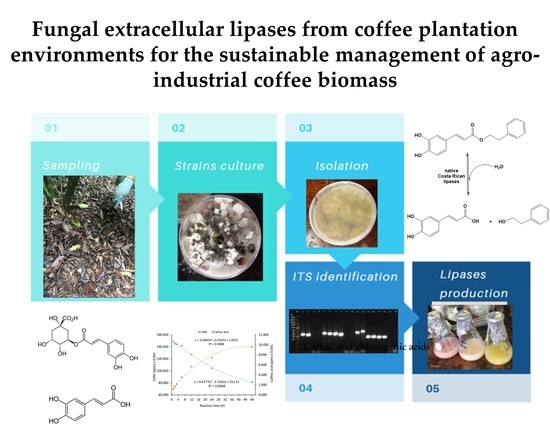
Fungal Extracellular Lipases from Coffee Plantation Environments for the Sustainable Management of Agro-Industrial Coffee Biomass
Abstract
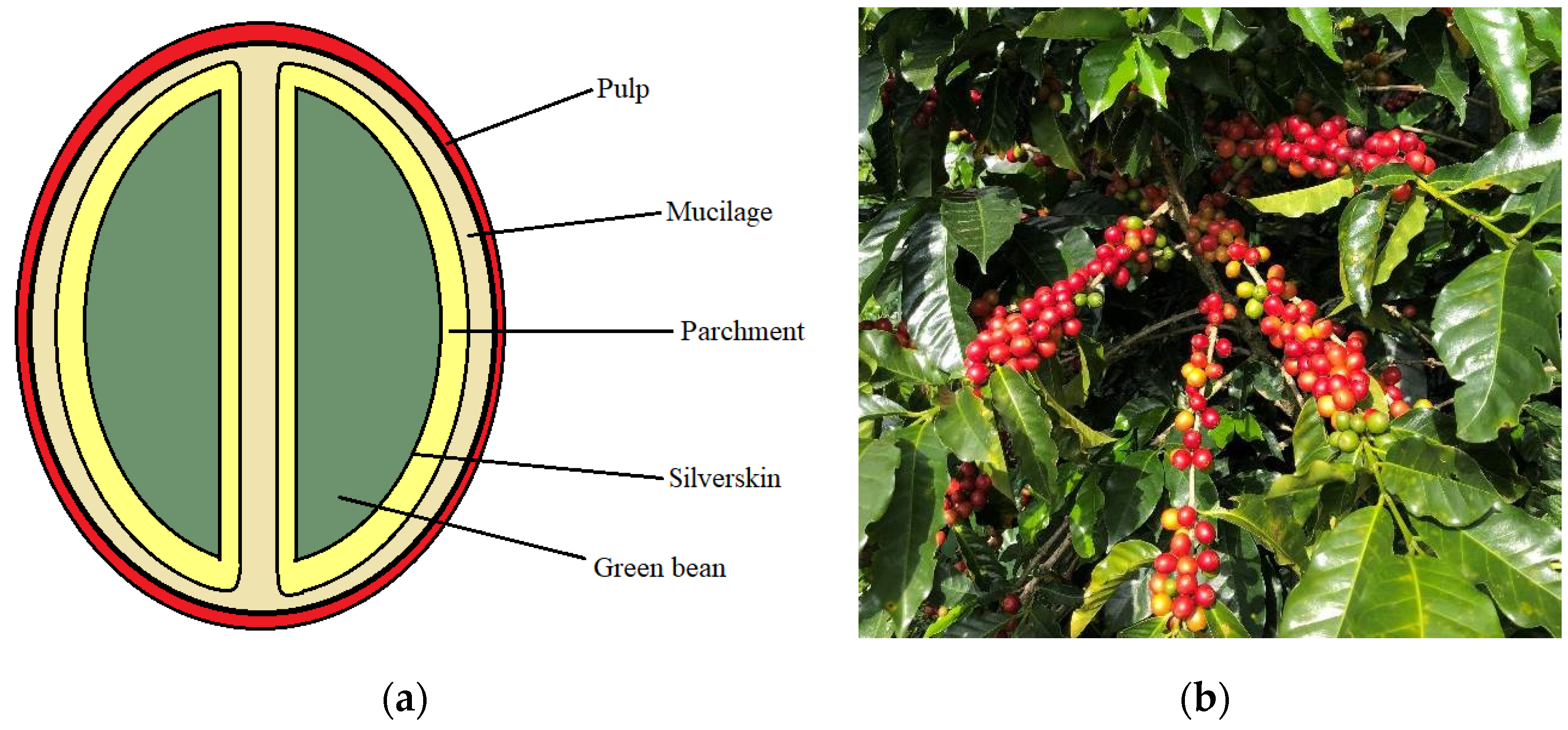
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bioprospecting Permits for Biodiversity Resources and Legal Frame
2.3. Culture Media and Growth Conditions
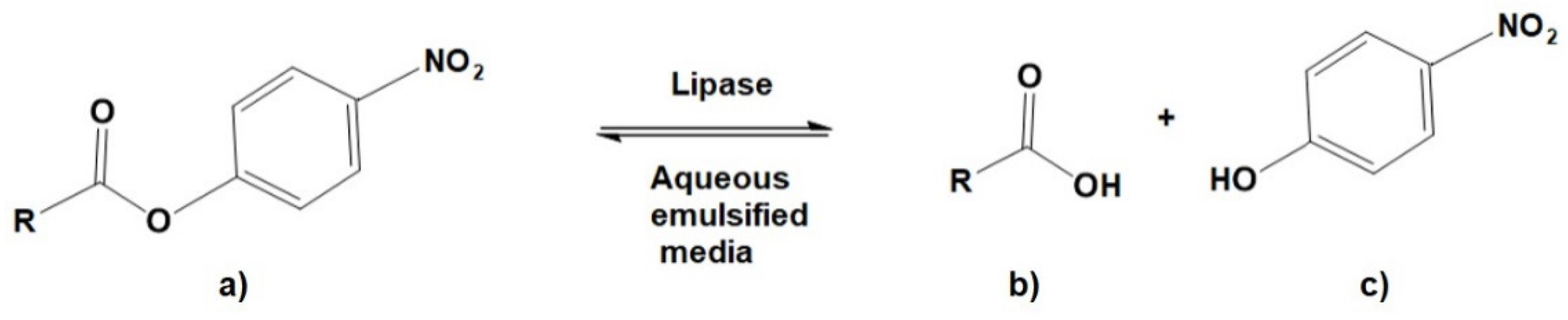
2.4. Protein Determinations and Lipase Activity Assays
2.5. Coffee Plantation Microorganism Sampling and Screening
2.6. Fungal Extracellular Lipases Production and Screening
2.7. Strain Characterization: DNA Extraction
2.8. Strain Characterization: DNA Barcoding and Lipase Sequencing
2.9. Substrate, pH, and Temperature Effect on Lipase Activity
2.10. Kinetic Parameters
2.11. Enzyme Stability for Freezing and Lyophilization
2.12. Determination of Ionic Strength Effect on Enzyme Activity
2.13. Lipase Application in Biomass Degradation
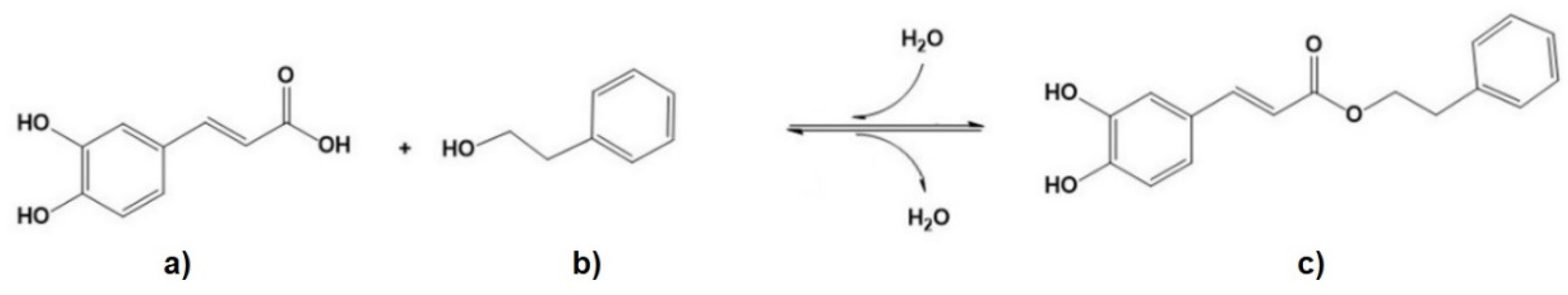
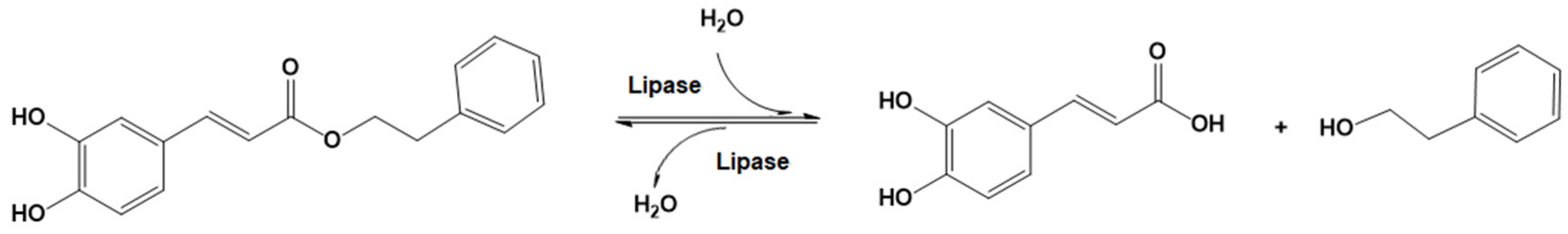
2.14. Lipase-Catalyzed CAPE Hydrolysis
2.15. Statistical Analysis
3. Results
3.1. Fungal DNA Barcoding and Lipase Sequencing
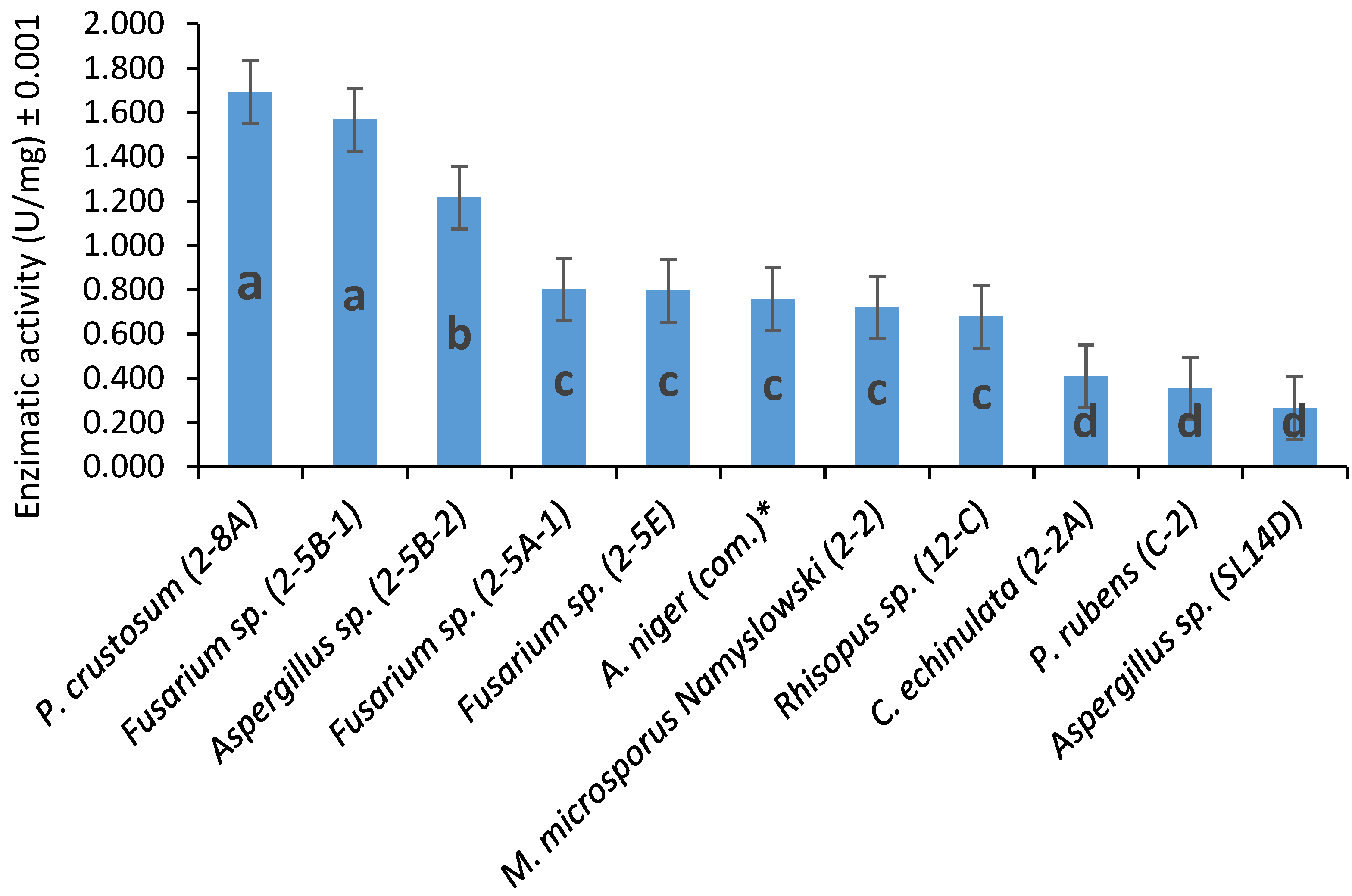
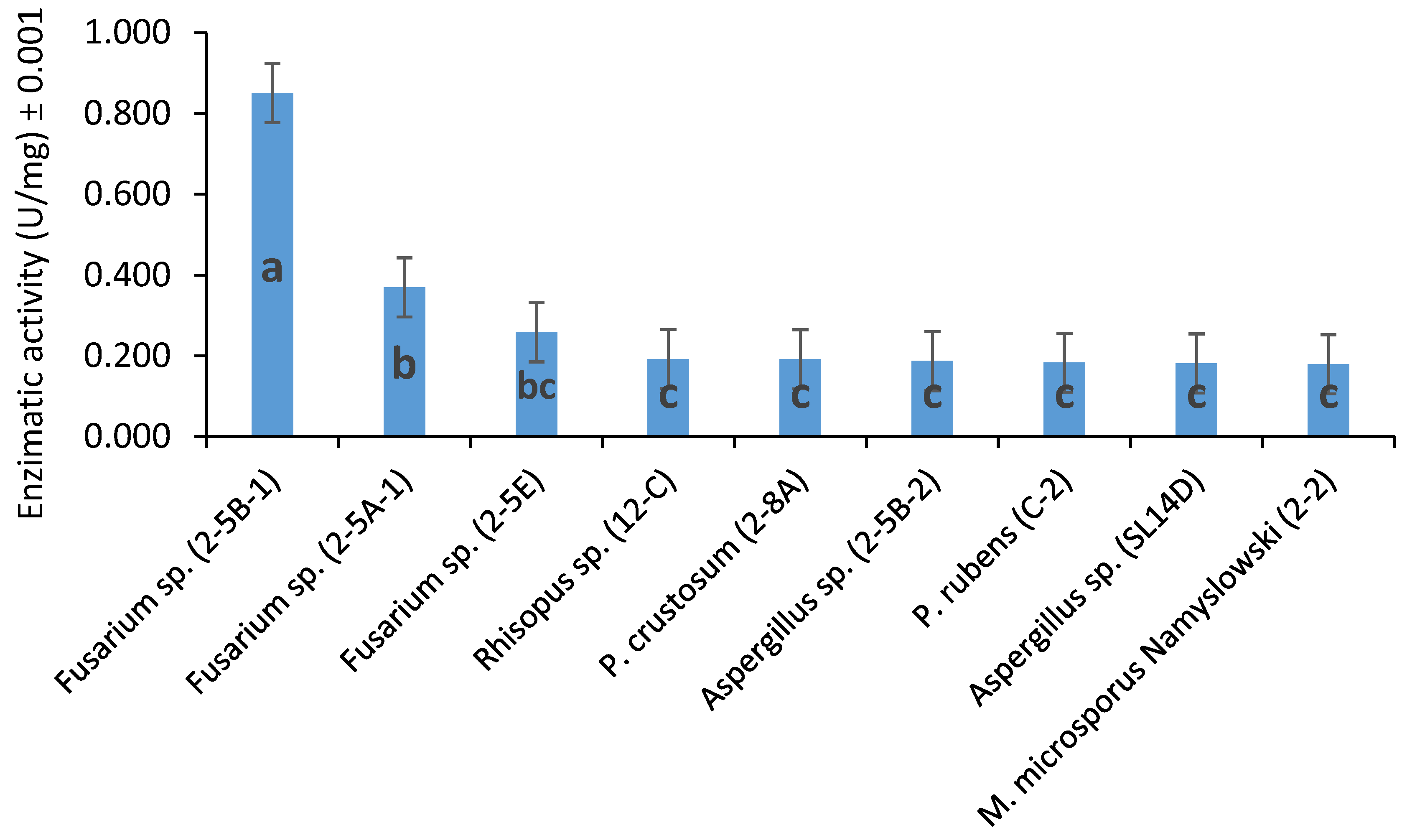
3.2. Fungal Extracellular Lipases and Esterases Screening
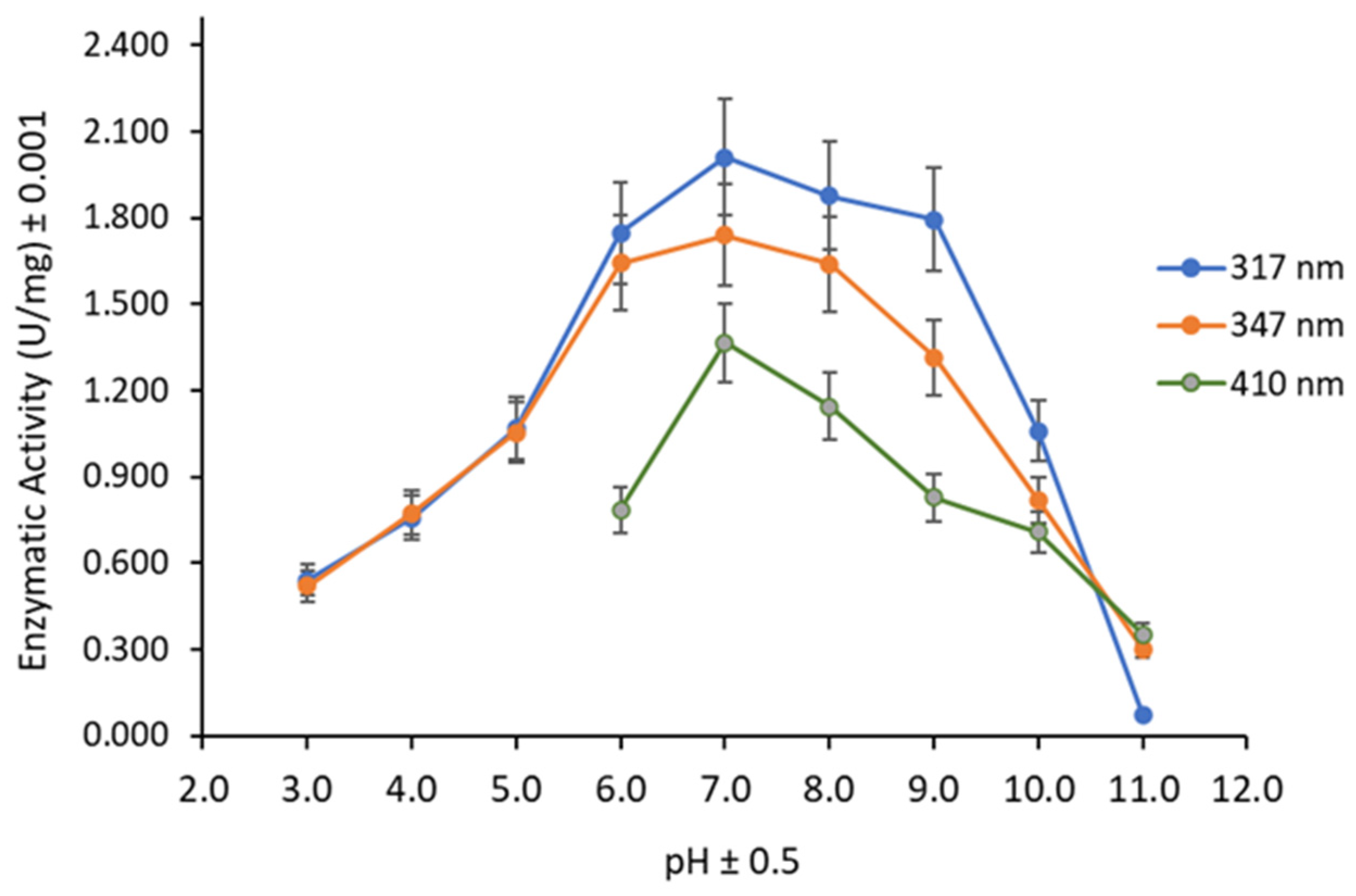
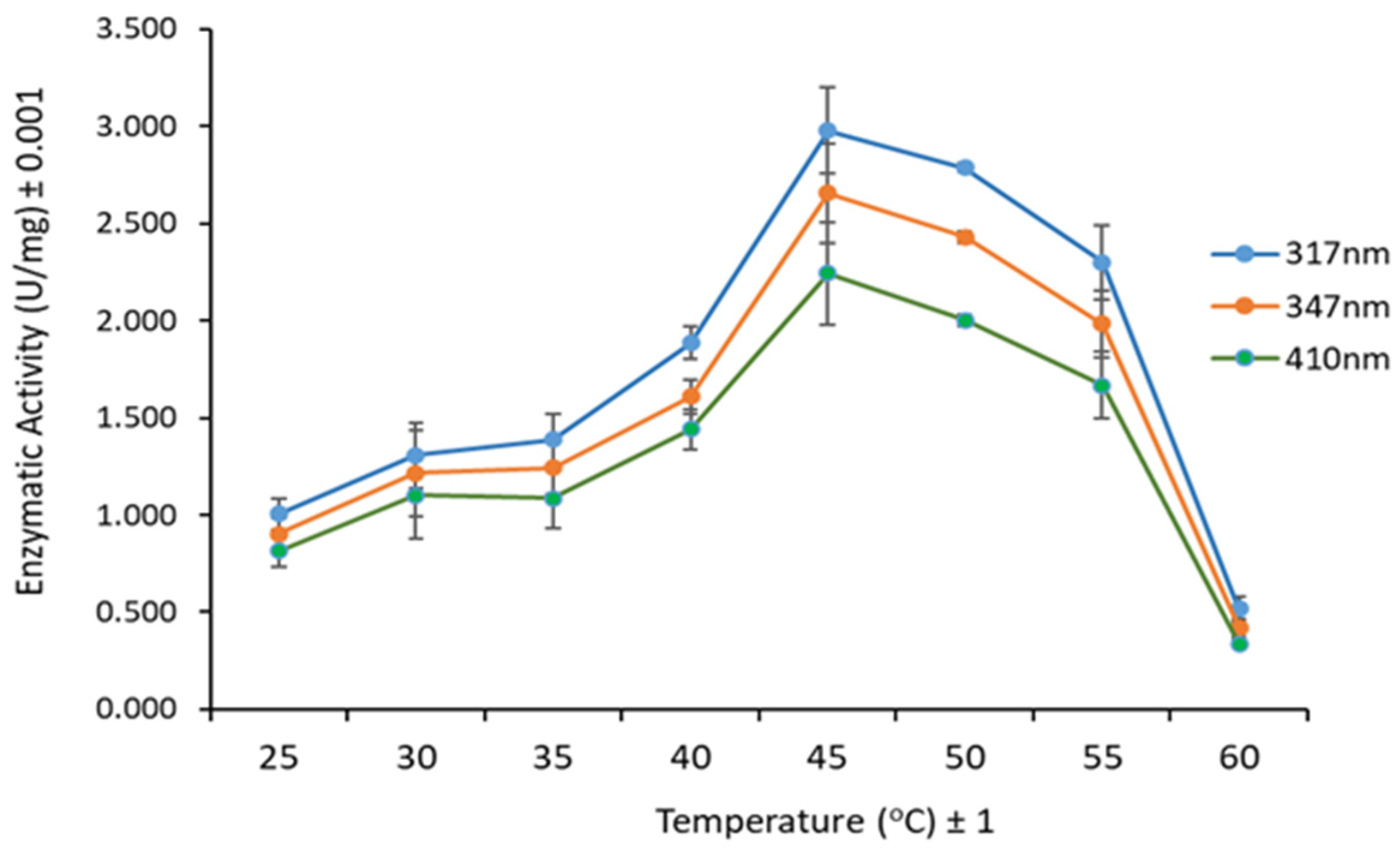
3.3. Substrate, pH, and Temperature Effect on Lipase Activity
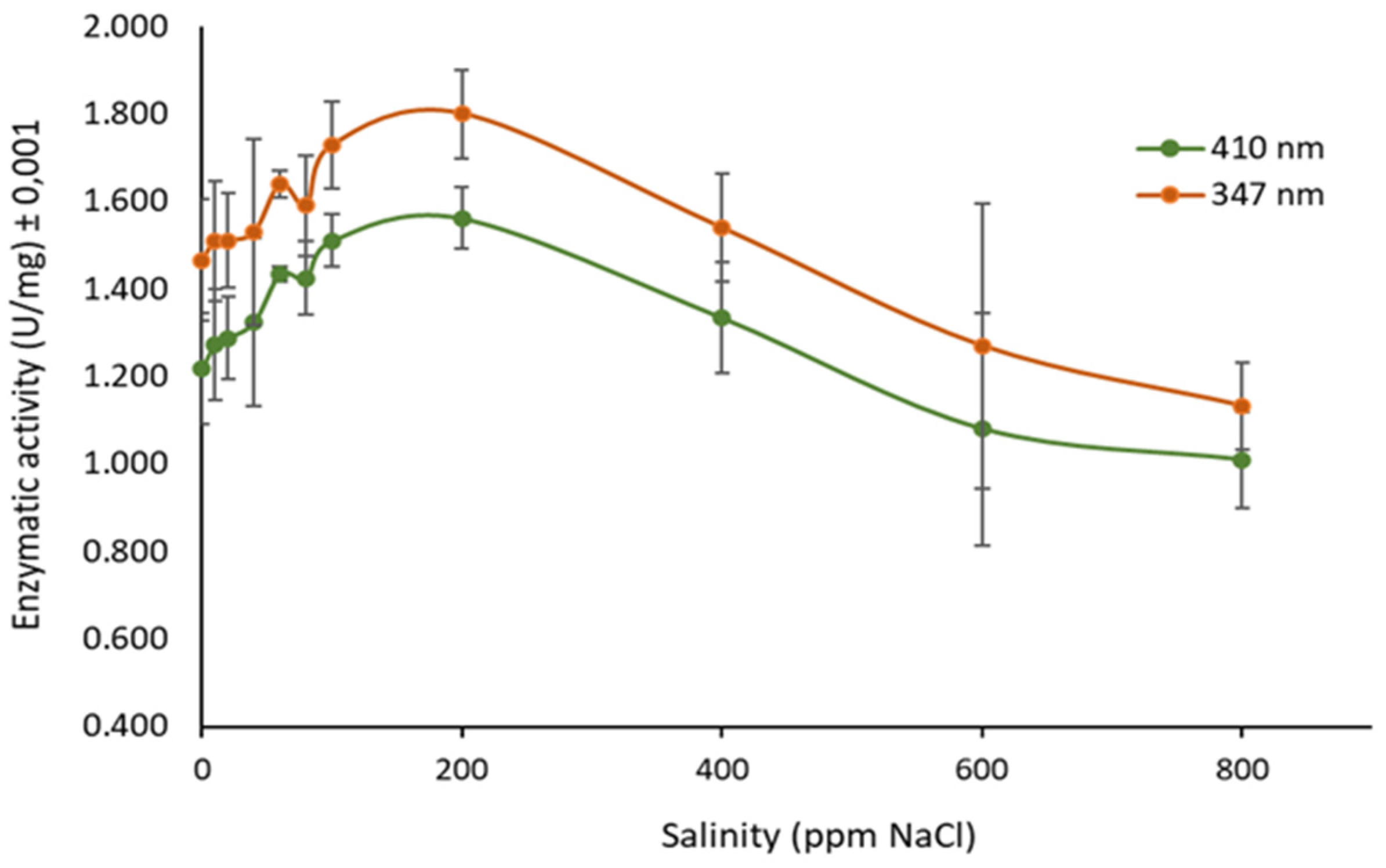
3.4. Determination of Ionic Strength Effect on Enzyme Activity
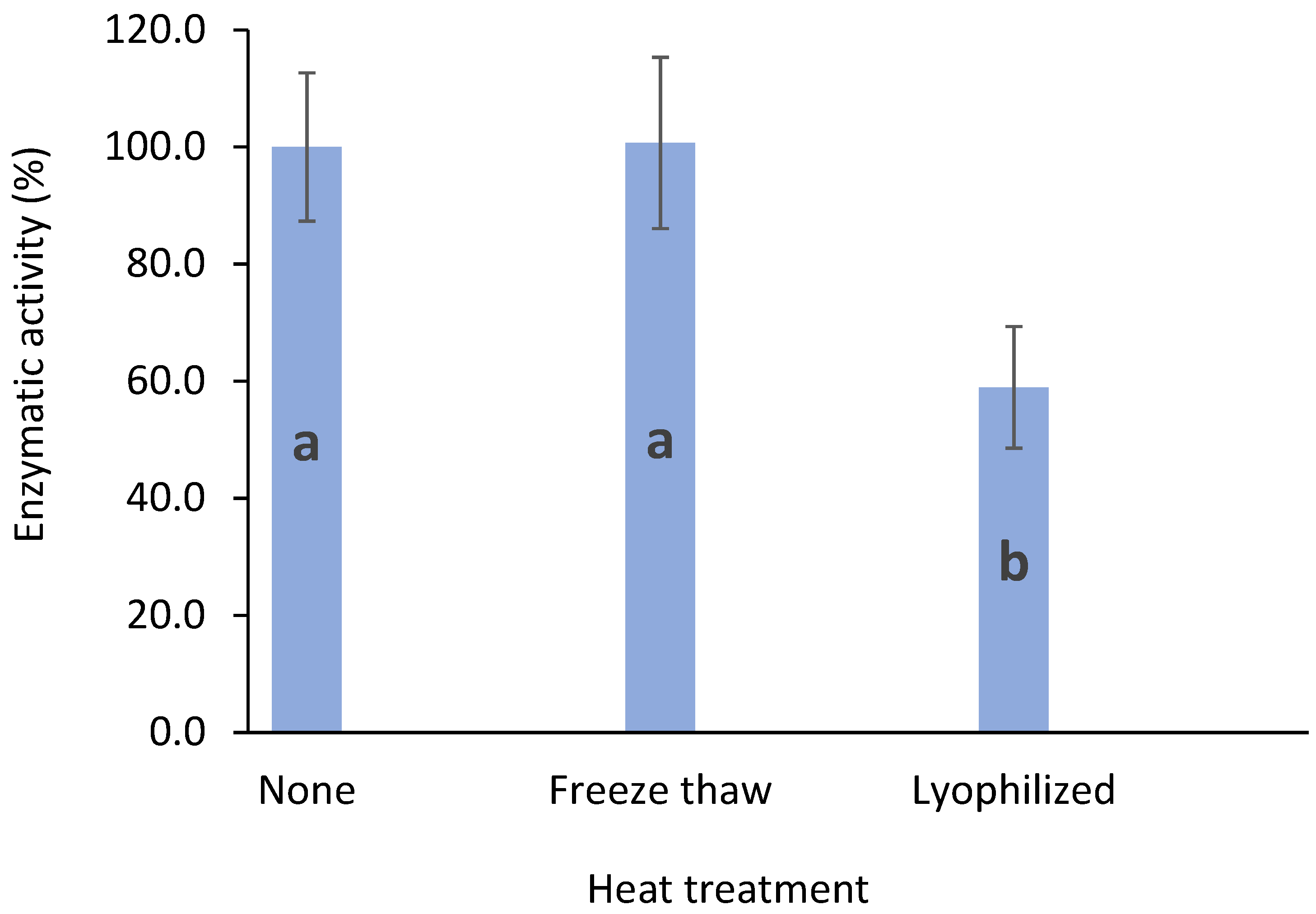
3.5. Enzyme Stability after Freeze/Thawing and Lyophilization
3.6. Lipase Applications in Biomass Degradation
3.7. Vmax and Km Kinetic Constants
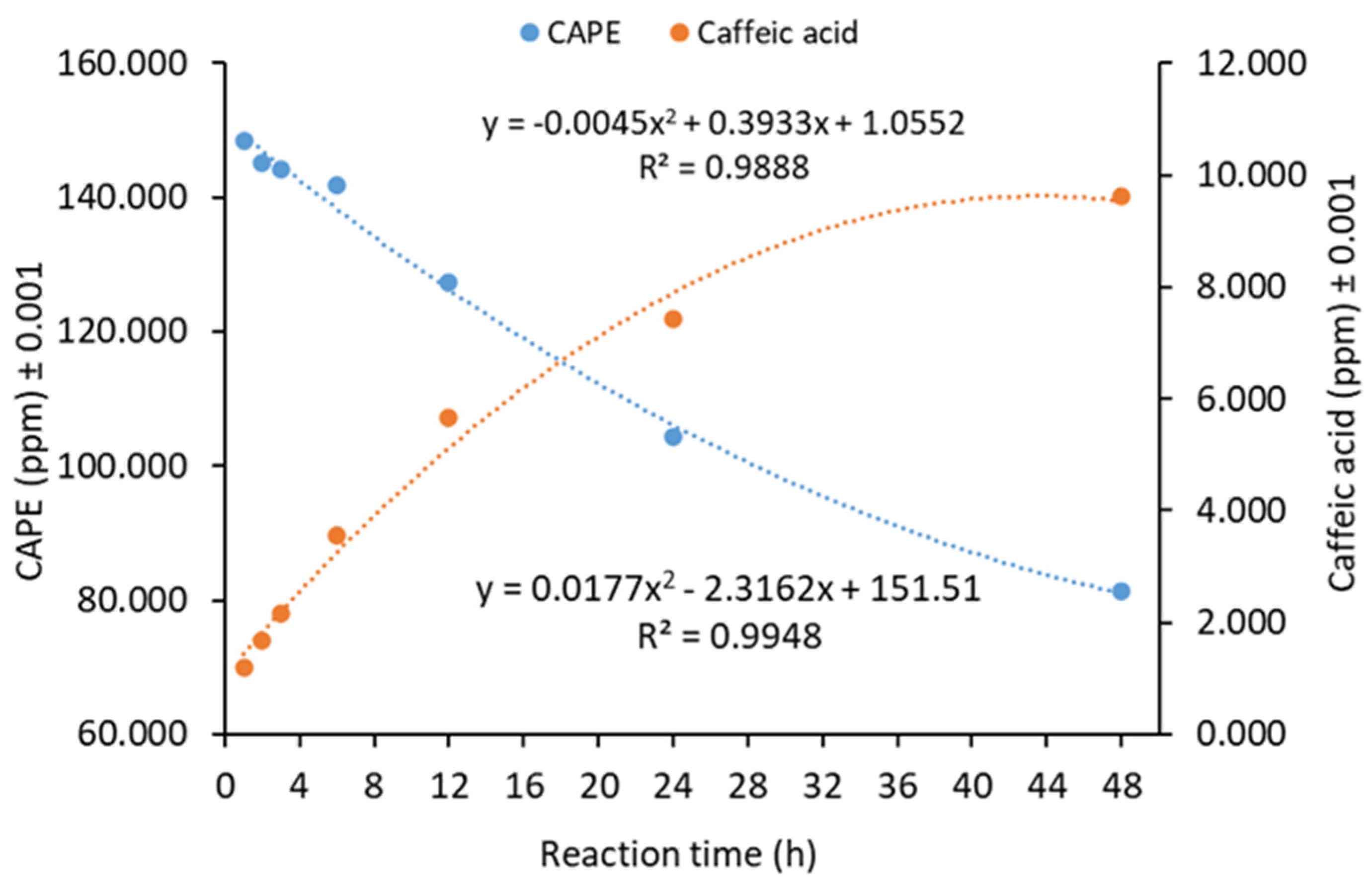
3.8. Kinetics of Lipase-Catalyzed CAPE Hydrolysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Coffee Organization. Trade Statistics. Total Production by All Exporting Countries. 2020. Available online: https://www.ico.org/prices/po-production.pdf (accessed on 14 December 2021).
- Esquivel, P.; Jiménez, V.M. Functional properties of coffee and coffee by-products. Food Res. Int. 2012, 46, 488–495. [Google Scholar] [CrossRef]
- Farah, A.; dos Santos, T.F. The Coffee Plant and Beans: An Introduction. Coffee Health Dis. Prev. 2015, 5–10. [Google Scholar] [CrossRef]
- Oliveira, L.S.; Franca, A.S. An Overview of the Potential Uses for Coffee Husks. In Coffee in Health and Disease Prevention; Elsevier: Amsterdam, The Netherlands, 2015; pp. 281–291. ISBN 9780124095175. [Google Scholar] [CrossRef]
- Low, J.H.; Rahman, W.A.W.A.; Jamaluddin, J. Structural elucidation of tannins of spent coffee grounds by CP-MAS 13C NMR and MALDI-TOF MS. Ind. Crops Prod. 2015, 69, 456–461. [Google Scholar] [CrossRef] [Green Version]
- Janissen, B.; Huynh, T. Chemical composition and value-adding applications of coffee industry by-products: A review. Resour. Conserv. Recycl. 2017, 128, 110–117. [Google Scholar] [CrossRef]
- Franklin, G.; Dias, A. Chlorogenic acid participates in the regulation of shoot, root and root hair development in Hypericum perforatum. Plant Physiol. Biochem. 2011, 49, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Bressani, R. The By-Products of Coffee Berries. In Coffee Pulp: Composition, Technology and Utilization; Braham, J.E., Bresani, R., Eds.; International Development Research Centre: Ottawa, ON, Canada, 1979; p. 95. [Google Scholar]
- Torres-Mancera, M.T.; Cordova-López, J.; Rodríguez-Serrano, G.; Roussos, S.; Ramírez-Coronel, M.A.; Favela-Torres, E.; Saucedo-Castañeda, G. Enzymatic extraction of hydroxycinnamic acids from coffee pulp. Food Technol. Biotechnol. 2011, 49, 369–373. [Google Scholar]
- Torres-Mancera, M.T.; Baqueiro-Peña, I.; Figueroa-Montero, A.; Rodríguez-Serrano, G.; González-Zamora, E.; Favela-Torres, E.; Saucedo-Castañeda, G. Biotransformation and improved enzymatic extraction of chlorogenic acid from coffee pulp by filamentous fungi. Biotechnol. Prog. 2013, 29, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Baeza, G.; Amigo-Benavent, M.; Sarriá, B.; Goya, L.; Mateos, R.; Bravo, L. Green coffee hydroxycinnamic acids but not caffeine protect human HepG2 cells against oxidative stress. Food Res. Int. 2014, 62, 1038–1046. [Google Scholar] [CrossRef] [Green Version]
- Cho, A.-S.; Jeon, S.-M.; Kim, M.-J.; Yeo, J.; Seo, K.-I.; Choi, M.-S.; Lee, M.-K. Chlorogenic acid exhibits anti-obesity property and improves lipid metabolism in high-fat diet-induced-obese mice. Food Chem. Toxicol. 2010, 48, 937–943. [Google Scholar] [CrossRef]
- dos Santos, M.D.; Almeida, M.C.; Lopes, N.P.; de Souza, G.E.P. Evaluation of the Anti-inflammatory, Analgesic and Antipyretic Activities of the Natural Polyphenol Chlorogenic Acid. Biol. Pharm. Bull. 2006, 29, 2236–2240. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Ruan, Z.; Zhou, L.; Shu, X.; Sun, X.; Mi, S.; Yang, Y.; Yin, Y. Chlorogenic acid ameliorates endotoxin-induced liver injury by promoting mitochondrial oxidative phosphorylation. Biochem. Biophys. Res. Commun. 2016, 469, 1083–1089. [Google Scholar] [CrossRef] [PubMed]
- Magoni, C.; Bruni, I.; Guzzetti, L.; Dell’Agli, M.; Sangiovanni, E.; Piazza, S.; Regonesi, M.E.; Maldini, M.; Spezzano, R.; Caruso, D.; et al. Valorizing coffee pulp by-products as anti-inflammatory ingredient of food supplements acting on IL-8 release. Food Res. Int. 2018, 112, 129–135. [Google Scholar] [CrossRef]
- Grunberger, D.; Banerjee, R.; Eisinger, K.; Oltz, E.M.; Efros, L.; Caldwell, M.; Estevez, V.; Nakanishi, K. Preferential cytotoxicity on tumor cells by caffeic acid phenethyl ester isolated from propolis. Experientia 1988, 44, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Wang, D.; He, Y.; Xie, J.; Zhong, Z.; Li, Z.; Xie, J. Caffeic acid phenethyl ester induces growth arrest and apoptosis of colon cancer cells via the β-catenin/T-cell factor signaling. Anti-Cancer Drugs 2006, 17, 753–762. [Google Scholar] [CrossRef]
- Omene, C.O.; Wu, J.; Frenkel, K. Caffeic Acid Phenethyl Ester (CAPE) derived from propolis, a honeybee product, inhibits growth of breast cancer stem cells. Investig. New Drugs 2011, 30, 1279–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, V.; Touraud, D.; Kunz, W. Eco-friendly one pot synthesis of caffeic acid phenethyl ester (CAPE) via an in-situ formed deep eutectic solvent. Sustain. Chem. Pharm. 2016, 4, 40–45. [Google Scholar] [CrossRef]
- Touaibia, M.; Guay, M. Natural Product Total Synthesis in the Organic Laboratory: Total Synthesis of Caffeic Acid Phenethyl Ester (CAPE), A Potent 5-Lipoxygenase Inhibitor from Honeybee Hives. J. Chem. Educ. 2011, 88, 473–475. [Google Scholar] [CrossRef]
- Zhang, P.; Tang, Y.; Li, N.-G.; Zhu, Y.; Duan, J.-A. Bioactivity and Chemical Synthesis of Caffeic Acid Phenethyl Ester and Its Derivatives. Molecules 2014, 19, 16458–16476. [Google Scholar] [CrossRef] [Green Version]
- A Bakar, P.N.M.; Gonawan, F.N.; Kamaruddin, A.H. Parameters study of lipase-transesterification reaction of ethyl caffeate and glycerol in deep eutectic solvent (DES). IOP Conf. Series Mater. Sci. Eng. 2018, 440, 012004. [Google Scholar] [CrossRef]
- Ha, S.H.; Van Anh, T.; Koo, Y.-M. Optimization of lipase-catalyzed synthesis of caffeic acid phenethyl ester in ionic liquids by response surface methodology. Bioprocess Biosyst. Eng. 2013, 36, 799–807. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Zhang, L.; Gu, S.; Wu, F. Lipase-catalyzed Synthesis of Caffeic Acid Phenethyl Ester in Ionic Liquids: Effect of Specific Ions and Reaction Parameters. Chin. J. Chem. Eng. 2013, 21, 1376–1385. [Google Scholar] [CrossRef]
- Gu, S.; Wang, J.; Wei, X.; Cui, H.; Wu, X.; Wu, F. Enhancement of Lipase-catalyzed Synthesis of Caffeic Acid Phenethyl Ester in Ionic Liquid with DMSO Co-solvent. Chin. J. Chem. Eng. 2014, 22, 1314–1321. [Google Scholar] [CrossRef]
- Kotogán, A.; Németh, B.; Vágvölgyi, C.; Papp, T.; Takó, M. Screening for extracellular lipase enzymes with transesterification capacity in Mucoromycotina strains. Food Technol. Biotechnol. 2014, 52, 73–82. [Google Scholar]
- Rajapriya, G.; Morya, V.; Mai, N.L.; Koo, Y.-M. Aspergillus niger whole-cell catalyzed synthesis of caffeic acid phenethyl ester in ionic liquids. Enzym. Microb. Technol. 2018, 111, 67–73. [Google Scholar] [CrossRef]
- Anastas, P.; Eghbali, N. Green Chemistry: Principles and Practice. Chem. Soc. Rev. 2010, 39, 301–312. [Google Scholar] [CrossRef]
- Sheldon, R.A. Fundamentals of green chemistry: Efficiency in reaction design. Chem. Soc. Rev. 2011, 41, 1437–1451. [Google Scholar] [CrossRef] [Green Version]
- Asamblea Legislativa de la República de Costa Rica. Ley de Biodiversidad N°7788. La Gaceta N° 101 del 27 de Mayo de 1998; San José, Costa Rica, 1998; p. 40. Available online: http://www.pgrweb.go.cr/scij/Busqueda/Normativa/Normas/nrm_texto_completo.aspx?param2=NRTC&nValor1=1&nValor2=39796&strTipM=TC (accessed on 14 December 2021).
- The Contracting Parties. Convention on Biological Diversity; Rio de Janeiro, Brazil, 1992; p. 31. Available online: https://www.cbd.int/doc/legal/cbd-en.pdf (accessed on 14 December 2021).
- Costa Rican National Board for Biodiversity Management (Comisión Nacional de Gestión de la Biodiversidad). Granted Biopropective Permits. Available online: https://www.conagebio.go.cr/Conagebio/public/permisosOtorgados.html (accessed on 14 December 2021).
- Syedd-León, R.; Sandoval-Barrantes, M.; Trimiño-Vásquez, H.; Villegas-Peñaranda, L.R.; Rodríguez-Rodríguez, G. Revisiting the fundamentals of p-nitrophenol analysis for its application in the quantification of lipases activity. A graphical update. Uniciencia 2020, 34, 31–43. [Google Scholar] [CrossRef]
- Peng, Y.; Fu, S.; Liu, H.; Lucia, L.A. Accurately Determining Esterase Activity via the Isosbestic Point of p-Nitrophenol. BioResources 2016, 11, 10099–10111. [Google Scholar] [CrossRef]
- Fourage, L.; Helbert, M.; Nicolet, P.; Colas, B. Temperature Dependence of the Ultraviolet–Visible Spectra of Ionized and Un-ionized Forms of Nitrophenol: Consequence for the Determination of Enzymatic Activities Using Nitrophenyl Derivatives—A Warning. Anal. Biochem. 1999, 270, 184–185. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Stewart, C.N., Jr. Rapid DNA extraction from plants. In Fingerprinting Methods Based on Arbitrarily Primed PCR; Springer: Berlin/Heidelberg, Germany, 1997. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Elsevier: Amsterdam, The Netherlands, 1990; pp. 315–322. [Google Scholar]
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 140. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, V.; Wang, Q.; Greenfield, P.; Charleston, M.; Porras-Alfaro, A.; Kuske, C.R.; Cole, J.R.; Midgley, D.J.; Tran-Dinh, N. Fungal identification using a Bayesian classifier and the Warcup training set of internal transcribed spacer sequences. Mycologia 2016, 108, 1–5. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; De Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Gadberry, M.D.; Malcomber, S.T.; Doust, A.N.; Kellogg, E. Primaclade--a flexible tool to find conserved PCR primers across multiple species. Bioinformatics 2004, 21, 1263–1264. [Google Scholar] [CrossRef] [Green Version]
- Fischer, M. The Lipase Engineering Database: A navigation and analysis tool for protein families. Nucleic Acids Res. 2003, 31, 319–321. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Stanke, M.; Steinkamp, R.; Waack, S.; Morgenstern, B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004, 32, W309–W312. [Google Scholar] [CrossRef] [Green Version]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armenteros, J.J.A.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef] [PubMed]
- Kohno, M.; Funatsu, J.; Mikami, B.; Kugimiya, W.; Matsuo, T.; Marita, Y. The Crystal Structure of Lipase II from Rhizopus niveus at 2.2 A Resolution. J. Biochem. 1996, 120, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ferraz, L.R.; Oliveira, D.D.S.D.; Silva, M.F.; Rigo, E.; Di Luccio, M.; Oliveira, J.V.; de Oliveira, D.; Treichel, H. Production and partial characterization of multifunctional lipases by Sporobolomyces ruberrimus using soybean meal, rice meal and sugarcane bagasse as substrates. Biocatal. Agric. Biotechnol. 2012, 1, 243–252. [Google Scholar] [CrossRef]
- Shu, Z.-Y.; Yang, J.-K.; Yan, Y.-J. Purification and characterization of a lipase from Aspergillus niger F044. Chin. J. Biotechnol. 2007, 23, 96–101. [Google Scholar] [CrossRef]
- Demir, B.S.; Tükel, S.S. Purification and characterization of lipase from Spirulina platensis. J. Mol. Catal. B Enzym. 2010, 64, 123–128. [Google Scholar] [CrossRef]
- Supakdamrongkul, P.; Bhumiratana, A.; Wiwat, C. Characterization of an extracellular lipase from the biocontrol fungus, Nomuraea rileyi MJ, and its toxicity toward Spodoptera litura. J. Invertebr. Pathol. 2010, 105, 228–235. [Google Scholar] [CrossRef]
- Maia, M. Effect of culture conditions on lipase production by Fusarium solani in batch fermentation. Bioresour. Technol. 2001, 76, 23–27. [Google Scholar] [CrossRef]
- Geoffry, K.; Achur, R.N. Screening and production of lipase from fungal organisms. Biocatal. Agric. Biotechnol. 2018, 14, 241–253. [Google Scholar] [CrossRef]
- Rashid, N.; Shimada, Y.; Ezaki, S.; Atomi, H.; Imanaka, T. Low-Temperature Lipase from Psychrotrophic Pseudomonas sp. Strain KB700A. Appl. Environ. Microbiol. 2001, 67, 4064–4069. [Google Scholar] [CrossRef] [Green Version]
- Treichel, H.; Oliveira, D.; Mazutti, M.; Di Luccio, M.; Oliveira, J.V. A Review on Microbial Lipases Production. Food Bioprocess Technol. 2009, 3, 182–196. [Google Scholar] [CrossRef]
- Kapoor, M.; Gupta, M.N. Lipase promiscuity and its biochemical applications. Process Biochem. 2012, 47, 555–569. [Google Scholar] [CrossRef]
- Wu, J.; Omene, C.; Karkoszka, J.; Bosland, M.; Eckard, J.; Klein, C.; Frenkel, K. Caffeic acid phenethyl ester (CAPE), derived from a honeybee product propolis, exhibits a diversity of anti-tumor effects in pre-clinical models of human breast cancer. Cancer Lett. 2011, 308, 43–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishida, Y.; Gao, R.; Shah, N.; Bhargava, P.; Furune, T.; Kaul, S.C.; Terao, K.; Wadhwa, R. Anticancer Activity in Honeybee Propolis: Functional Insights to the Role of Caffeic Acid Phenethyl Ester and Its Complex With γ-Cyclodextrin. Integr. Cancer Ther. 2018, 17, 867–873. [Google Scholar] [CrossRef] [Green Version]
- Gülçin, I. Antioxidant activity of caffeic acid (3,4-dihydroxycinnamic acid). Toxicology 2006, 217, 213–220. [Google Scholar] [CrossRef]
- Sato, Y.; Itagaki, S.; Kurokawa, T.; Ogura, J.; Kobayashi, M.; Hirano, T.; Sugawara, M.; Iseki, K. In vitro and in vivo antioxidant properties of chlorogenic acid and caffeic acid. Int. J. Pharm. 2010, 403, 136–138. [Google Scholar] [CrossRef]
- Norata, G.D.; Marchesi, P.; Passamonti, S.; Pirillo, A.; Violi, F.; Catapano, A.L. Anti-inflammatory and anti-atherogenic effects of cathechin, caffeic acid and trans-resveratrol in apolipoprotein E deficient mice. Atherosclerosis 2007, 191, 265–271. [Google Scholar] [CrossRef]
- Huang, M.T.; Smart, R.C.; Wong, C.Q.; Conney, A.H. Inhibitory effect of curcumin, chlorogenic acid, caffeic acid, and ferulic acid on tumor promotion in mouse skin by 12-O-tetradecanoylphorbol-13-acetate. Cancer Res. 1988, 48, 5941–5946. Available online: http://www.ncbi.nlm.nih.gov/pubmed/3139287 (accessed on 19 November 2018). [PubMed]
- Chen, H.-C.; Chen, J.-H.; Chang, C.; Shieh, C.-J. Optimization of ultrasound-accelerated synthesis of enzymatic caffeic acid phenethyl ester by response surface methodology. Ultrason. Sonochem. 2011, 18, 455–459. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syedd-León, R.; Solano-Campos, F.; Campos-Rodríguez, J.; Pereira-Arce, D.; Villegas-Peñaranda, L.R.; Sandoval-Barrantes, M. Fungal Extracellular Lipases from Coffee Plantation Environments for the Sustainable Management of Agro-Industrial Coffee Biomass. Biomass 2022, 2, 62-79. https://doi.org/10.3390/biomass2020005
Syedd-León R, Solano-Campos F, Campos-Rodríguez J, Pereira-Arce D, Villegas-Peñaranda LR, Sandoval-Barrantes M. Fungal Extracellular Lipases from Coffee Plantation Environments for the Sustainable Management of Agro-Industrial Coffee Biomass. Biomass. 2022; 2(2):62-79. https://doi.org/10.3390/biomass2020005
Chicago/Turabian StyleSyedd-León, Randall, Frank Solano-Campos, Jorge Campos-Rodríguez, Daniela Pereira-Arce, Luis Roberto Villegas-Peñaranda, and Manuel Sandoval-Barrantes. 2022. "Fungal Extracellular Lipases from Coffee Plantation Environments for the Sustainable Management of Agro-Industrial Coffee Biomass" Biomass 2, no. 2: 62-79. https://doi.org/10.3390/biomass2020005