Preimplantation Endometrial Transcriptomics in Natural Conception Cycle of the Rhesus Monkey
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Procedures
2.2. Animal Groups and Tissue Collection
2.3. RNA Extraction
2.4. Whole-Genome Expression Array
2.5. Array Data Analysis and Post-Hoc Enrichment Analysis
2.6. Quantification of Candidate Gene Expression by Real-Time RT-PCR
2.7. Immunohistochemical Localisation of Candidate Proteins for Orthogonal Validation
3. Results
3.1. General
3.2. Expression Array
3.2.1. Descriptive Data Analysis
3.2.2. Differentially Expressed (DE) Genes
3.2.3. Group-Specific Expression of Non-Matching Genes
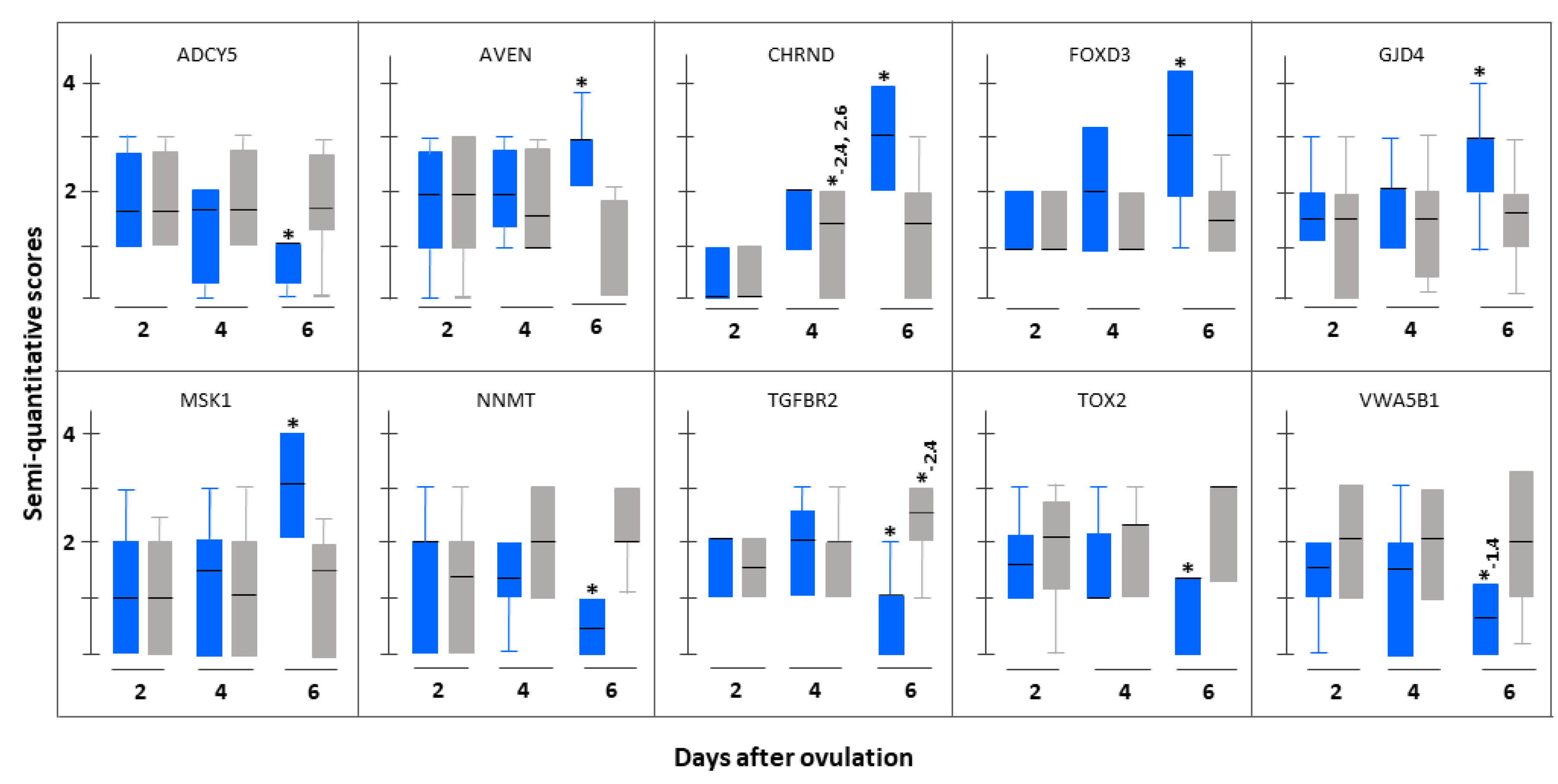
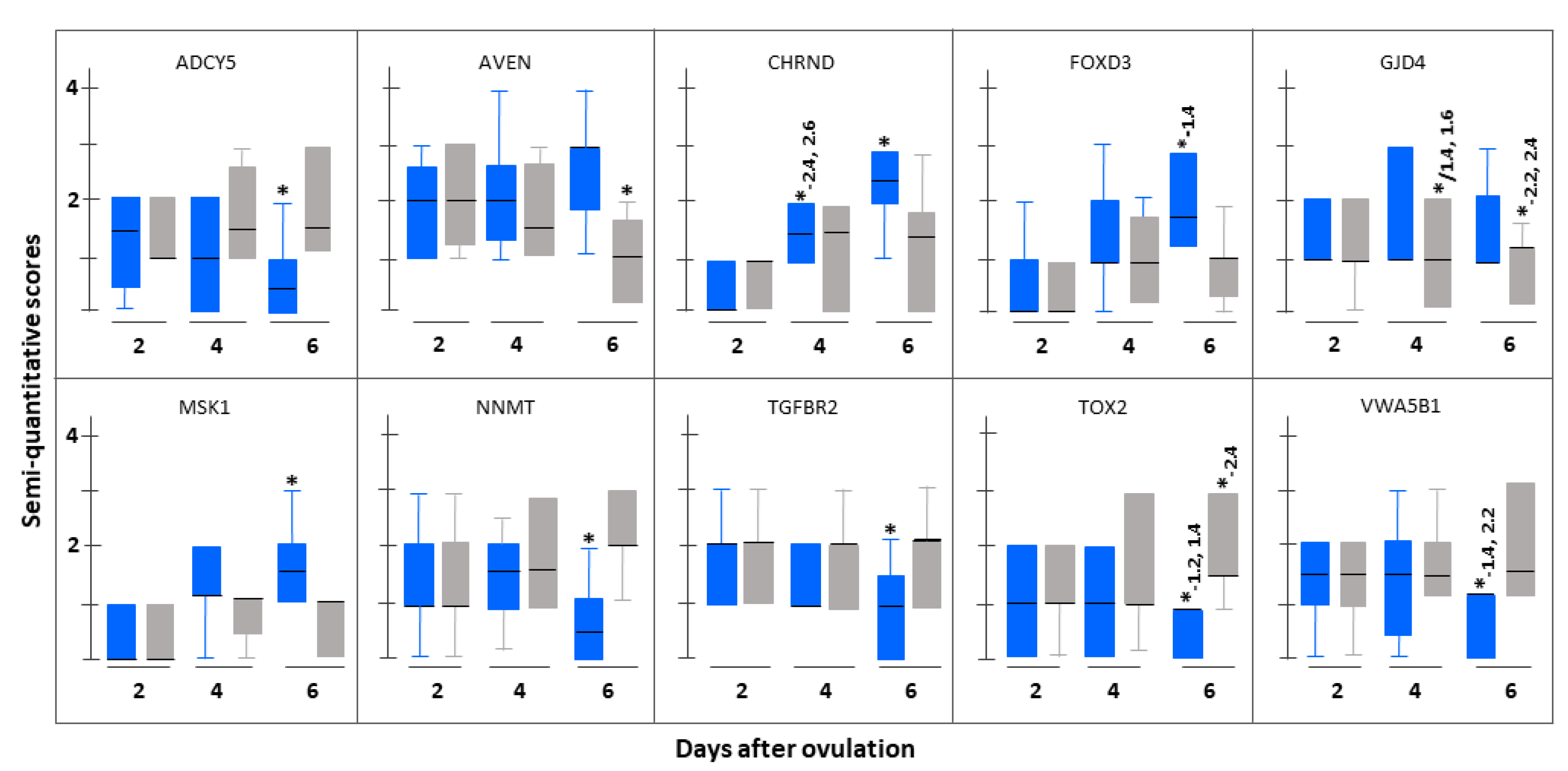
3.3. Immunohistochemistrical Validation
4. Discussion
4.1. Genes Expressed in Both Groups and Involved in Protein Synthesis and Secretion along with Metabolic Processes Might Be Regulated by Progesterone
4.2. Genes Uniquely Expressed in Natural Conception Cycle and Involved in Induction of Immunotolerance to Non-Self Antigens Might Be Regulated by Factors in Seminal Plasma
- Immunotolerance to ovo-implantation in a natural pregnancy cycle is a primary process triggered in receptive endometrium, and it is not exclusively dependent on trophoblastic input, although it has—in all likelihood—reinforcing action to the process for successful establishment of pregnancy [52,53].
- The underlying genomic expression of the immunotolerance process in secretory phase endometrium is unlikely to be regulated by the preimplantation stage embryo because, in primates including the rhesus macaque, embryos seldom reach uterine lumen on Day 2 after ovulation [56]. Still, in the present study, we observed from transcriptomic expressions, endometrium geared towards immune tolerance to non-self antigens on Day 2 post-fertilization onwards.
- A plausible explanation for the observed differentiation towards immunotolerance in the secretory phase endometrium of mated and proven fecund group of animals could be due to the presence of seminal plasma inducing the observed genomic expression. In fact, evidence based on a series of elegant experiments is now available across various mammalian species to indicate that seminal plasma exerts substantial modifying action on endometrium towards receptivity, ovo-implantation, immunotolerance, embryo development, and establishment of pregnancy [38,52,57,58,59,60,61,62]. To our knowledge, this is the first report supporting the notion that seminal plasma could induce at the genomic expression level a process bias for immunotolerance to non-self antigens in the secretory phase endometrium of natural conception cycle in the rhesus macaque.
4.3. Genes Involved in Cell Polarity, Differentiation, Signaling, Fate, and Immunomodulation Might Be Regulated by Embryo-Derived Signals
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macklon, N.S.; Geraedts, J.P.M.; Fauser, B.C.J.M. Conception to ongoing pregnancy: The ‘black box’ of early pregnancy loss. Hum. Reprod. Update 2002, 8, 333–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sengupta, J.; Ghosh, D. Multi-level and multi-scale integrative approach to the understanding of human blastocyst implantation. Prog. Biophys. Mol. Biol. 2014, 114, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Diedrich, K.; Fauser, B.C.; Devroey, P.; Griesinger, G. The role of the endometrium and embryo in human implantation. Hum. Reprod. Update 2007, 13, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Monad, M.; Marsh, C.; Schumacher, K.; Nothnick, W. Secretory phase of menstruation and implantation. Front. Womens Health 2018, 3, 1–5. [Google Scholar] [CrossRef]
- Licht, P.; Lösch, A.; Dittrich, R.; Neuwinger, J.; Siebzehnrübl, E.; Wildt, L. Novel insights into human endometrial paracrinology and embryo-maternal communication by intrauterine microdialysis. Hum. Reprod. Update 1998, 4, 532–538. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.M.; Kim, J.S. A review of mechanisms of implantation. Dev. Reprod. 2017, 21, 351–359. [Google Scholar] [CrossRef] [Green Version]
- Ochoa-Bernal, M.A.; Fazleabas, A.T. Physiologic events of embryo implantation and decidualization in human and non-human primates. Int. J. Mol. Sci. 2020, 21, 1973. [Google Scholar] [CrossRef] [Green Version]
- Roberts, R.M.; Xie, S.; Mathialagan, N. Maternal recognition of pregnancy. Biol. Reprod. 1996, 54, 294–302. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.Y.; DeMayo, F.J. Animal models of implantation. Reproduction 2004, 128, 679–695. [Google Scholar] [CrossRef] [Green Version]
- Reese, J.; Wang, H.; Ding, T.; Paria, B.C. The hamster as a model for embryo implantation: Insights into a multifaceted process. Semin. Cell Dev. Biol. 2008, 19, 194–203. [Google Scholar] [CrossRef] [Green Version]
- Bazer, F.W. Pregnancy recognition signaling mechanisms in ruminants and pigs. J. Anim. Sci. Biotechnol. 2013, 4, 23. [Google Scholar] [CrossRef] [Green Version]
- Ka, H.; Heewon, H.; Choi, Y.; Yoo, I.; Han, J. Endometrial response to conceptus derived estrogen and interleukin-1β at the time of implantation in pigs. J. Anim. Sci. Biotechnol. 2018, 9, 44. [Google Scholar] [CrossRef]
- Harding, J.D. Nonhuman primates and translational research: Progress, opportunities, and challenges. ILAR J. 2017, 58, 141–150. [Google Scholar] [CrossRef] [Green Version]
- Friedman, H.; Ator, N.; Haigwood, N.; Newsome, W.; Allan, J.S.; Golos, T.G.; Kordower, J.H.; Shade, R.E.; Goldberg, M.E.; Bailey, M.R.; et al. The critical role of nonhuman primates in medical research. Pathog. Immun. 2017, 2, 352–365. [Google Scholar] [CrossRef] [Green Version]
- Neill, J.D.; Johansson, E.D.; Knobil, E. Patterns of circulating progesterone concentrations during the fertile menstrual cycle and the remainder of gestation in the rhesus monkey. Endocrinology 1969, 84, 45–48. [Google Scholar] [CrossRef]
- Lenton, E.A.; Sulaiman, R.; Sobowale, O.; Cooke, I.D. The human menstrual cycle: Plasma concentrations of prolactin, LH, FSH, oestradiol and progesterone in conceiving and non-conceiving women. J. Reprod. Fertil. 1982, 65, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, D.; De, P.; Sengupta, J. Luteal phase ovarian oestrogen is not essential for implantation and maintenance of pregnancy from surrogate embryo transfer in the rhesus monkey. Hum. Reprod. 1994, 9, 629–637. [Google Scholar] [CrossRef]
- Zegers-Hochschild, F.; Altieri, E. Luteal estrogen is not required for the establishment of pregnancy in the human. J. Assist. Reprod. Genet. 1995, 12, 224–228. [Google Scholar] [CrossRef]
- Ghosh, D.; Sengupta, J. Recent developments in endocrinology and paracrinology of blastocyst implantation in the primate. Hum. Reprod. Update 1998, 4, 153–168. [Google Scholar] [CrossRef] [Green Version]
- Ng, E.H.Y.; Yeung, W.S.B.; Lau, E.Y.L.; So, W.W.K.; Ho, P.C. A rapid decline in serum oestradiol concentrations around the mid-luteal phase had no adverse effect on outcome in 763 assisted reproduction cycles. Hum. Reprod. 2000, 15, 1903–1908. [Google Scholar] [CrossRef] [Green Version]
- Fishel, S.B.; Edwards, R.G.; Evans, C.J. Human chorionic gonadotropin secreted by preimplantation embryos cultured in vitro. Science 1984, 223, 816–818. [Google Scholar] [CrossRef] [Green Version]
- Hearn, J.P.; Webley, G.E.; Gidley-Baird, A.A. Chorionic gonadotrophin and embryo-maternal recognition during the peri-implantation period in primates. J. Reprod. Fertil. 1991, 92, 497–509. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, D.; Stewart, D.R.; Nayak, N.R.; Lasley, B.L.; Overstreet, J.W.; Hendrickx, A.G.; Sengupta, J. Serum concentrations of oestradiol-17β, progesterone, relaxin and chorionic gonadotrophin during blastocyst implantation in natural pregnancy cycle and in embryo transfer cycle in the rhesus monkey. Hum. Reprod. 1997, 12, 914–920. [Google Scholar] [CrossRef] [Green Version]
- Hertig, A.T.; Rock, J.; Adams, E.C. A description of 34 human ova within the first 17 days of development. Am. J. Anat. 1956, 98, 435–493. [Google Scholar] [CrossRef]
- Enders, A.C.; Welsh, A.O.; Schlafke, S. Implantation in the rhesus monkey: Endometrial responses. Am. J. Anat. 1985, 173, 147–169. [Google Scholar] [CrossRef]
- Enders, A.C. Trophoblast differentiation during the transition from trophoblastic plate to lacunar stage of implantation in the rhesus monkey and human. Am. J. Anat. 1989, 186, 85–98. [Google Scholar] [CrossRef]
- Sengupta, J.; Ghosh, D. Blastocyst-endometrium interaction at implantation in the rhesus monkey. J. Reprod. Immunol. 2002, 53, 227–239. [Google Scholar] [CrossRef]
- Carter, A.M.; Allen, C.; Enders, A.C.; Pijnenborg, R. The role of invasive trophoblast in implantation and placentation of primates. Philos. Trans. R Soc. B Biol. Sci. 2015, 370, 20140070. [Google Scholar] [CrossRef]
- Enders, A.C.; Welsh, A.O. Structural interactions of trophoblast and uterus during hemochorial placenta formation. J. Exp. Zool. 1993, 266, 578–587. [Google Scholar] [CrossRef]
- Dhara, S.; Lalitkumar, P.G.L.; Sengupta, J.; Ghosh, D. Immunohistochemical localization of insulin-like growth factors I and II at the primary implantation site in the Rhesus monkey. Mol. Hum. Reprod. 2001, 7, 365–371. [Google Scholar] [CrossRef] [Green Version]
- Benirschke, K.; Burton, G.J.; Baergen, R.N. Nonvillous parts and trophoblast invasion. In Pathology of the Human Placenta, 6th ed.; Springer: New York, NY, USA, 2012; pp. 157–240. [Google Scholar]
- Ghosh, D.; Sengupta, J.; Hendrickx, A.G. Effect of single-dose, early luteal phase administration of mifepristone (RU486) on implantation stage endometrium in the rhesus monkey. Hum. Reprod. 1996, 11, 2026–2035. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Sharkey, A.M.; Charnock-Jones, D.S.; Smith, S.K.; Sengupta, J. Effect of low-dose mifepristone administration on Day 2 after ovulation on transcript profiles in implantation-stage endometrium of rhesus monkeys. Reproduction 2009, 138, 357–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrell, R.E. RNA Methodologies—A Laboratory Guide for Isolation and Characterization, 4th ed.; Academic Press: San Diego, CA, USA, 2009; pp. 179–216. [Google Scholar]
- Khan, M.A.; Sengupta, J.; Mittal, S.; Ghosh, D. Genome-wide expressions in autologous eutopic and ectopic endometrium of fertile women with endometriosis. Reprod. Biol. Endocrinol. 2012, 10, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, A.; Sengupta, J.; Kriplani, A.; Roy, K.K.; Ghosh, D. Profiles of cytokines secreted by isolated human endometrial cells under the influence of chorionic gonadotropin during the window of embryo implantation. Reprod. Biol. Endocrinol. 2013, 11, 116. [Google Scholar] [CrossRef] [Green Version]
- Bhat, M.A.; Sharma, J.B.; Roy, K.K.; Sengupta, J.; Ghosh, D. Genomic evidence of Y chromosome microchimerism in the endometrium during endometriosis and in cases of infertility. Reprod. Biol. Endocrinol. 2019, 17, 22. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Adams, E.J.; Green, J.A.; Clark, A.H.; Youngson, J.H. Comparison of different scoring systems for immunohistochemical staining. Clin. Pathol. 1999, 52, 75–77. [Google Scholar] [CrossRef] [Green Version]
- Bondarenko, A.; Angrisani, N.; Meyer-Lindenberg, A.; Seitz, J.M.; Waizy, H.; Reifenrath, J. Magnesium-based bone implants: Immunohistochemical analysis of peri-implant osteogenesis by evaluation of osteopontin and osteocalcin expression. J. Biomed. Mater. Res. Part A 2014, 102, 1449–1457. [Google Scholar] [CrossRef]
- Sim, J.; Wright, C.C. The kappa statistic in reliability studies: Use, interpretation, and sample size requirements. Phys. Ther. 2005, 85, 257–268. [Google Scholar] [CrossRef] [Green Version]
- Sokal, R.R.; Rohlf, F.J. Biometry: The principles and practice of statistics. In Biological Research, 3rd ed.; Freeman: New York, NY, USA, 1995; pp. 445–447. [Google Scholar]
- Dytham, C. Choosing and Using Statistics: A Biologist’s Guide, 3rd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2010; pp. 145–150. [Google Scholar]
- Cummings, A.M.; Yochim, J.M. Differentiation of the uterus in preparation for gestation: A model for the action of progesterone. J. Ther. Biol. 1984, 106, 353–374. [Google Scholar] [CrossRef]
- Talbi, S.; Hamilton, A.E.; Vo, K.C.; Tulac, S.; Overgaard, M.T.; Dosiou, C.; Le Shay, N.; Nezhat, C.N.; Kempson, R.; Lessey, B.A.; et al. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology 2006, 147, 1097–1121. [Google Scholar] [CrossRef] [Green Version]
- Strowitzki, T.; Germeyer, A.; Popovici, R.; von Wolff, A. The human endometrium as a fertility-determining factor. Hum. Reprod. Update 2006, 12, 617–630. [Google Scholar] [CrossRef] [Green Version]
- Greening, D.W.; Nguyen, H.P.; Evans, J.; Simpson, R.J.; Salamonsen, L.A. Modulating the endometrial epithelial proteome and secretome in preparation for pregnancy: The role of ovarian steroid and pregnancy hormones. J. Proteom. 2016, 144, 99–112. [Google Scholar] [CrossRef]
- Edwards, R.G. Conception in the Human Female; Academic Press: London, UK, 1980; pp. 767–826. [Google Scholar]
- Daza-Vamenta, R.; Glusman, G.; Rowen, L.; Guthrie, B.; Geraghty, D.E. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004, 14, 1501–1515. [Google Scholar] [CrossRef] [Green Version]
- Maccari, G.; Robinson, J.; Bontrop, R.E.; Otting, N.; Natasja, G.; de Groot, N.G.; Ho, C.S.; Ballingall, K.T.; Marsh, S.G.E.; Hammond, J.A. IPD-MHC: Nomenclature requirements for the non-human major histocompatibility complex in the next-generation sequencing era. Immunogenetics 2018, 70, 619–623. [Google Scholar] [CrossRef] [Green Version]
- Robertson, S.A.; Moldenhauer, L.M. Immunological determinants of implantation success. Int. J. Dev. Biol. 2014, 58, 205–217. [Google Scholar] [CrossRef] [Green Version]
- PrabhuDas, M.; Bonney, E.; Caron, K.; Caron, K.; Dey, S.; Erlebacher, A.; Fazleabas, A.; Fisher, S.; Golos, T.; Matzuk, M.; et al. Immune mechanisms at the maternal-fetal interface: Perspectives and challenges. Nat. Immunol. 2015, 16, 328–334. [Google Scholar] [CrossRef]
- Enders, A.C.; Hendrickx, A.G. Implantation in non-human primates-I. In Non-Human Primate Models for Study of Human Reproduction; Anand Kumar, T.C., Ed.; Karger: Basel, Switzerland, 1980; pp. 99–108. [Google Scholar]
- Ghosh, D.; Sengupta, J. Delineating the prime mover action of progesterone for endometrial receptivity in primates. Indian J Med Res. 2014, 140, S130–S136. [Google Scholar]
- Croxatto, B.H.; Diaz, S.; Fuentealba, B.; Croxatto, H.D.; Carrillo, D.; Fabres, C. Studies on the duration of egg transport in the human oviduct. I. The time interval between ovulation and egg recovery from the uterus in normal women. Fertil. Steril. 1972, 27, 447–458. [Google Scholar] [CrossRef]
- Bromfield, J.J. Seminal fluid and reproduction: Much more than previously thought. J. Assist. Reprod. Genet. 2014, 31, 627–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nederlof, I.; Meulemana, T.; van der Hoorn, M.L.P.; Claas, F.H.J.; Eikmans, M. The seed to success: The role of seminal plasma in pregnancy. J. Reprod. Immunol. 2017, 123, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Martinez, C.A.; Cambra, J.M.; Parrilla, I.; Roca, J.; Ferreira-Dias, G.; Pallares, F.J.; Lucas, X.; Vazquez, J.M.; Martinez, E.A.; Gil, M.A.; et al. Seminal plasma modifies the transcriptional pattern of the endometrium and advances embryo development in pigs. Front. Vet. Sci. 2019, 6, 465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, H.L.; Watkins, A.J. The influence of seminal plasma on offspring development and health. Semin. Cell Dev. Biol. 2020, 97, 131–137. [Google Scholar] [CrossRef]
- Mayoral Andrade, G.; Vásquez Martínez, G.; Pérez-Campos Mayoral, L.; Hernández-Huerta, M.T.; Zenteno, E.; Pérez-Campos Mayoral, E.; Martínez Cruz, M.; Martínez Cruz, R.; Matias-Cervantes, C.A.; Meraz Cruz, N.; et al. Molecules and prostaglandins related to embryo tolerance. Front. Immunol. 2020, 11, 555414. [Google Scholar] [CrossRef]
- Mateo-Otero, Y.; Sánchez, J.M.; Recuero, S.; Bagés-Arnal, S.; McDonald, M.; Kenny, D.A.; Yeste, M.; Lonerga, P.; Fernandez-Fuertes, B. Effect of exposure to seminal plasma through natural mating in cattle on conceptus length and gene expression. Front. Cell Dev. Biol. 2020, 8, 341. [Google Scholar] [CrossRef]
- Mueller, D.L. Mechanisms maintaining peripheral tolerance. Nat Immunol. 2010, 11, 21–27. [Google Scholar] [CrossRef]
- Parham, P.; Moffett, A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat. Rev. Immunol. 2013, 13, 133–144. [Google Scholar] [CrossRef] [Green Version]
- De Groot, N.; Blokhuis, J.H.; Doxiadis, G.G.M.; Bontrop, R.E. Co-evolution of the MHC class I and KIR gene families in rhesus macaques: Ancestry and plasticity. Immunol. Rev. 2015, 267, 228–245. [Google Scholar] [CrossRef]
- Tanaka, N.; Miyazaki, K.; Tashiro, H.; Mizutani, H.; Okamura, H. Changes in adenylyl cyclase activity in human endometrium during the menstrual cycle and in human decidua during pregnancy. J. Reprod. Fertil. 1993, 98, 33–39. [Google Scholar] [CrossRef]
- Jones, R.L.; Stoikos, C.; Findlay, J.K.; Salamonsen, L.A. TGF-beta superfamily expression and actions in the endometrium and placenta. Reproduction 2006, 132, 217–232. [Google Scholar] [CrossRef]
- Kuokkanen, S.; Chen, B.; Ojalvo, L.; Benard, L.; Santoro, N.; Pollard, J.W. Genomic profiling of microRNAs and messenger RNAs reveals hormonal regulation in microRNA expression in human endometrium. Biol. Reprod. 2010, 82, 791–801. [Google Scholar] [CrossRef] [Green Version]
- Sales, K.J.; Grant, V.; Catalano, R.D.; Jabbour, H.N. Chorionic gonadotrophin regulates CXCR4 expression in human endometrium via E-series prostanoid receptor 2 signalling to PI3K-ERK1/2: Implications for fetal-maternal crosstalk for embryo implantation. Mol. Hum. Reprod. 2011, 17, 22–32. [Google Scholar] [CrossRef] [Green Version]
- Emera, D.; Romero, R.; Wagner, G. The evolution of menstruation: A new model for genetic assimilation: Explaining molecular origins of maternal responses to fetal invasiveness. BioEssays 2012, 34, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Altmäe, S.; Martinez-Conejero, J.A.; Esteban, F.J.; Ruiz-Alonso, M.; Stavreus-Evers, A.; Horcajadas, J.A.; Salumets, A. MicroRNAs miR-30b, miR-30d, and miR-494 regulate human endometrial receptivity. Reprod. Sci. 2013, 20, 308–317. [Google Scholar] [CrossRef] [Green Version]
- Kusama, K.; Yoshie, M.; Tamura, K.; Kodaka, Y.; Hirata, A.; Sakurai, T.; Bai, H.; Imakawa, K.; Nishi, H.; Isaka, K.; et al. Regulation of decidualization in human endometrial stromal cells through exchange protein directly activated by cyclic AMP (Epac). Placenta 2013, 34, 212–221. [Google Scholar] [CrossRef]
- Aplin, J.D.; Ruane, P.T. Embryo-epithelium interactions during implantation at a glance. J. Cell Sci. 2017, 130, 15–22. [Google Scholar] [CrossRef] [Green Version]
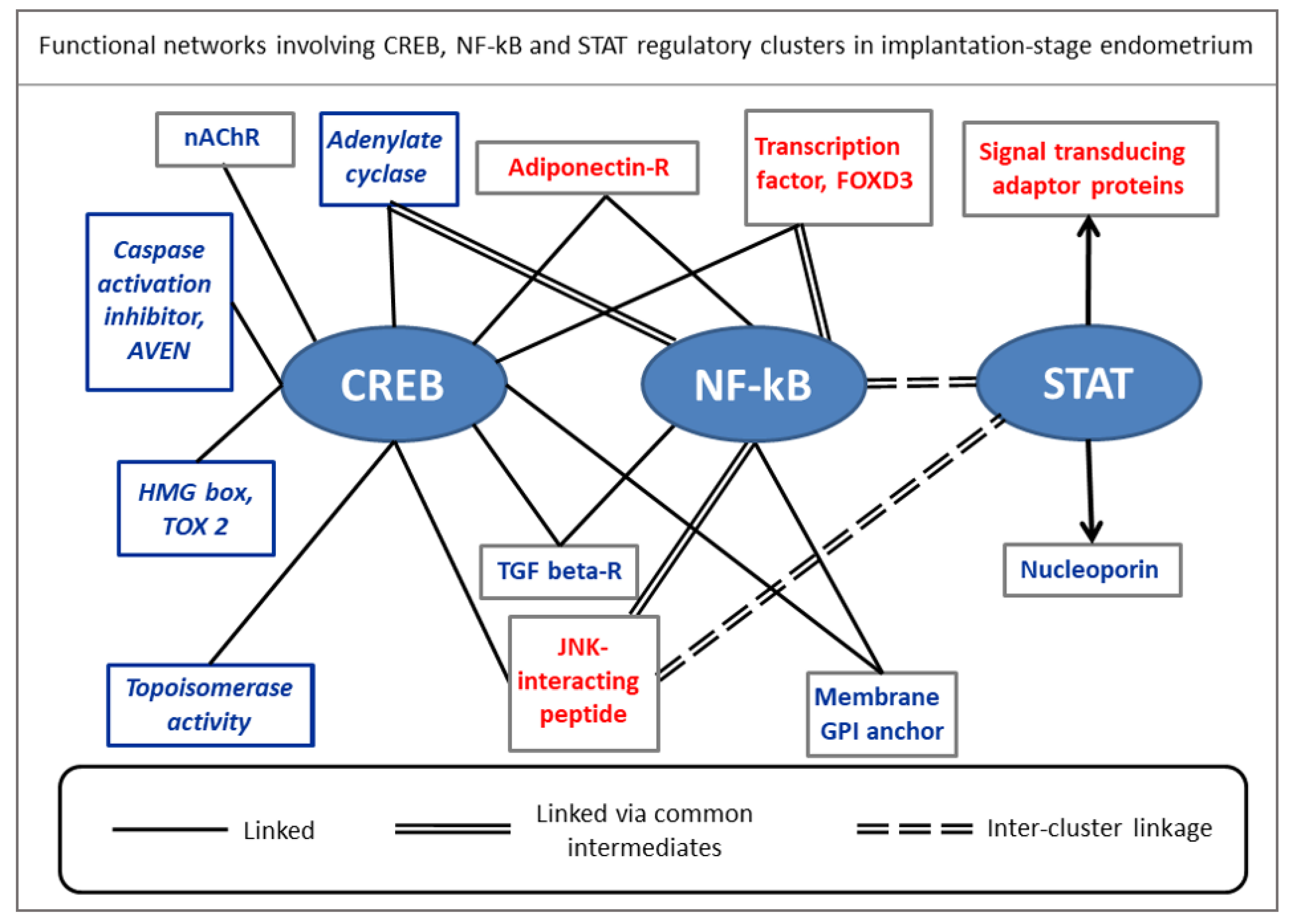
- Zanin, M. Simplifying functional network representation and interpretation through causality clustering. Sci. Rep. 2021, 11, 15378. [Google Scholar] [CrossRef]
- Page, M.; Tuckerman, E.M.; Li, T.C.; Laird, S.M. Expression of nuclear factor kappa B components in human endometrium. J. Reprod. Immunol. 2002, 54, 1–13. [Google Scholar] [CrossRef]
- Ersahin, A.; Acet, M.; Acet, T.; Yavuz, Y. Disturbed endometrial NF-kB expression in women with recurrent implantation failure. Eur. Rev. Med. Pharm. Sci. 2016, 20, 5037–5040. [Google Scholar]
- Sakowicz, A. The role of NFκB in the three stages of pregnancy—Implantation, maintenance, and labour: A review article. BJOG 2018, 125, 1379–1387. [Google Scholar] [CrossRef] [Green Version]
- Massimiani, M.; Lacconi, V.; Civita, F.L.; Ticconi, C.; Rago, R.; Campagnolo, L. Molecular signaling regulating endometrium–blastocyst crosstalk. Int. J. Mol. Sci. 2019, 21, 23. [Google Scholar] [CrossRef] [Green Version]
- Mrozikiewicz, A.E.; Ozarowski, M.; Jedrzejczak, P. Biomolecular markers of recurrent implantation failure—A review. Int. J. Mol. Sci. 2021, 22, 10082. [Google Scholar] [CrossRef]
- Shen-Orr, S.S.; Gaujoux, R. Computational deconvolution: Extracting cell type-specific information from heterogeneous samples. Curr. Opin. Immunol. 2013, 25, 571–578. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Breschi, A.; Muñoz-Aguirre, M.; Wucher, V.; Davis, C.A.; Garrido-Martín, D.; Djebali, S.; Gillis, J.; Pervouchine, D.D.; Vlasova, A.; Dobin, A.; et al. A limited set of transcriptional programs define major cell types. Genom. Res. 2020, 30, 1047–1059. [Google Scholar] [CrossRef]
- Gellersen, B.; Brosens, J.J. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr. Rev. 2014, 35, 851–905. [Google Scholar] [CrossRef]
- Thandapani, P.; O’Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG motif. Mol. Cell. 2013, 50, 613–623. [Google Scholar] [CrossRef] [Green Version]
- Stokes, C.; Treinin, M.; Papke, R.L. Looking below the surface of nicotinic acetylcholine receptors. Trends Pharm. Sci. 2015, 36, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Saez, J.C.; Berthoud, V.M.; Branes, M.C.; Martinez, A.D.; Beyer, E.C. Plasma membrane channels formed by connexins: Their regulation and functions. Physiol. Rev. 2003, 83, 1359–1400. [Google Scholar] [CrossRef] [Green Version]
- Krishnakumar, R.; Chen, A.F.; Pantovich, M.G.; Danial, M.; Parchem, R.J.; Labosky, P.A.; Blelloch, R. FOXD3 regulates pluripotent stem cell potential by simultaneously initiating and repressing enhancer activity. Cell Stem Cell. 2016, 18, 104–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, J.; Mlodzik, M. Planar cell polarity signaling: Coordination of cellular orientation across tissues. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 479–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulanovskaya, O.A.; Zuhl, A.M.; Cravatt, B.F. NNMT promotes epigenetic remodelling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 2013, 9, 300–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vong, Q.P.; Leung, W.H.; Houston, J.; Li, Y.; Rooney, B.; Holladay, M.; Oostendorp, R.A.; Leung, W. TOX2 regulates human natural killer cell development by controlling T-BET expression. Blood 2014, 124, 3905–3913. [Google Scholar] [CrossRef]
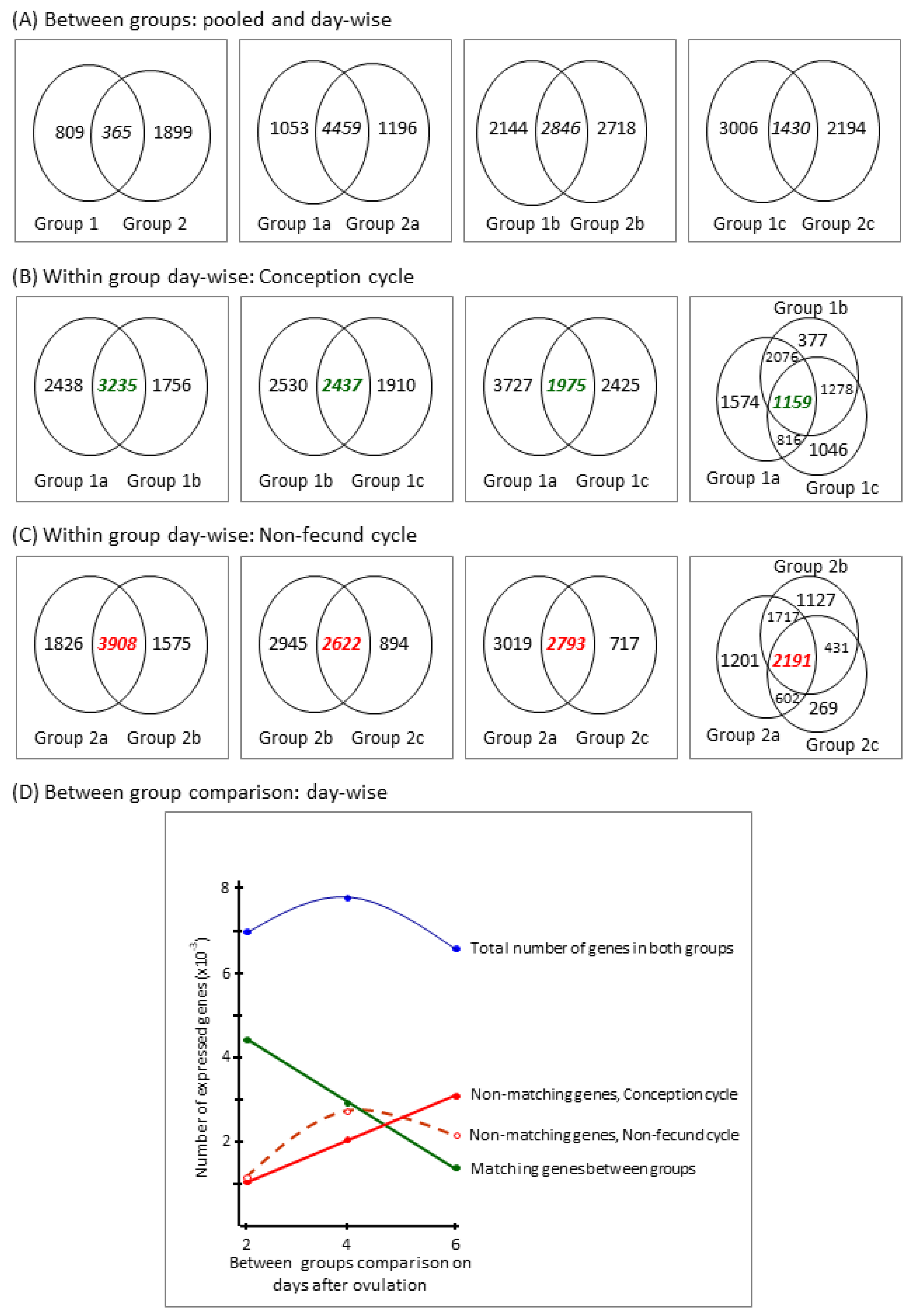


| Group a (Subgroup) | Day after Ovulation b | Number of | ||||
|---|---|---|---|---|---|---|
| Animals Recruited | Ovulated | RNA Samples Obtained/Extracted c | Used in Array/Qrtpcr d | Tissue Samples Used Immunohistochemistry e | ||
| 1(A) | 2 | 12 | 10 | 5 | 4 | 4 |
| 1(B) | 4 | 12 | 11 | 5 | 4 | 4 |
| 1(C) | 6 | 11 | 10 | 5 | 4 | 4 |
| Total | 35 | 31 | 15 | 12 | 12 | |
| 2(A) | 2 | 12 | 10 | 6 | 4 | 4 |
| 2(B) | 4 | 12 | 10 | 5 | 4 | 4 |
| 2(C) | 6 | 12 | 10 | 5 | 4 | 4 |
| Total | 36 | 30 | 16 | 12 | 12 | |
| 1. GEO accession number | GSE108760 | ||
| 2. Microarray details | |||
| Product name: | Rhesus (V2) Macaque gene expression microarray | ||
| Product format per slide: | 4 × 44 K; 60-mer printed oligonucleotides | ||
| Total number of probes/chip: | 45,018 | ||
| Total number of genes/chip: | 17,987 | ||
| Content source: | RefSeq Release 37, UCSC mRNA, | ||
| UniGene Build 13,ENSEMBL Release | |||
| 56, rheMac2 | |||
| 3. Descriptive details of microarray data | |||
| Total number of hybridized a | |||
| Probes: | 20,277 | ||
| Genes: | 12,489 | ||
| 4. Details of hybridized probes (genes), Group-wise | |||
| Group details | Number of samples arrayed | Quantitative description of hybridized a probes (genes) | |
| Estimate b | Per cent | ||
| Group 1(a) | 4 | 11,218 ± 376 (7922 ± 214) | 55.3 (63.4) |
| Group 1(b) | 4 | 9574 ± 372 (7645 ± 85) | 47.2 (61.2) |
| Group 1(c) | 4 | 12,603 ± 592 (7394 ± 216) | 62.2 (59.2) |
| Group 2(a) | 4 | 11,386 ± 337 (8805 ± 261) | 56.2 (70.5) |
| Group 2(b) | 4 | 11,230 ± 462 (8294 ± 298) | 55.4 (66.4) |
| Group 2(c) | 4 | 11,684 ± 334 (8374 ± 260) | 57.6 (67.0) |
| Serial Number | Gene Name (Gene Symbol) | Fold Change Based on (pFDR) | |
|---|---|---|---|
| Expression Arrays | RT-PCR | ||
| Between Day 6 and Day 2 after ovulation in conception cycle group (Group 1c vs. Group 1a) | |||
| Up-regulated | |||
| 1. | Cholinergic receptor, nicotinic, delta (CHRND) | 18.68 (0.046) | 3.80 (0.011) |
| 2. | Forkhead box D3 (FOXD3) | 5.91 (0.024) | 5.76 (0.006) |
| 3. | Gap Junction Protein, delta 4, 40.1 kDa (GJD4) | 3.79 (0.019) | 26.29 (0.006) |
| 4. | Mitogen-activated protein kinase 8 interacting protein 3 (MAPK8IP3) | 5.39 (0.020) | 3.99 (0.014) |
| 5. | Meckel syndrome, type 1 (MKS1) | 6.29 (0.023) | 3.98 (0.029) |
| 6. | Nucleoporin 50 kDa (NUP50) | 2.18 (0.021) | 2.39 (0.011) |
| Down-regulated | |||
| 1. | Adenylate cyclase 5 (ADCY5) | 5.95 (0.028) | 4.13 (0.026) |
| 2. | Adiponectin receptor 1 (ADIPOR1) | 4.45 (0.018) | 2.12 (0.030) |
| 3. | Nicotinamide N-methyltransferase (NNMT) | 16.77 (0.046) | 3.08 (0.018) |
| 4. | Protein associated with topoisomerase II (PATL1) | 2.01 (0.001) | 2.65 (0.012) |
| 5. | Phosphatidylinositol glycan anchor biosynthesis, class V (PIGV) | 2.01 (0.003) | 3.86 (0.007) |
| 6. | Transforming growth factor, beta receptor II (TGFBR2) | 3.24 (0.009) | 2.16 (0.013) |
| 7. | TOX high mobility group box family member 2 (TOX2) | 18.92 (0.015) | 13.85 (0.015) |
| 8. | von Willebrand factor A domain containing 5B1 (VWA5B1) | 10.58 (0.003) | 19.48 (0.001) |
| Between conception cycle and non-fecund cycle groups on Day 6 after ovulation (Group 1c vs. Group 2c) | |||
| Up-regulated | |||
| 1. | Apoptosis, caspase activation inhibitor (AVEN) | 2.02 (0.011) | 17.20 (0.012) |
| Down-regulated | |||
| 1. | Adenylate cyclase 5 (ADCY5) | 6.14 (0.011) | 18.00 (0.007) |
| 2. | Nicotinamide N-methyltransferase (NNMT) | 18.02 (0.015) | 4.74 (0.002) |
| 3. | TOX high mobility group box family member 2 (TOX2) | 20.54 (0.039) | 2.10 (0.012) |
| 4. | von Willebrand factor A domain containing 5B1 (VWA5B1) | 11.27 (0.036) | 52.00 (0.001) |
| Group Number (Group Description)(Number of Genes Group Specifically Expressed) | GO Process (p-Value) | Number of Network Objects | |
|---|---|---|---|
| Total | Input Data | ||
| Group 1 (Conception cycle) [809] | Positive regulation of tolerance induction to non-self antigen (2.4 × 10−31) | 25 | 23 a |
| Group 2 (Non-fecund cycle) [1899] | Metabolic process 5.6 × 10−55 | 11,851 | 1275 a |
| Groups 1 and 2 [370] | Protein targeting to membrane (1.1 × 10−21)/and endoplasmic reticulum (3.7 × 10−21) | 109/114 | 25 a |
| Metabolic process 4.0 × 10−21 | 155 | 15 a | |
| Marker Proteins (Alias) | Physiological Functions | Reference |
|---|---|---|
| Adenylate cyclase 5 (ADCY5) | Class III adenylate cyclase gene family expressed as transmembrane protein catalyzing the formation of cAMP in response to G-protein signaling. Required for stromal cell decidualization in mammals. | Gellersen and Brosens [83] |
| Apoptosis, caspase activation (AVEN) | Involved in cell fate regulation, stabilizes pro-survival protein BclxL, inhibits pro-apoptosis protein Apaf-1 function, exhibits cell cycle regulatory function. | Thandapani et al. [84] |
| Cholinergic receptor nicotinic, delta (CHRND) | Family of pentameric acetylcholine-gated ion channels expressed in both neuronal, non-neuronal cells. Mediates signal transduction independent of generating ion currents in non-neuronal cells. Suppresses pro-inflammatory cytokines in the cholinergic anti-inflammatory pathway (CAP) in epithelial and endothelial cells. | Stokes et al. [85] |
| Connexin 40.1 (GJD4) | Involved in formation of gap junctional intercellular of communication (GJIC) for direct intercellular transfer ions and small molecules, cAMP, and cGMP and propagation of inositol trisphosphate-dependent calcium waves, involved in growth control, differentiation, and apoptosis of normal cells. | Saez et al. [86] |
| Foxhead box D3 (FOXD3) | Tumor suppressor. Member of forkhead box (FOX) family of transcription factors bearing helix-turn-helix DNA binding motif that interacts with DNA consensus sequences for nuclear localization and transcriptional regulation. Required for maintenance of cellular pluripotency and epigenetic priming of enhancers. | Krishnakumar et al. [87] |
| Meckel syndrome1 (MKS1) | Structural protein required for ciliogenesis. Also involved in planar cell polarity of epithelium and directional cell migration. | Singh and Mlodzik [88] |
| Nicotinamide-N-methyl (NNMT) | Catalyses S-adenosyl-L-methionine (SAM) to generate S-adenosyl-homocysteine (SAH). SAM donates methyl groups to substrates including histones. High NNMT expression results in hypomethylated histone proteins. | Ulanovskaya et al. [89] |
| Transforming growth receptor 2 (TGFBR2) | Mediates pleiotropic functions of TGFb (beta). isoforms 1–3. Involved in implantation stage immune surveillance through functioning of endometrium and placenta including their immuno-modulatory/anti-inflammatory actions. | Jones et al. [64] |
| TOX high mobility group, box family member 2 (TOX2) | Transcription factor belonging to TOX family domain, involved in differentiation and maturation of different cell types. | Vong et al. [90] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhat, M.A.; Khan, M.A.; Lalitkumar, P.G.L.; Poorasamy, J.; Sengupta, J.; Ghosh, D. Preimplantation Endometrial Transcriptomics in Natural Conception Cycle of the Rhesus Monkey. Reprod. Med. 2022, 3, 16-35. https://doi.org/10.3390/reprodmed3010003
Bhat MA, Khan MA, Lalitkumar PGL, Poorasamy J, Sengupta J, Ghosh D. Preimplantation Endometrial Transcriptomics in Natural Conception Cycle of the Rhesus Monkey. Reproductive Medicine. 2022; 3(1):16-35. https://doi.org/10.3390/reprodmed3010003
Chicago/Turabian StyleBhat, Muzaffer A., Meraj A. Khan, P.G.L. Lalitkumar, Jeevitha Poorasamy, Jayasree Sengupta, and Debabrata Ghosh. 2022. "Preimplantation Endometrial Transcriptomics in Natural Conception Cycle of the Rhesus Monkey" Reproductive Medicine 3, no. 1: 16-35. https://doi.org/10.3390/reprodmed3010003






