Mapping m6A Sites on HIV-1 RNA Using Oligonucleotide LC-MS/MS
Abstract
:1. Introduction
2. Materials and Methods
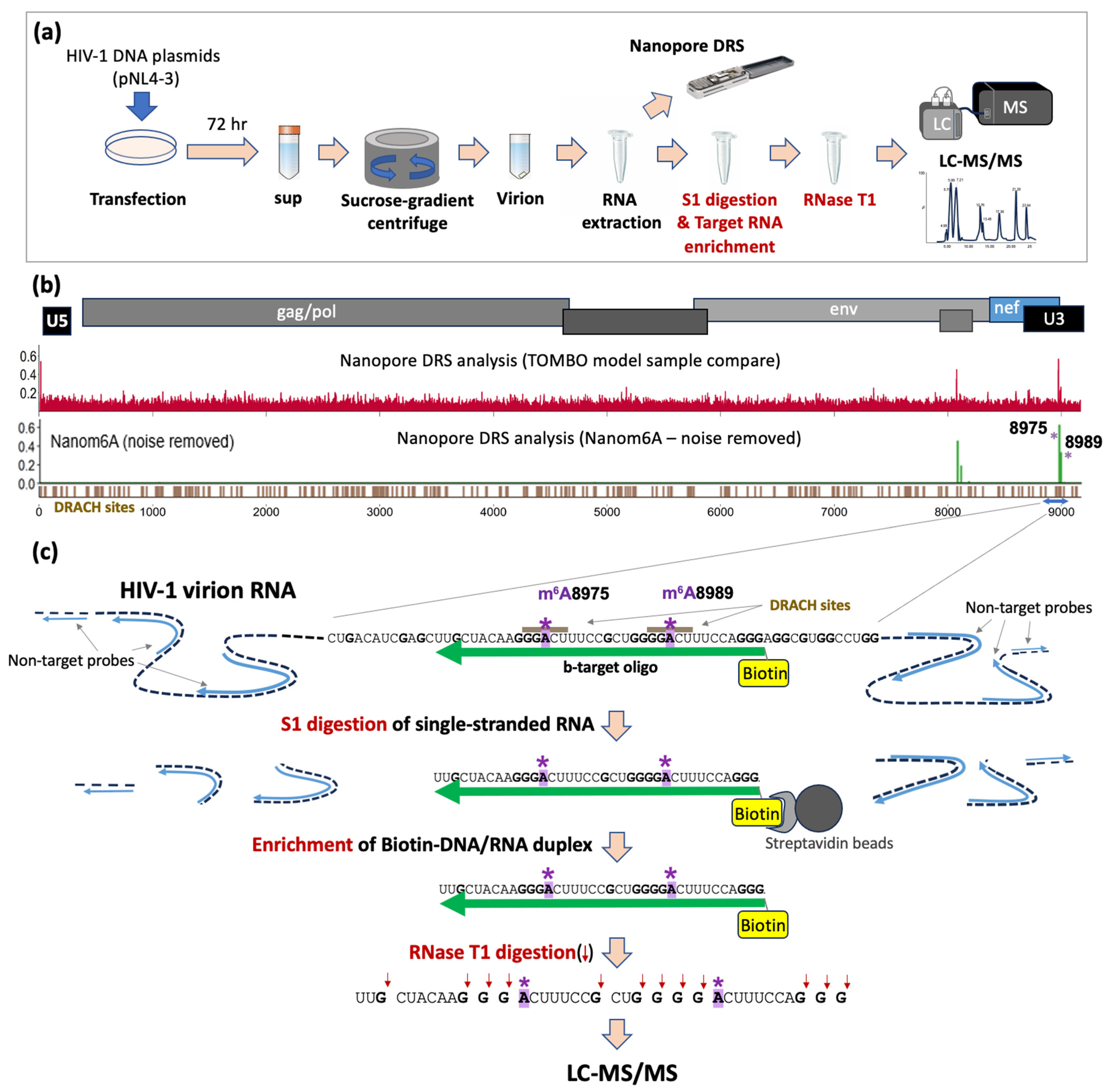
2.1. HIV-1 Viral RNA Extraction
2.2. HIV-1-Specific Oligos
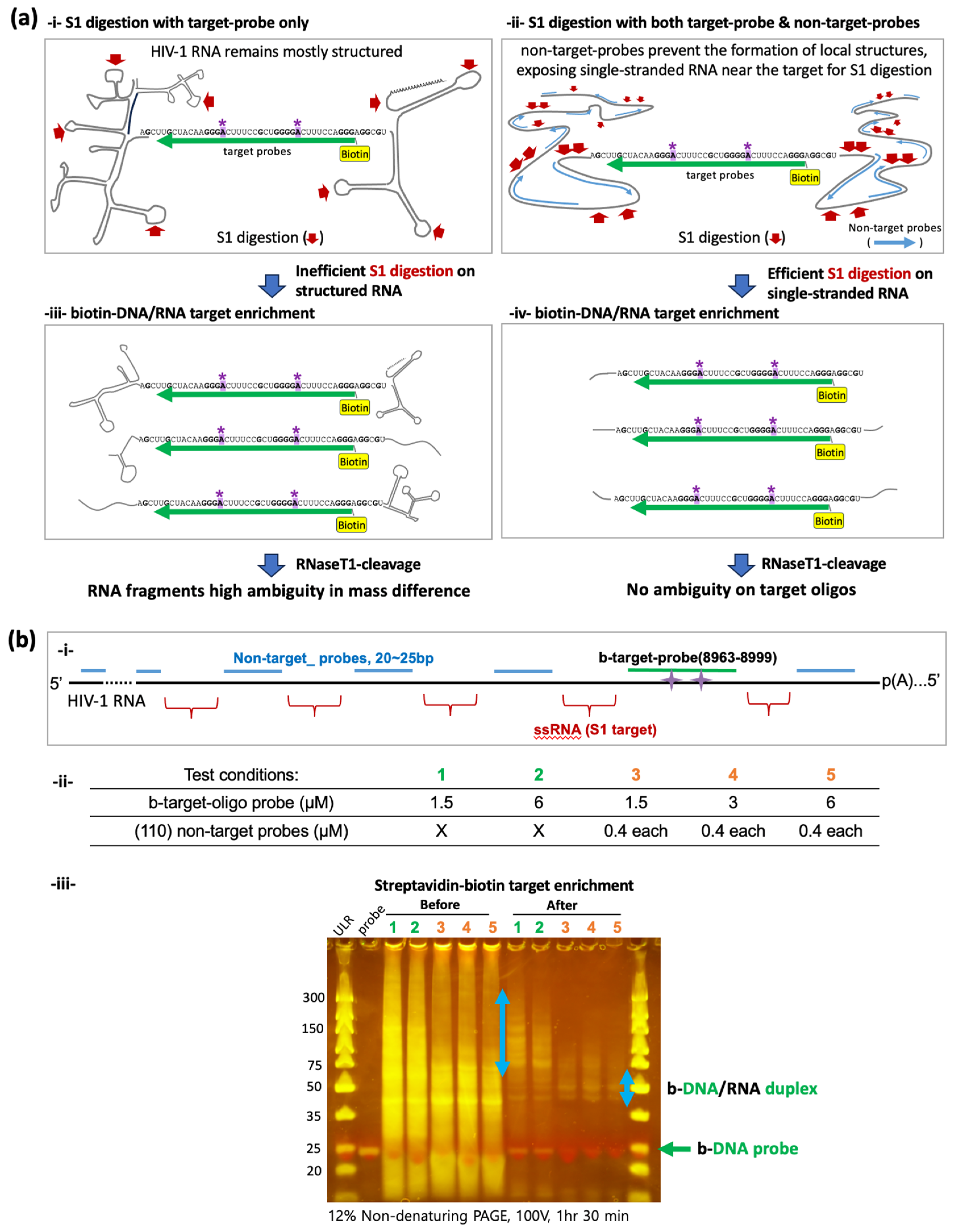
2.3. S1 Nuclease Digestion and the Recovery of Biotinylated-DNA/RNA Duplex
2.4. Oligonucleotide LC-MS/MS of HIV-1 RNA Fragment
2.5. Synthetic RNA Control
2.6. LC-MS/MS Data Processing
3. Results
3.1. Identification of Potential Site-Specific m6A Modifications on HIV-1 Viral RNA
3.2. Enriching Target RNA Fragments of HIV-1 RNA for Oligonucleotide LC-MS/MS
3.3. Enhanced Enrichment of More Homogeneous Target RNA Fragments Using Non-Target Probes
3.4. Oligonucleotide LC-MS/MS of Synthetic RNA Oligos with m6As at A8975 or A8989
3.5. Oligonucleotide LC-MS/MS Confirms the Presence of m6As at A8975 and A8989 of HIV-1 RNA
3.6. Detection of Both Methylated and Unmethylated Versions of the Same Oligomer Indicates Partial Methylation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murakami, S.; Jaffrey, S.R. Hidden codes in mRNA: Control of gene expression by m6A. Mol. Cell 2022, 82, 2236–2251. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2021, 50, D231–D235. [Google Scholar] [CrossRef] [PubMed]
- Abebe, J.S.; Verstraten, R.; Depledge, D.P. Nanopore-Based Detection of Viral RNA Modifications. mBio 2022, 13, e0370221. [Google Scholar] [CrossRef] [PubMed]
- Davis, F.F.; Allen, F.W. Ribonucleic acids from yeast which contain a fifth nucleotide. J. Biol. Chem. 1957, 227, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, L.; Li, X. Detection technologies for RNA modifications. Exp. Mol. Med. 2022, 54, 1601–1616. [Google Scholar] [CrossRef]
- Helm, M.; Motorin, Y. Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 2017, 18, 275–291. [Google Scholar] [CrossRef]
- Alfonzo, J.D.; Brown, J.A.; Byers, P.H.; Cheung, V.G.; Maraia, R.J.; Ross, R.L. A call for direct sequencing of full-length RNAs to identify all modifications. Nat. Genet. 2021, 53, 1113–1116. [Google Scholar] [CrossRef]
- Baquero-Perez, B.; Geers, D.; Diez, J. From A to m6A: The Emerging Viral Epitranscriptome. Viruses 2021, 13, 1049. [Google Scholar] [CrossRef]
- Courtney, D.G.; Tsai, K.; Bogerd, H.P.; Kennedy, E.M.; Law, B.A.; Emery, A.; Swanstrom, R.; Holley, C.L.; Cullen, B.R. Epitranscriptomic Addition of m(5)C to HIV-1 Transcripts Regulates Viral Gene Expression. Cell Host Microbe 2019, 26, 217–227.e6. [Google Scholar] [CrossRef]
- Tsai, K.; Vasudevan, A.A.J.; Campos, C.M.; Emery, A.; Swanstrom, R.; Cullen, B.R. Acetylation of Cytidine Residues Boosts HIV-1 Gene Expression by Increasing Viral RNA Stability. Cell Host Microbe 2020, 28, 306–312.e6. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.M.; Bogerd, H.P.; Kornepati, A.V.R.; Kang, D.; Ghoshal, D.; Marshall, J.B.; Poling, B.C.; Tsai, K.; Gokhale, N.S.; Horner, S.M.; et al. Posttranscriptional m6A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe 2016, 19, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Ringeard, M.; Marchand, V.; Decroly, E.; Motorin, Y.; Bennasser, Y. FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 2019, 565, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Lichinchi, G.; Gao, S.; Saletore, Y.; Gonzalez, G.M.; Bansal, V.; Wang, Y.; Mason, C.E.; Rana, T.M. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016, 1, 16011. [Google Scholar] [CrossRef] [PubMed]
- Tirumuru, N.; Zhao, B.S.; Lu, W.; Lu, Z.; He, C.; Wu, L. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 2016, 5, e15528. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- König, J.; Zarnack, K.; Rot, G.; Curk, T.; Kayikci, M.; Zupan, B.; Turner, D.J.; Luscombe, N.M.; Ule, J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010, 17, 909–915. [Google Scholar] [CrossRef]
- Llibre, J.M.; Pulido, F.; García, F.; Deltoro, M.G.; Blanco, J.L.; Delgado, R. Genetic barrier to resistance for dolutegravir. AIDS Rev. 2015, 17, 56–64. [Google Scholar]
- Sun, L.; Fazal, F.M.; Li, P.; Broughton, J.P.; Lee, B.; Tang, L.; Huang, W.; Kool, E.T.; Chang, H.Y.; Zhang, Q.C. RNA structure maps across mammalian cellular compartments. Nat. Struct. Mol. Biol. 2019, 26, 322–330. [Google Scholar] [CrossRef]
- Wu, B.; Su, S.; Patil, D.P.; Liu, H.; Gan, J.; Jaffrey, S.R. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018, 9, 420. [Google Scholar] [CrossRef] [PubMed]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol. Cell 2019, 74, 494–507.e8. [Google Scholar] [CrossRef] [PubMed]
- Gaston, K.W.; Limbach, P.A. The identification and characterization of non-coding and coding RNAs and their modified nucleosides by mass spectrometry. RNA Biol. 2014, 11, 1568–1585. [Google Scholar] [CrossRef] [PubMed]
- Thakur, P.; Estevez, M.; Lobue, P.A.; Limbach, P.A.; Addepalli, B. Improved RNA modification mapping of cellular non-coding RNAs using C- and U-specific RNases. Analyst 2020, 145, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Jora, M.; Borland, K.; Abernathy, S.; Zhao, R.; Kelley, M.; Kellner, S.; Addepalli, B.; Limbach, P.A. Chemical Amination/Imination of Carbonothiolated Nucleosides during RNA Hydrolysis. Angew. Chem. Int. Ed. 2020, 60, 3961–3966. [Google Scholar] [CrossRef] [PubMed]
- Addepalli, B.; Lesner, N.P.; Limbach, P.A. Detection of RNA nucleoside modifications with the uridine-specific ribonuclease MC1 from Momordica charantia. RNA 2015, 21, 1746–1756. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, W.; Netzband, R.; Bonenfant, G.; Biegel, J.M.; Miller, C.; Fuchs, G.; Henderson, E.; Arra, M.; Canki, M.; Fabris, D.; et al. Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 2018, 46, 5776–5791. [Google Scholar] [CrossRef]
- Pereira-Montecinos, C.; Toro-Ascuy, D.; Ananías-Sáez, C.; Gaete-Argel, A.; Rojas-Fuentes, C.; Riquelme-Barrios, S.; Rojas-Araya, B.; García-De-Gracia, F.; Aguilera-Cortés, P.; Chnaiderman, J.; et al. Epitranscriptomic regulation of HIV-1 full-length RNA packaging. Nucleic Acids Res. 2022, 50, 2302–2318. [Google Scholar] [CrossRef]
- Konan, S.N.; Ségéral, E.; Bejjani, F.; Bendoumou, M.; Said, M.A.; Gallois-Montbrun, S. YTHDC1 regulates distinct post-integration steps of HIV-1 replication and is important for viral infectivity. Retrovirology 2022, 19, 4. [Google Scholar] [CrossRef]
- Tsai, K.; Bogerd, H.P.; Kennedy, E.M.; Emery, A.; Swanstrom, R.; Cullen, B.R. Epitranscriptomic addition of m6A regulates HIV-1 RNA stability and alternative splicing. Minerva Anestesiol. 2021, 35, 992–1004. [Google Scholar] [CrossRef]
- Lu, W.; Tirumuru, N.; Gelais, C.S.; Koneru, P.C.; Liu, C.; Kvaratskhelia, M.; He, C.; Wu, L. N6-Methyladenosine–binding proteins suppress HIV-1 infectivity and viral production. J. Biol. Chem. 2018, 293, 12992–13005. [Google Scholar] [CrossRef] [PubMed]
- D’ascenzo, L.; Popova, A.M.; Abernathy, S.; Sheng, K.; Limbach, P.A.; Williamson, J.R. Pytheas: A software package for the automated analysis of RNA sequences and modifications via tandem mass spectrometry. Nat. Commun. 2022, 13, 2424. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, C.G.; Crain, P.F.; Gupta, R.; Hashizume, T.; Hocart, C.H.; A Kowalak, J.; Pomerantz, S.C.; Stetter, K.O.; McCloskey, J.A. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria). J. Bacteriol. 1991, 173, 3138–3148. [Google Scholar] [CrossRef] [PubMed]
- Kowalak, J.A.; Dalluge, J.J.; McCloskey, J.A.; Stetter, K.O. The Role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 1994, 33, 7869–7876. [Google Scholar] [CrossRef] [PubMed]
- Bakin, A.; Kowalak, J.A.; McCloskey, J.A.; Ofengand, J. The single pseudouridine residue inEscherichia coli16S RNA is located at position 516. Nucleic Acids Res. 1994, 22, 3681–3684. [Google Scholar] [CrossRef]
- A Kowalak, J.; A McCloskey, J.; O Stetter, K.; Crain, P.F.; Bruenger, E.; Kowalak, J.A.; Kuchino, Y.; McCloskey, J.A.; Mizushima, H.; Stetter, K.O.; et al. 5S rRNA modification in the hyperthermophilic archaea Sulfolobus solfataricus and Pyrodictium occultum. FASEB J. 1993, 7, 196–200. [Google Scholar] [CrossRef]
- Kowalak, J.A.; Bruenger, E.; Hashizume, T.; Peltier, J.M.; Ofengand, J.; McCloskey, J.A. Structural characterization of U*-1915 in domain IV from Escherichia coli 23S ribosomal RNA as 3-methylpseudouridine. Nucleic Acids Res. 1996, 24, 688–693. [Google Scholar] [CrossRef]
- Kowalak, J.A.; Bruenger, E.; McCloskey, J.A. Posttranscriptional modification of the central loop of domain V in Escherichia coli 23 S Ribosomal RNA. J. Biol. Chem. 1995, 270, 17758–17764. [Google Scholar] [CrossRef]
- Phillips, S.; Baek, A.; Kim, S.; Chen, S.; Wu, L. Protocol for the generation of HIV-1 genomic RNA with altered levels of N (6)-methyladenosine. STAR Protoc. 2022, 3, 101616. [Google Scholar] [CrossRef]
- Alice Baek, G.-E.L.; Golconda, S.; Rayhan, A.; Manganaris, A.A.; Chen, S.; Tirumuru, N.; Yu, H.; Kim, S.; Kimmel, C.; Zablocki, O.; et al. Single-RNA-Level Analysis of Full-Length HIV-1 RNAs Reveals Functional Redundancy of m6As. Preprint (Version 1) Research Square. 2023. Available online: https://assets.researchsquare.com/files/rs-2679540/v1/47af6f79-e742-4fdc-8ed6-77e46ea8cfd8.pdf?c=1686941532 (accessed on 15 December 2023).
- Sambrook, J.; Russell, D.W. Purification of nucleic acids by extraction with phenol:chloroform. Cold Spring Harb. Protoc. 2006, 2006, pdb.prot4455. [Google Scholar] [CrossRef]
- Tirumuru, N.; Wu, L. HIV-1 envelope proteins up-regulate N6-methyladenosine levels of cellular RNA independently of viral replication. J. Biol. Chem. 2019, 294, 3249–3260. [Google Scholar] [CrossRef] [PubMed]
- Hendra, C.; Pratanwanich, P.N.; Wan, Y.K.; Goh, W.S.S.; Thiery, A.; Göke, J. Detection of m6A from direct RNA sequencing using a multiple instance learning framework. Nat. Methods 2022, 19, 1590–1598. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, X.; Wu, B.; Wang, H.; Xi, F.; Kohnen, M.V.; Reddy, A.S.N.; Gu, L. Quantitative profiling of N6-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using Nanopore direct RNA sequencing. Genome Biol. 2021, 22, 22. [Google Scholar] [CrossRef] [PubMed]
- Tomezsko, P.J.; Corbin, V.D.A.; Gupta, P.; Swaminathan, H.; Glasgow, M.; Persad, S.; Edwards, M.D.; Mcintosh, L.; Papenfuss, A.T.; Emery, A.; et al. Determination of RNA structural diversity and its role in HIV-1 RNA splicing. Nature 2020, 582, 438–442. [Google Scholar] [CrossRef]
- Watts, J.M.; Dang, K.K.; Gorelick, R.J.; Leonard, C.W.; Bess, J.W., Jr.; Swanstrom, R.; Burch, C.L.; Weeks, K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 2009, 460, 711–716. [Google Scholar] [CrossRef]
- Desai, N.A.; Shankar, V. Single-strand-specific nucleases. FEMS Microbiol. Rev. 2003, 26, 457–491. [Google Scholar] [CrossRef]

| m6A Position | Oligomer | Retention Time | Peak Area Counts | Total Area | Relative Ratio of Modified vs. Total |
|---|---|---|---|---|---|
| 8975 | ACUUUCCGp | 23.82 | 434 | 500 | 69/569 = 0.1213 |
| ACUUUCCG>p | 20.12 | 66 | |||
| m6ACUUUCCGp | 27.12 | 49 | 69 | ||
| m6ACUUUCCG>p | 23.91 | 20 | |||
| 8989 | ACUUUCCAGp | 27.36 | 102 | 102 | 174/276 = 0.6304 |
| ACUUUCCAG>p | 0 | 0 | |||
| m6ACUUUCCAGp | 30.22 | 174 | 174 | ||
| m6ACUUUCCAG>p | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, A.; Rayhan, A.; Lee, G.-E.; Golconda, S.; Yu, H.; Kim, S.; Limbach, P.A.; Addepalli, B.; Kim, S. Mapping m6A Sites on HIV-1 RNA Using Oligonucleotide LC-MS/MS. Methods Protoc. 2024, 7, 7. https://doi.org/10.3390/mps7010007
Baek A, Rayhan A, Lee G-E, Golconda S, Yu H, Kim S, Limbach PA, Addepalli B, Kim S. Mapping m6A Sites on HIV-1 RNA Using Oligonucleotide LC-MS/MS. Methods and Protocols. 2024; 7(1):7. https://doi.org/10.3390/mps7010007
Chicago/Turabian StyleBaek, Alice, Asif Rayhan, Ga-Eun Lee, Sarah Golconda, Hannah Yu, Shihyoung Kim, Patrick A. Limbach, Balasubrahmanyam Addepalli, and Sanggu Kim. 2024. "Mapping m6A Sites on HIV-1 RNA Using Oligonucleotide LC-MS/MS" Methods and Protocols 7, no. 1: 7. https://doi.org/10.3390/mps7010007






