun, L.; Cao, J.; Yuen, T.; Lu, P.; Bab, I.; Leu, N.A.; Srinivasan, S.; Wagage, S.; Hunter, C.A.; et al. Smoke carcinogens cause bone loss through the aryl hydrocarbon receptor and induction of Cyp1 enzymes. Proc. Natl. Acad. Sci. USA 2013, 110, 11115–11120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furue, M.; Fuyuno, Y.; Mitoma, C.; Uchi, H.; Tsuji, G. Therapeutic Agents with AHR Inhibiting and NRF2 Activating Activity for Managing Chloracne. Antioxidants 2018, 7, 90. [Google Scholar] [CrossRef]
- Lewis, J.E.; Brameld, J.M.; Jethwa, P.H. Neuroendocrine Role for VGF. Front. Endocrinol. (Lausanne) 2015, 6, 3. [Google Scholar] [CrossRef]
- Zhao, Z.; Lange, D.J.; Ho, L.; Bonini, S.; Shao, B.; Salton, S.R.; Thomas, S.; Pasinetti, G.M. Vgf is a novel biomarker associated with muscle weakness in amyotrophic lateral sclerosis (ALS), with a potential role in disease pathogenesis. Int. J. Med. Sci. 2008, 5, 92–99. [Google Scholar] [CrossRef]
- Jiang, H.; Chen, S.; Lu, N.; Yue, Y.; Yin, Y.; Zhang, Y.; Jiang, W.; Liang, J.; Yuan, Y. Reduced serum VGF levels were reversed by antidepressant treatment in depressed patients. World J. Biol. Psychiatry 2017, 18, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Saavedra, M.; De Repentigny, Y.; Yang, D.; O’Meara, R.W.; Yan, K.; Hashem, L.E.; Racacho, L.; Ioshikhes, I.; Bulman, D.E.; Parks, R.J.; et al. Voluntary Running Triggers VGF-Mediated Oligodendrogenesis to Prolong the Lifespan of Snf2h-Null Ataxic Mice. Cell Rep. 2016, 17, 862–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephens, S.B.; Edwards, R.J.; Sadahiro, M.; Lin, W.J.; Jiang, C.; Salton, S.R.; Newgard, C.B. The Prohormone VGF Regulates beta Cell Function via Insulin Secretory Granule Biogenesis. Cell Rep. 2017, 20, 2480–2489. [Google Scholar] [CrossRef] [PubMed]


| Primer | Sequence |
|---|---|
| GAPDH, forward | 5′ GGACCTGACCTGCCGTCTAG 3′ |
| GAPDH, reverse | 5′ GAGGAGTGGGTGTCGCTGTT 3′ |
| HMOX1, forward | 5′ GACAGCATGCCCCAGGATT 3′ |
| HMOX1, reverse | 5′ GTGGTACAGGGAGGCCATCA 3′ |
| GCLM, forward | 5′ TTGCCTCCTGCTGTGTGATG 3′ |
| GCLM, reverse | 5′ GTGCGCTTGAATGTCAGGAA 3′ |
| SLC7A11, forward | 5′ TGGGCTGATTTATCTTCGATACAA 3′ |
| SLC7A11, reverse | 5′ ATGACGAAGCCAATCCCTGTAC 3′ |
| Gene | Gene Name | Fold Change by Microarray | Fold Change by qPCR | Fold Change by RNA-seq |
|---|---|---|---|---|
| HMOX1 | heme oxygenase (decycling) 1 | 2.6 | 2.6 | 10.6 ± 0.3 |
| GCLM | glutamate-cysteine ligase, catalytic subunit | 5.4 | 8.5 | 9.2 ± 0.1 |
| SLC7A11 | solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 | 4.4 | 8.6 | 9.5 ± 0.3 |
| Gene | Gene Description | Nrf2 Activators | |||
|---|---|---|---|---|---|
| PB125 | PB123 | Protandim | DBM | ||
| C9orf72 | C9orf72 | 3.1 | 3.9 | 1.8 | 1.2 |
| CCPG1 | Cell cycle progression 1 | 5.2 | 3.6 | 4.8 | 2.3 |
| CTH | Cystathionine gamma-lyase | 7.2 | 8.0 | 4.5 | 1.4 |
| GCKR | Glucokinase (hexokinase 4) regulator | 4.0 | 23.6 | 1.8 | 2.2 |
| LRP10 | Low density lipoprotein receptor-related protein 10 | 2.5 | 5.1 | 2.4 | 1.3 |
| NCF2 | Neutrophil cytosolic factor 2 | 2.4 | 1.5 | 1.5 | 1.3 |
| DKK1 | Dickkopf WNT signaling pathway inhibitor 1 | −9.5 | −21.4 | −2.1 | −1.4 |
| FABP1 | fatty acid binding protein 1, liver | −6.5 | −16.7 | −7.7 | −2.7 |
| FMO5 | Flavin containing monooxygenase 5 | −3.3 | −14.3 | −3.2 | −1.5 |
| HMGCR | 3-Hydroxy-3-methylglutaryl-CoA reductase | −2.8 | −6.3 | −2.0 | −1.5 |
| LEAP2 | Liver expressed antimicrobial peptide 2 | −5.8 | −1.75 | −4.8 | −2.3 |
| PCSK9 | Proprotein convertase subtilisin/kexin type 9 | −4.4 | −1.6 | −5.3 | −1.6 |
| Gene | Gene Description | Nrf2 Activators | |||
|---|---|---|---|---|---|
| PB125 | PB123 | Protandim | DBM | ||
| APOL1 | Apolipoprotein L1 | −1.4 | −1.3 | 1.5 | 1.3 |
| BHMT | Betaine-homocysteine S-methyltransferase | 2.5 | 6.9 | 1.0 | 1.4 |
| CBS | Cystathione beta-synthase | 1.4 | 2.4 | −2.1 | 1.0 |
| CYP1A1 | Cytochrome P450 family 1 subfamily A member 1 | −1.4 | −1.6 | 9.2 | 6.7 |
| IFIT1 | Interferon induced protein with tetratricopeptide repeats 1 | −2.1 | −1.4 | 3.1 | −1.3 |
| MAT1A | Methionine adenosyltransferase I, alpha | 1.3 | 2.3 | −1.8 | 1.7 |
| NOS3 | Nitric oxide synthase 3 | 1.7 | 1.2 | 1.0 | 1.6 |
| PLAU | Plasminogen activator, urokinase | 2.9 | 1.7 | −3.8 | −1.1 |
| TP53INP1 | Tumor protein p53 inducible nuclear protein 1 | −1.9 | −5.3 | 1.2 | −1.3 |
| VGF | VGF nerve growth factor inducible | 5.0 | 2.0 | 1.0 | 1.2 |
| YPEL3 | Yippee like 3 | −1.4 | −2.0 | 1.2 | −1.2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hybertson, B.M.; Gao, B.; Bose, S.; McCord, J.M. Phytochemical Combination PB125 Activates the Nrf2 Pathway and Induces Cellular Protection against Oxidative Injury. Antioxidants 2019, 8, 119. https://doi.org/10.3390/antiox8050119
Hybertson BM, Gao B, Bose S, McCord JM. Phytochemical Combination PB125 Activates the Nrf2 Pathway and Induces Cellular Protection against Oxidative Injury. Antioxidants. 2019; 8(5):119. https://doi.org/10.3390/antiox8050119
Chicago/Turabian StyleHybertson, Brooks M., Bifeng Gao, Swapan Bose, and Joe M. McCord. 2019. "Phytochemical Combination PB125 Activates the Nrf2 Pathway and Induces Cellular Protection against Oxidative Injury" Antioxidants 8, no. 5: 119. https://doi.org/10.3390/antiox8050119





