Improvement of the Collection, Maintenance, and Analysis of Neoplastic Cells from Urine Specimens with the Use of CytoMatrix
Abstract
:1. Introduction
2. Materials and Reagents
2.1. Materials
- I.
- SuperFrost Plus microscope slides (Thermo Fisher Scientific, Waltham, MA, USA, Cat. no.: 10149870)
- II.
- Square Cover Slip (18 × 18 mm) (Thermo Fisher Scientific, Cat. no.: 18 × 18-1.5)
- III.
- Microscope Cover Slip Menzel (12 × 12 mm) (Thermo Fisher Scientific Menzel, Waltham, MA, USA, Cat. no.: 11708701)
- IV.
- Conical centrifuge tubes 50 mL (Thermo Fisher Scientific, Cat. no.: 10788561)
- V.
- Conical centrifuge tubes 15 mL (Thermo Fisher Scientific, Cat. no.: 10136120)
- VI.
- Eppendorf Safe-Lock Tubes 1.5 mL (Eppendorf, Hamburg, Germany, Cat. no.: 0030120086)
- VII.
- Filter tips for Gilson PIPETMAN (Gilson, city, state abbrev if USA, country, models: P20, P200, P1000)
- VIII.
- SafeCapsule—Blue screwcap container, prefilled with buffer solution (Diapath, Martinengo, Italy, Cat. no.: SC041)
- IX.
- SafeCapsule—Safety red capsule, prefilled with formalin (Diapath, Cat. no.: SC022) to freshly prepare 10% neutral buffered formalin fixative
- X.
- Bond Polymer Refine Detection (Leica Biosystems, 1700 Leider Lane Buffalo Grove, IL, USA, Cat. no.: DS9800)
- XI.
- Primary Antibody Diluent (Leica Biosystems, Cat. no.: AR9352)
- XII.
- BOND Epitope Retrieval Solution 1 (Leica Biosystems, Cat. no.: AR9961)
- XIII.
- Dewax Solution (Leica Biosystems, Cat. no.: AR9222)
- XIV.
- Wash Solution 10×Concentrate (Leica Biosystems, Cat. no.: AR9590)
- XV.
- BOND Open Containers 30 mL (Leica Biosystems, Cat. no.: OP309700)
- XVI.
- BOND Universal Covertile (Leica Biosystems, Cat. no.: S21.2001)
- XVII.
- BOND Slide Tray (Leica Biosystems, United States, Cat. no.: S21.4586.A)
- XVIII.
- BOND™ Ready-to-Use Primary Antibody Ki67 (MM1) (Leica Biosystems, Cat. no.: PA0118)
- XIX.
- BOND™ Ready-to-Use Primary Antibody Cytokeratin 20 (Ks20.8) (Leica Biosystems, Cat. no.: PA0037)
- XX.
- BOND™ Ready-to-Use Primary Antibody p53 (DO-7) (Leica Biosystems, Cat. no.: PA0057)
- XXI.
- Vysis FISH Pretreatment Reagent Kit (Abbott Molecular, Thermo Fisher Scientific, Cat. no.: 02J03-032)
- XXII
- UroVysion Bladder Cancer Kit (Abbott Molecular, Thermo Fisher Scientific, Cat. no.: 02J27-020)
- XXIII.
- ProbeChek UroVysion Bladder Cancer Kit Control Slides (Abbott Molecular, Thermo Fisher Scientific, Cat. no.: 02J27-011)
- XXIV.
- Rubber cement Fixogum 125 mg (Marabu, Tamm, Germany, Cat. no.: 02J27-011Marabu-125)
- XXV.
- CytoFoam Core (Bioptica, Milano, Italy, Cat. no.: CFC1)
- XXVI.
- CytoFoam Disk (Bioptica, Cat. no.: CFD1)
- XXVII.
- CytoMatrix (UCS Diagnostic, Roche Diagnostics SPA, Basel, Switzerland, Cat. no.: nd)
2.2. Equipment
- XXVIII.
- Gilson™ PIPETMAN Classic™ Pipets (Gilson, models: P20, P200, P1000, Cat. no.: F123600, F123601, F123602)
- XXIX.
- Plastic Coplin Staining Jars 40 mm (Thermo Fisher Scientific, Cat. no.: 19-4)
- XXX.
- Glass Coplin Jar holds 5 slides (10 back to back) (Thermo Fisher Scientific, Cat. no.: E94)
- XXXI.
- HistoCore PELORIS 3 Premium Tissue Processing System (Leica Biosystems, model/Cat. no.: 45.0001)
- XXXII.
- Slide microtome for biological applications (Microm, Thermo Fisher Scientific, Cat. no.: model/Cat. no.: HM400R)
- XXXIII.
- Cold Plate (Kaltek, Padova, Italy, Cat. no.: CP 503)
- XXXIV.
- Histological section water bath (Weinkauf Medizintechnik, Hallerndorf, Germany, model/Cat. no.: WBRL 20)
- XXXV.
- Heated Paraffin Embedding Module (Leica Biosystems, model/Cat. no.: EG1150 H)
- XXXVI.
- Fully Automated IHC and ISH Stainer (Leica Biosystems, model/Cat. no.: BOND-III)
- XXXVII.
- Centrifuge Heraeus Primo Biofuge Primo (Thermo Fisher Scientific, Cat. no.: 75005181)
- XXXVIII.
- Water baths (37 °C and 100 °C) (Clifton, Nickel Electro Ltd, Weston-super-Mare, UK, model/Cat. no.: NE2-22D)
- XXXIX.
- Automatic Slide Hybridizer ThermoBrite (TopBrite, Locarno, Switzerland, model/Cat. no.: AS-05010-00)
- XL.
- Optical microscope (Nikon, Tokyo, Japan, model/Cat. no.: Eclipse 50i)
- XLI.
- Fluorescence microscope (Carl Zeiss, Oberkochen, Germany, model/Cat. no.: Axio Imager M1), equipped with appropriate excitation and emission filters allowing visualization of the intense red, green, aqua, and gold fluorescent signals
2.3. Software
- XLII.
- AxionVision 4V, 4.8.2.0 (Carl Zeiss Micro Imaging, cOberkochen, Germany)
- XLIII.
- Imaging Software NIS-Elements F Ver4.60.00 for 64bit edition (Nikon)
- XLIV.
- ImageJ64 software (National Institutes of Health, Bethesda, MD, USA)
3. Procedure
3.1. Patients–Study Cohort
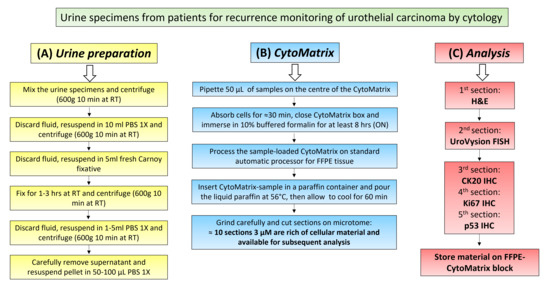
3.2. Urine Sample Preparation
- a
- Prepare Methanol-Carnoy fixative (3:1 mixture of methanol and glacial acetic acid).
- b.
- Prepare PBS Solution 1X, if not available.
- After urine cytology, mix all the remaining urine specimens (stored a 4 °C) of each patient in a 50 mL centrifuge tube.
- Centrifuge to 600 g for 10 min at room temperature (RT) (15–30 °C).
- Discard the fluid, leaving about 1 mL of liquid with cellular pellet on the bottom.
- Resuspend the pellet in the remaining 1 mL.OPTIONAL STEP Pellets from the urine of three consecutive days from the same patient can be mixed.
- Add PBS 1X up to 10 mL to rinse the pellet.
- Centrifuge to 600 g for 10 min at RT.
- Discard the fluid, leaving about 0.5 mL of liquid with cellular pellet on the bottom.
- Add slowly 5 mL of fresh Methanol-Carnoy fixative and mix.
- Leave cell to fix for 1–3 h at RT.
PAUSE STEP: After this step, samples can be stored at −20 °C for the night or for alonger time.
- Centrifuge to 600 g for 10 min at RT.
- Carefully remove the supernatant and resuspend in 1–5 mL of PBS 1X to wash.OPTIONAL STEP: If the cellular pellet is very small and difficult to see, carefully remove the maximum amount of Carnoy, leaving about 50–100 μL of fixative solution to directly pipette on the matrix during CytoMatrix preparation.
- Centrifuge to 600 g for 10 min at RT.
- Carefully remove the supernatant and resuspend in 50–100 µL of PBS 1X.
3.3. CytoMatrix
- Pipette 50 µL of resuspended cellular pellet onto the center of the CytoMatrix (Figure 1A).
 CRITICAL STEP: The deposition of 50 µL of resuspended cellular pellet could lead to the formation of a small drop, which could reduce the absorption of the sample by the matrix. It is suggested to spread the drop over the entire Cytomatrix surface with a needle.
CRITICAL STEP: The deposition of 50 µL of resuspended cellular pellet could lead to the formation of a small drop, which could reduce the absorption of the sample by the matrix. It is suggested to spread the drop over the entire Cytomatrix surface with a needle.- 2.
- Let the cell suspension to absorb for 10–30 min, or until the matrix has completely absorbed the fluid.
- 3.
- Close the biocassette (previously labelled with the patient’s id number) and immerse it in 10% buffered formalin for at least 8 h (Figure 1B).
 CRITICAL STEP: It is important to recognize the side of the matrix on which cells were pipetted; the correct side is indicated by the presence of a frame.
CRITICAL STEP: It is important to recognize the side of the matrix on which cells were pipetted; the correct side is indicated by the presence of a frame.- 4.
- Process the CytoMatrix-sample complex with the biocassette in the automatic tissue processor for FFPE, as any tissue sample (Figure 1C).
 CRITICAL STEP: Processing steps are 70% ethanol 1 h, 95% 1 h, first absolute ethanol 1 h, second absolute ethanol 1 h, third absolute ethanol 1 h, fourth absolute ethanol 2 h, first clearing agent (xylene) 1 h, second xylene 1 h.
CRITICAL STEP: Processing steps are 70% ethanol 1 h, 95% 1 h, first absolute ethanol 1 h, second absolute ethanol 1 h, third absolute ethanol 1 h, fourth absolute ethanol 2 h, first clearing agent (xylene) 1 h, second xylene 1 h.- 5.
- Remove the CytoMatrix-sample from the processor and insert it in the paraffin-block preparation tray for paraffin infiltration (Figure 1D).
 CRITICAL STEP: During this phase, remember to orient the part facing the frame towards the bottom of the tray.
CRITICAL STEP: During this phase, remember to orient the part facing the frame towards the bottom of the tray.- 6.
- Add the molten paraffin at 56 °C in the tray with CytoMatrix-sample, dispensing from the Paraffin Embedding Module.
- 7.
- Press gently the matrix flat to the bottom with a metallic stick, giving the correct orientation (Figure 1E).
- 8.
- Transfer the tray to the cold plate and allow the paraffin to cool for 60 min.
- 9.
- When the wax is completely cooled and hardened, extract the paraffin block from the tray (Figure 1F). The FFPE-CytoMatrix block is ready to cut the sections on microtome, as a standard FFPE block.
- 10.
- Grind very carefully the FFPE-CytoMatrix block until it reaches the surface of the matrix.
- 11.
- Pick the section up with forceps or a fine paint brush and float it on the surface of a water bath.
- 12.
- Float the section on the surface of clean positively charged slides.
- 13.
- Place the slide with the paraffin section on a 50 °C water bath for few seconds to bind and spread the CytoMatrix section to the glass.
- 14.
- Use the first CytoMatrix slide for H&E staining.
 CRITICAL STEP: For H&E staining, place slides in oven at 70 °C for 10 min, deparaffinize and rehydrate sections (3 × 3 min xylene and 3 × 3 min 100% ethanol, 1 × 3 min 95% ethanol, 1 × 3 min 70% ethanol 1 × 5 min deionized H2O), Hematoxylin staining 1 × 2 min, rinse with tap deionized water 1 × 5 min, Eosin staining 1 × 30 s, dehydration (1 × 2 min 70% ethanol, 1 × 2 min 90% ethanol, 3 × 2 min 100% ethanol), 3 × 5 min xylene, Coverslip slides using Permount (xylene based).
CRITICAL STEP: For H&E staining, place slides in oven at 70 °C for 10 min, deparaffinize and rehydrate sections (3 × 3 min xylene and 3 × 3 min 100% ethanol, 1 × 3 min 95% ethanol, 1 × 3 min 70% ethanol 1 × 5 min deionized H2O), Hematoxylin staining 1 × 2 min, rinse with tap deionized water 1 × 5 min, Eosin staining 1 × 30 s, dehydration (1 × 2 min 70% ethanol, 1 × 2 min 90% ethanol, 3 × 2 min 100% ethanol), 3 × 5 min xylene, Coverslip slides using Permount (xylene based).- 15.
- Observe the H&E on optical microscope in order to evaluate the morphology and the amount of cells for each patient.
- 16.
- Only in cases with sufficient material, cut the second 5 µm section for UroVysion FISH.
- 17.
- Cut the third, fourth and fifth 3 µm sections for immunohistochemistry.
3.4. UroVysion FISH
- a.
- The UroVysion Bladder Cancer Kit (Abbott Molecular) which contains DNA Probe Mixture (Fluorophore-labelled DNA probes for chromosomes 3, 7, and 17, and locus 9p21 in hybridization buffer), DAPI II Counterstain, 20X SSC, NP-40 (non-ionic detergent).
- b.
- The Vysis FISH Pretreatment Reagent Kit (Abbott Molecular) which includes Vysis Protease solution (3 × 25 mg) with Pepsin Activity, Vysis Pepsin Buffer (3 × 50 mL) 10 mM HCl, Vysis PBS 1X (2 × 250 mL), Vysis 20X SSC (66 g), 10% neutral buffered formalin and 4’,6-Diamidino-2-phenylindole (DAPI) solution.Before starting:
- c.
- In each session, include a Probe Check Bladder Cells Control Slides, containing both a positive control and a negative control on the same slide.
- a.
- Prepare Ethanol solutions at 70%, 90%, and 100% with purified waters in coplin jars.
- b.
- Prepare solutions 20X SCC (pH: 5.3), 2X SCC, 0.4X SSC/0.3% NP-40, 2X SSC/0.1% NP-40.
- c.
- Add protease solution to Coplin jar and place in a 39 °C water bath for at least 30 min before use, or until the solution temperature reaches 37 ± 1 °C. Verify the solution temperature before use with a calibrated thermometer.
- d.
- Add 2X SCC solution to Coplin jar and place in a 94 °C water bath for at least 30 min before use, or until the solution temperature reaches 94 ± 1 °C. Verify the solution temperature before use with a calibrated thermometer.
- Select the hybridization area with a circle in the 5 µm sections cut from FFPE-CytoMatrix blocks for FISH analysis.
- Place slides in oven at 70 °C for 30 min.
- Deparaffinize in xylene for 3 × 10 min.
- Rehydrate sections in 100% ethanol 2 × 10 min, 90% ethanol 1 × 5 min, 70% ethanol 1 × 3 min, and deionized H2O 1 × 3 min.
- Allow slides to completely dry at RT.
- Immerse slides in 2X SSC for 10 min at 94 ± 1 °C.
- Wash with deionized H2O for 3 min.
- Immerse slides in protease solution for 30 min at 37 ± 1 °C.
- Wash slides in 1X PBS for 3 min.
- Dehydrate sections in 70% ethanol 2 min, 90% ethanol 2 min, 100% ethanol 2 min.
- Allow slides to completely dry at RT.
- Probes preparation:
- a.
- Remove the UroVysion probe from −20 °C storage and allow to warm to RT.
- b.
- Vortex to mix and then spin the tubes briefly (3 s) in a microcentrifuge to bring the contents to the bottom of the tube.
- c.
- Apply 3–5 μL of probes solution to the selected target area of each slide.
- d.
- Immediately, place a 12–18 mm round glass coverslip over the probe.
- e.
- Carefully apply light pressure on the coverslip to allow the probe solution to spread evenly under the coverslip (Note: air bubbles can interfere with hybridization and should be avoided).
- f.
- Seal coverslip with rubber cement around the periphery of the coverslip.
- Add around 75 mL of distilled or deionized water in each humid chamber of the Automatic Slide Hybridizer ThermoBrite.
- Set the program for denaturation temperature 76 °C 5 min (Denaturation Step) and for hybridization temperature 39 °C 14–18 h (Hybridization Step) and run it.
- Post-Hybridization washes:
- a.
- Fill a Coplin jar with 0.4X SSC/0.3% NP-40 solution and place it in a 74 °C water bath for at least 30 min before use, or until the solution temperature reaches 73 ± 1 °C. Verify the solution temperature before use with a calibrated thermometer.
- b.
- Fill a second Coplin jar with 2X SSC/0.1% NP-40 solution and place it at RT.
- c.
- Remove the rubber cement and the coverslip from the slides with a metallic tweezer (Note: if the coverslip does not come off easily, immerse the slides in 2X SSC/0.1% NP-40 solution for a few seconds, in order to easily remove it).
- d.
- Immediately after removing the coverslip, place the slides in the Coplin jar with 0.4X SSC/0.3% NP-40 solution at 73 ± 1 °C and incubate for 2 min.
- e.
- Remove the slides from the wash solution, place the slides in the Coplin jar containing 2X SSC/0.1% NP-40 at RT and incubate for 1 min.
- f.
- Remove the slides from the second wash solution and place vertically in a dark area on a paper towel to dry completely.
- Apply 10 μL of DAPI II onto the target area and place a coverslip (18 mm square is recommended), avoiding air bubbles.
- Store the slides in the dark prior to signal enumeration under the fluorescence microscope.
3.5. Immunohistochemistry
- Place the three labelled slides with 3 µm sections in oven at 70 °C for 15 min.
- Set the program on the automatic stainer according to manufacturer’s instructions. The protocol used for each antibody is shown in Table S1.
- Load the slides onto the BOND Slide Tray, apply new BOND Universal Covertiles, place it in the Leica BOND III and start the run.
- When the run is complete, remove Covertiles, rinse the slides in deionized water, dehydrate in ethanol, clear in xylene and mount sections with coverslips.
- Store the slides in a slide-box prior to evaluate under the optical microscope.
4. Results
- 12/14 (85%) tested urine specimens, obtained from different patients, showed sufficient cellular material after the CytoMatrix protocol and H&E staining (Figure 2A). The two cases with insufficient materials were originally diagnosed as “inadequate sample”, given that not enough cells were found in the first urine sample analyzed by cytology, suggesting that cells were absent or too few to be captured and stored by CytoMatrix as well. For all other samples, even starting from a limited number of cells, it was possible to obtain from each single CytoMatrix several slides to perform the FISH analysis for UroVysion and the panel of immunostainings for various markers.
- Our tests showed that the first ten 3 μm sections are the richest in cellular material; on average, the cells penetrate inside the matrix for about 30 μm. In subsequent cuts, the cells are poor and lose a “tissue-like” structure.
- The fibers of the CytoMatrix are stained fuchsia by eosin and can be clearly distinguished.
- H&E staining showed the power of this tool to confer a 3D-structure of cells from urine. CytoMatrix-FFPE block from patient number 4, stained with H&E, clearly showed the presence of papillary clusters, small vessels and a fibrous-connective axis (Figure 2B); in this case, the property of CytoMatrix allowed to capture and store cells and also micro-cellular aggregates in its three-dimensional structure, helping the diagnosis of HGUC.
- CytoFoam is a 12-mm disc previously described in literature and used to adsorb various cytological samples, as fine needle aspiration of thyroid carcinomas or plasma for the evaluation of circulating tumor cells [27,28]. It is a diagnostic tool similar to CytoMatrix, since it is processed as a tissue fragment, formalin-fixed, paraffin-embedded, and used for IHC or other molecular analyses. We used CytoFoam, as previously described, to absorb our cell suspensions from urine specimens in order to compare the results obtained with CytoMatrix [27,28]. Interestingly, when compared with this CytoFoam adsorbent support, CytoMatrix can improve the amount of cells and can also confer an architecture or a "tissue-like" structure (Figure 2C).
4.1. CytoMatrix-FFPE UroVysion Evaluation
- UroVysion probe signals and DAPI counterstain should be viewed under fluorescence microscope with the following filters: DAPI single-bandpass, Aqua single-bandpass (chromosome 17), Yellow (Gold) single-bandpass (9p21 locus), Red/Green dual-bandpass (chromosomes 3 and 7).
- Begin analysis in the upper left quadrant of the target area. Scan fields from left to right and top to bottom, without rescanning the same areas.
- Use the following criteria to select cells suspicious for malignancy: large nuclear size, irregular nuclear shape, “patchy” DAPI staining, and cell clusters.
- Determine the number of signals for all 4 probes in a minimum of 200 cells and/or 25 morphologically abnormal neoplastic cells.
- Record the chromosome pattern only if:
- There is a gain (3 or more signals) of 2 or more of chromosomes 3 (red), 7 (green), or 17 (aqua).
- There is a loss of both copies of LSI 9p21 (Gold).
- Other situations should be considered uninterpretable due to hybridization failure.
- Take images using a Zeiss fluorescence microscope (Axio Imager M1), equipped with appropriate excitation and emission filters, at magnification 20x and 100x within the area of interest.
- Analyze the images with Zeiss AxionVision 4V, 4.8.2.0 Software, manually counting the number of cells in which signals colocalize with DAPI nuclei.
- If no abnormalities are detected, the remaining cells are counted until a sufficient number of cells without chromosomal abnormalities are visualized (minimum 200 cells evaluated).
- A positive result is indicated by the presence of ≥4 cells with gains of two or more of chromosomes 3, 7 and 17 on 25 neoplastic cells analyzed. In the case of chromosome 9p21, a positive result is considered when ≥12 cells show absence of 9p21 signals on 25 neoplastic cells analyzed.Our results showed that:
- The UroVysion test can be used with CytoMatrix samples and can improve the accuracy of diagnosis for urothelial carcinoma.
- All non-neoplastic samples (5/5, 100%) showed normal copy number of chromosomes 3, 7, 17 and 9p21 (Figure 3A).
- All LGUC and HGUC (7/7, 100%) showed the presence of nuclei with 3 or more signals for chr 3, 7, and 17 in selected areas; only 2 HGUC showed homozygous deletion of 9p21, not present in LGUC, suggesting that p16/INK4 deletion could be associated higher malignancy and aggressiveness of urothelial tumors (Figure 3A).
- The fibers of the CytoMatrix exhibited an autofluorescence that can be clearly distinguished under the microscope with all the filters (Figure 3B).
4.2. CytoMatrix-FFPE IHC Evaluation
- The slides are scored on the scale 0–3+ on the basis of DAB intensity and are quantified by counting at least 200 stained cells.
- Take images using a Nikon optical microscope (Eclipse 50i) at magnification 10x, 20x and 40x within the area of interest (Note: all microscope and camera settings (e.g., light level, exposure, gain, etc.) should be identical for all images).
- Analyze the images with Nikon Imaging Software NIS-Elements and quantify the DAB signal with ImageJ64 software.
- Manually count the number of positive cells with the expression of the antigen of interest.
- Staining intensity is scored as “no expression” (0), “weak expression” (1+), “moderate expression” (2+) and “strong expression” (3+).
- Ki67 is scored from 0% to 30% of nuclei, to evaluate proliferation rate.
- CK20 is scored as positive when neoplastic cells show cytoplasmatic/membrane staining intensity >1, showing the presence of cells with epithelial origin.
- p53 is scored as positive when neoplastic cells show nuclear staining intensity >1, suggesting the presence of a TP53 mutation.
- The Automatic IHC can be used with CytoMatrix samples and can improve the accuracy of diagnosis for urothelial carcinoma.
- The fibers of the CytoMatrix did not show any staining in IHC under the microscope.
- Ki67 staining was positive in neoplastic cells of two HGUC (N° 2, 4), while the other HGUC (N° 1, 3) showed negative staining. Staining of Ki67 was negative in LGUC and non-neoplastic cases; however, two non-neoplastic samples (N° 10, 11) showed weak positivity for Ki67 (<5% of cells) in granulocytes and immune cells (Figure 4).
- CK20 staining was positive in HGUC (N° 1, 2, 3) that maintain epithelial differentiation and it can be useful to identify neoplastic cells; however, immuno-staining for CK20 can also be negative in HGUC (N° 4) with loss of epithelial differentiation. Staining of CK20 was negative in all LGUC and non-neoplastic cases (Figure 4).
- p53 staining was negative in all analyzed cases, both in neoplastic and non-neoplastic samples, suggesting a low utility of this biomarker in supporting the diagnosis of urothelial carcinoma.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barkan, G.A.; Wojcik, E.M.; Nayar, R.; Savic-Prince, S.; Quek, M.L.; Kurtycz, D.F.; Rosenthal, D.L. The Paris System for Reporting Urinary Cytology: The Quest to Develop a Standardized Terminology. Acta Cytol. 2016. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Zhang, S.; MacLennan, G.T.; Williamson, S.R.; Lopez-Beltran, A.; Montironi, R. Bladder cancer: Translating molecular genetic insights into clinical practice. Hum. Pathol. 2011. [Google Scholar] [CrossRef]
- Knowles, M.A.; Hurst, C.D. Molecular biology of bladder cancer: New insights into pathogenesis and clinical diversity. Nat. Rev. Cancer 2015. [Google Scholar] [CrossRef]
- Netto, G.J. Molecular biomarkers in urothelial carcinoma of the bladder: Are we there yet? Nat. Rev. Urol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Rai, S.; Lali, B.S.; Venkataramana, C.G.; Philipose, C.S.; Rao, R.; Prabhu, G.L. A quest for accuracy: Evaluation of the Paris system in diagnosis of urothelial carcinomas. J. Cytol. 2019. [Google Scholar] [CrossRef]
- Yafi, F.A.; Brimo, F.; Steinberg, J.; Aprikian, A.G.; Tanguay, S.; Kassouf, W. Prospective analysis of sensitivity and specificity of urinary cytology and other urinary biomarkers for bladder cancer. Urol. Oncol. Semin. Orig. Investig. 2015. [Google Scholar] [CrossRef] [PubMed]
- Babjuk, M.; Burger, M.; Compérat, E.M.; Gontero, P.; Mostafid, A.H.; Palou, J.; van Rhijn, B.W.G.; Rouprêt, M.; Shariat, S.F.; Sylvester, R.; et al. European Association of Urology Guidelines on Non-muscle-invasive Bladder Cancer (TaT1 and Carcinoma In Situ)—2019 Update. Eur. Urol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Cajulis, R.S.; Haines, G.K.; Frias-Hidvegi, D.; McVary, K. Interphase cytogenetics as an adjunct in the cytodiagnosis of urinary bladder carcinoma: A comparative study of cytology, flow cytometry and interphase cytogenetics in bladder washes. Anal. Quant. Cytol. Histol. 1994, 16, 1–10. [Google Scholar]
- Reuter, V.E. The pathology of bladder cancer. Urology 2006. [Google Scholar] [CrossRef]
- Mao, L.; Schoenberg, M.P.; Scicchitano, M.; Erozan, Y.S.; Merlo, A.; Schwab, D.; Sidransky, D. Molecular detection of primary bladder cancer by microsatellite analysis. Science 1996. [Google Scholar] [CrossRef]
- McCroskey, Z.; Pambuccian, S.E.; Kleitherms, S.; Antic, T.; Cohen, M.B.; Barkan, G.A.; Wojcik, E.M. Accuracy and interobserver variability of the cytologic diagnosis of low-grade urothelial carcinoma in instrumented urinary tract cytology specimens. Am. J. Clin. Pathol. 2015. [Google Scholar] [CrossRef] [Green Version]
- Nagai, T.; Naiki, T.; Etani, T.; Iida, K.; Noda, Y.; Shimizu, N.; Isobe, T.; Nozaki, S.; Okamura, T.; Ando, R.; et al. UroVysion fluorescence in situ hybridization in urothelial carcinoma: A narrative review and future perspectives. Transl. Androl. Urol. 2021. [Google Scholar] [CrossRef]
- Dimashkieh, H.; Wolff, D.J.; Smith, T.M.; Houser, P.M.; Nietert, P.J.; Yang, J. Evaluation of urovysion and cytology for bladder cancer detection: A study of 1835 paired urine samples with clinical and histologic correlation. Cancer Cytopathol. 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halling, K.C.; Kipp, B.R. Bladder cancer detection using FISH (UroVysion assay). Adv. Anat. Pathol. 2008. [Google Scholar] [CrossRef]
- Ho, C.C.K.; Tan, W.P.; Pathmanathan, R.; Tan, W.K.; Tan, H.M. Fluorescence-in-situ-hybridization in the surveillance of urothelial cancers: Can use of cystoscopy or ureteroscopy be deferred? Asian Pac. J. Cancer Prev. 2013. [Google Scholar] [CrossRef] [Green Version]
- Kojima, T.; Nishiyama, H.; Ozono, S.; Hinotsu, S.; Keino, N.; Yamaguchi, A.; Sakai, H.; Enomoto, Y.; Horie, S.; Fujimoto, K.; et al. Clinical evaluation of two consecutive UroVysion fluorescence in situ hybridization tests to detect intravesical recurrence of bladder cancer: A prospective blinded comparative study in Japan. Int. J. Clin. Oncol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Lavery, H.J.; Zaharieva, B.; McFaddin, A.; Heerema, N.; Pohar, K.S. A prospective comparison of UroVysion FISH and urine cytology in bladder cancer detection. BMC Cancer 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapia, C.; Glatz, K.; Obermann, E.C.; Grilli, B.; Barascud, A.; Herzog, M.; Schönegg, R.; Savic, S.; Bubendorf, L. Evaluation of chromosomal aberrations in patients with benign conditions and reactive changes in urinary cytology. Cancer Cytopathol. 2011. [Google Scholar] [CrossRef]
- Wilkerson, M.L.; Lin, F.; Liu, H.; Cheng, L. The application of immunohistochemical biomarkers in urologic surgical pathology. Arch. Pathol. Lab. Med. 2014. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Swellam, M.; Amin, A.; Balbaa, M.E.; Yacout, G.A.; El-Zayat, T.M. The clinical relevance of urine-based markers for diagnosis of bladder cancer. Med. Oncol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.; Propping, C.; Siow, W.Y.; Lohse-Fischer, A.; Toma, M.; Baldauf-Twelker, A.; Hakenberg, O.W.; Wirth, M.P.; Fuessel, S. Diagnostic and prognostic value of bladder cancer-related transcript markers in urine. J. Cancer Res. Clin. Oncol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, N.; Diwaker, P.; Lotha, N.; Arora, V.K.; Singh, N. Cytokeratin 20 immunocytochemistry on urine sediments: A potential low-cost adjunct to cytology in the diagnosis of low-grade urothelial carcinoma. Cytopathology 2017. [Google Scholar] [CrossRef] [PubMed]
- Bruschini, S.; di Martino, S.; Pisanu, M.E.; Fattore, L.; De Vitis, C.; Laquintana, V.; Buglioni, S.; Tabbì, E.; Cerri, A.; Visca, P.; et al. CytoMatrix for a reliable and simple characterization of lung cancer stem cells from malignant pleural effusions. J. Cell. Physiol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Scarpino, S.; Taccogna, S.; Pepe, G.; Papini, E.; D’Angelo, M.; Cascone, F.; Nicoletti, D.; Guglielmi, R.; Palermo, A.; Trombetta, M.; et al. Morphological and Molecular Assessment in Thyroid Cytology Using Cell-Capturing Scaffolds. Horm. Metab. Res. 2020. [Google Scholar] [CrossRef]
- Spugnini, E.P.; Menicagli, F.; Giaconella, R.; Zanni, F.; Camponi, C.; De Luca, A.; Santoro, A.; Baldi, A. Filling the gap between histology and cytology: Description of an innovative technology (Cytomatrix) to increase the diagnostic effectiveness of fine needle aspirates data. J. Clin. Pathol. 2021. [Google Scholar] [CrossRef]
- Stigliano, S.; Crescenzi, A.; Taffon, C.; Covotta, F.; Hassan, C.; Antonelli, G.; Verri, M.; Biasutto, D.; Scarpa, R.M.; Di Matteo, F.M. Role of fluorescence confocal microscopy for rapid evaluation of EUS fine-needle biopsy sampling in pancreatic solid lesions. Gastrointest. Endosc. 2021. [Google Scholar] [CrossRef]
- Taccogna, S.; Guglielmi, R.; Persichetti, A.; Morano, C.; Angelini, F.; Ienzi, S.; Scarpino, S.; Liverani, A.; Annovazzi, A.; Papini, E. Carcinomas of the Thyroid with Ewing Family Tumor Elements (CEFTEs): A Diagnostic Challenge Before Surgery. Head Neck Pathol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Mayall, F.G.; Pepperell, J.; Bodger, I.; Higbee, D.; Stevanato, L.; Hustler, A.; Mumford, K.M. Cytology and cell-block immunohistochemistry of circulating tumour cells. Cytopathology 2019. [Google Scholar] [CrossRef] [Green Version]




| n° | Sex | Age at Diagnosis | Cytological Diagnosis | CK20 | Ki67 | p53 | UroVysion FISH | CytoMatrix Diagnosis |
|---|---|---|---|---|---|---|---|---|
| 1 | M | 90 y | HGUC | Pos > 10% | neg | neg | gain of chr 3 (red), 7 (green) | HGUC confirmed |
| 2 | M | 75 y | HGUC | Pos > 10% | Pos 5% | neg | gain of chr 3, 17 (aqua), homozygous loss 9p21 (gold) | HGUC confirmed |
| 3 | M | 64 y | HGUC | Pos > 10% | neg | neg | gain of chr 3, 7 | HGUC confirmed |
| 4 | M | 62 y | HGUC | neg | Pos 10% | neg | gain of chr 3, 7, 17, homozygous loss 9p21 | HGUC confirmed |
| 5 | F | 88 y | LGUC | neg | neg | neg | gain of chr 3, 17 | LGUC confirmed |
| 6 | M | 68 y | LGUC | neg | neg | neg | gain of chr 3, 17 | LGUC confirmed |
| 7 | F | 70 y | AUC | neg | neg | neg | normal copy number chr 3, 7, 17, 9p21 | Non neoplastic confirmed |
| 8 | M | 67 y | AUC | neg | neg | neg | normal copy number chr 3, 7, 17, 9p21 | Non neoplastic confirmed |
| 9 | M | 66 y | Non evaluable | Pos 10% | neg | neg | gain of chr 3, 17 | LGUC newly diagnosed |
| 10 | M | 83 y | Non neoplastic | neg | Pos immune cells | neg | normal copy number chr 3, 7, 17, 9p21 | Non neoplastic confirmed |
| 11 | F | 66 y | Non neoplastic | neg | Pos immune cells | neg | normal copy number chr 3, 7, 17, 9p21 | Non neoplastic confirmed |
| 12 | M | 74 y | Non neoplastic | neg | neg | neg | normal copy number chr 3, 7, 17, 9p21 | Non neoplastic confirmed |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minasi, S.; Bosco, D.; Moretti, B.; Giangaspero, F.; Santoro, A.; Buttarelli, F.R. Improvement of the Collection, Maintenance, and Analysis of Neoplastic Cells from Urine Specimens with the Use of CytoMatrix. Methods Protoc. 2021, 4, 65. https://doi.org/10.3390/mps4030065
Minasi S, Bosco D, Moretti B, Giangaspero F, Santoro A, Buttarelli FR. Improvement of the Collection, Maintenance, and Analysis of Neoplastic Cells from Urine Specimens with the Use of CytoMatrix. Methods and Protocols. 2021; 4(3):65. https://doi.org/10.3390/mps4030065
Chicago/Turabian StyleMinasi, Simone, Daniela Bosco, Bernardo Moretti, Felice Giangaspero, Antonio Santoro, and Francesca Romana Buttarelli. 2021. "Improvement of the Collection, Maintenance, and Analysis of Neoplastic Cells from Urine Specimens with the Use of CytoMatrix" Methods and Protocols 4, no. 3: 65. https://doi.org/10.3390/mps4030065