What’s Wrong in a Jump? Prediction and Validation of Splice Site Variants
Abstract
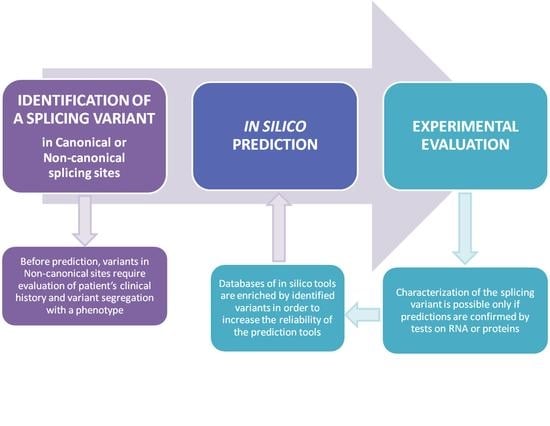
:1. Introduction
2. Constitutive Splicing vs. Alternative Splicing
3. Genomic Variants Affecting Splicing Process
3.1. DNA Variants in Canonical Splicing Sites
3.1.1. Variants in Splicing Acceptor and Donor Sites
3.1.2. Variants Affecting Branch Point and Polypyrimidine Tract
3.2. Exonic Variants Affecting Splicing
3.3. Deep Intronic Variants
4. Identification of Splice Variants in NGS Era
5. Predictive Tools for Splice Variant Identification
5.1. Early Computational Methodologies for Splice Variant Prediction
5.1.1. Input Sequences
5.1.2. Statistical Models
5.1.3. Tools Combining Multiple Algorithms
| Tool Name | Analyzed Regions | Predictive Model | URL | Ref. |
|---|---|---|---|---|
| Canonical Splice Sites | ||||
| MaxEntScan | 5′ and 3′ SSMs | PWM, MDD, MM, and MED | http://hollywood.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html (accessed on 3 September 2021) | [48] |
| SpliceView | 5′ and 3′ SSMs | PWM | http://bioinfo.itb.cnr.it/~webgene/wwwspliceview.html (accessed on 3 September 2021) | [42] |
| GeneSplicer | 5′ and 3′ SSMs | MDD | https://www.cbcb.umd.edu/software/GeneSplicer/gene_spl.shtml (accessed on 3 September 2021) | [43] |
| Spliceport | 5′ and 3′ SSMs | SVM | http://spliceport.cbcb.umd.edu/ (accessed on 3 September 2021) | [44] |
| GENSCAN | 5′ and 3′ SSMs | MDD | http://hollywood.mit.edu/GENSCAN.html (accessed on 3 September 2021) | [45] |
| NetGene2 | 5′ and 3′ SSMs | NN | http://www.cbs.dtu.dk/services/NetGene2/(accessed on 3 September 2021) | [46] |
| NNSplice | 5′ and 3′ SSMs | NN | https://www.fruitfly.org/seq_tools/splice.html (accessed on 3 September 2021) | [47] |
| SVM-BP Finder | BPs + PPT | SVM | http://regulatorygenomics.upf.edu/Software/SVM_BP/ (accessed on 3 September 2021) | [49] |
| IntSplice | BPs + PPT | SVM | https://www.med.nagoya-u.ac.jp/neurogenetics/IntSplice_v1.0/index.php (accessed on 3 September 2021) | [50] |
| Cryptic sites | ||||
| CRYP-SKIP | exons + flanking intronic sequences | multiple logistic regression | https://cryp-skip.img.cas.cz/ (accessed on 3 September 2021) | [51] |
| Spliceman | variant + flanking nucleotides | L1 distance metric | http://fairbrother.biomed.brown.edu/spliceman/ (accessed on 3 September 2021) | [52] |
| Exonic Sequences | ||||
| ESE Finder | ESE | PWM | http://krainer01.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home (accessed on 3 September 2021) | [53,54] |
| RESCUE-ESE | SREs | experimental + computational approach | http://hollywood.mit.edu/burgelab/rescue-ese/ (accessed on 3 September 2021) | [58] |
| ESRseq | ESE + ESS | PWM | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3149502/ (accessed on 3 September 2021) | [55] |
| Hexplorer | SREs | experimental + computational approach | https://www2.hhu.de/rna/html/hexplorer_score.php (accessed on 3 September 2021) | [57] |
| FAS-ESS | ESS | MED | http://hollywood.mit.edu/fas-ess/(accessed on 3 September 2021) | [56] |
| SpliceAid | ESE + ESS + ISE + ISS | scanning against validated splicing sequences | http://www.introni.it/splicing.html (accessed on 3 September 2021) | [59] |
| Conservation | ||||
| RBPmap | RBP sites | Weighted-Rank (WR) | http://rbpmap.technion.ac.il/ (accessed on 3 September 2021) | [60] |
| Splicing Factor Finder | RBP sites | WR | https://pubmed.ncbi.nlm.nih.gov/19296853/ (accessed on 3 September 2021) | [61] |
| RNA Secondary Structure | ||||
| pFold | RNA sequence | stochastic context-free grammar (SCFG) | https://pubmed.ncbi.nlm.nih.gov/12824339/ (accessed on 3 September 2021) | [62] |
| UNAFold | RNA sequence | free energy minimization, partition function calculations, and stochastic sampling | http://www.unafold.org/ (accessed on 3 September 2021) | [63] |
| Combined Analysis | ||||
| EX-SKIP | ESEs + ESSs | four algorithms | https://ex-skip.img.cas.cz/ (accessed on 3 September 2021) | [68] |
| HOT-SKIP | ESEs + ESSs | four algorithms | https://hot-skip.img.cas.cz/ (accessed on 3 September 2021) | [68] |
| Sroogle | SSM + BP + PPT + SRE | nine algorithms | http://sroogle.tau.ac.il/ (accessed on 3 September 2021) | [66] |
| Human Splicing Finder (*) | SREs, splice sites or branch sites | PWM and MED | http://www.umd.be/HSF3/ (accessed on 3 September 2021) | [65] |
5.2. Machine Learning-Based Tools
| Tool Name | Prediction | Model | Datasets | Key-Points | Ref |
|---|---|---|---|---|---|
| CADD | Score of pathogenicity | Rirst version: linear SVM Later versions: L2-regularized logistic regression | Training datasets: Benign: evolutionarily neutral variants; Pathogenic: simulated de novo pathogenic variants Testing datasets: Benign: benign variants; Pathogenic: pathogenic ClinVar variants, somatic cancer mutation frequencies | Effective tool for protein-coding impact prediction; may not be informative for poorly-conserved regions | [74,75] |
| CryptSplice | Impact of variants on existing splice sites, cryptic splice site prediction | SVM with RBF kernel | True and false splice sites from GenBank-derived datasets | Identify creation of cryprtic acceptor/donor site; use of a quite obsolete database | [78] |
| DARTS | Prediction of alternative splicing using both cis sequence features and mRNA levels of trans RBPs | DNN and Bayesian Hypothesis Testing | RNA-seq data (*) | Evaluation of RBP impact on splicing | [82] |
| MMSplice | Multiple predictions: exon skipping, competitive interactions, changes in splicing effciency, and pathogenicity | Modular NN, linear and logistic regression | Donor/acceptor modules: GENCODE v24 true (known sites) and false (random sequences) splice sites Exon/intron modules: MPRA data from [83] | Easily clinically applicable training set; contains false positive/unverified sites | [84] |
| MutPred Splice | Impact of coding region substitutions on disruption of pre-mRNA splicing | Linear SVM | Positive: HGMD exonic disease-causing/disease-associated variants Negative: HGMD disease-causing missense, not reported to disrupt exon splicing, high frequency exonic SNPs (SNP- from 1000 Genomes Project [85] | Suitable for use in an NGS high-throughput setting to identify and prioritize potentially splice-altering variants | [86] |
| PEPSI | Prediction of coding and noncoding variant impact on pre-mRNA splicing based on sequence conservation, RNA secondary structure, and regulatory sequence elements | Random forest regression model | Data obtained form the Vex-seq experiment (measurement of the ΔPSI of 2055 variants from the Exome Aggregation Consortium (ExAC; [Kircher et al., 2014]) v24 a selection of chromosomes as training set, the remaining ones as testing set (*) | Indels and intronic variants included | [80] |
| S-CAP | Score of variant pathogenicity using compartmentalization of genomic regions | DNN | Pathogenic variants selected from HGMD and ClinVar; benign variants from gnomAD | Evaluation of intronic pathogenic variants; variants lying more than 50 bp into the intron are not covered by the model | [79] |
| SPANR | Cassette exon skipping prediction | NN modeled on Bayesian framework | PSI values for all human exons across 16 tissues, based on the Illumina Human Body Map project (*) | Web server easy to use, availability of a dataset of pre-computed scores for all eligible variants in the genome; evaluation of exon sequence only | [87] |
| SpliceAI | Prediction of variant impact on loss or gain of acceptor/donor sites | 32-layer DNN | Protein-coding transcripts from GENCODE v24 (a selection of chromosomes as training set, the remaining ones as testing set) (*) | Very powerful tool able to use a “near-agnostic” approach | [81] |
| SpliceFinder | Classification of variants based on impact on donor site, acceptor site or non-splice-site | CNN | Sequences of donor, acceptor, and non-splice-site, randomly selected from human reference genome (90% for training, 10% for testing, and then 20% of the training data for validation) | Non-canonical splice sites can also be predicted correctly; decreased number of false positives | [88] |
| TraP | Quantification of impact of variant on transcripts | Random forest | Benign: De novo mutations in healthy individuals Pathogenic: selected synonymous variants associated with rare disease (*) | High performance in distinguishing pathogenic and benign variants, both intronic and synonymous; evaluation of potential impact of variants across multiple transcripts | [76] |
6. Interpretation and Evaluation
7. Validation of Predicted Splicing Variants
7.1. Transcriptomics Functional Testing
7.2. Proteomics Analysis
8. Splice Variant Characterization in Diagnostics
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gilbert, W. Why genes in pieces? Nat. Cell Biol. 1978, 271, 501. [Google Scholar] [CrossRef]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nat. Cell Biol. 2010, 463, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, J.; Huang, B.O.; Xu, Y.-M.; Li, J.; Zhang, J.; Min, Q.-H.; Yang, W.-M.; Wang, X.-Z. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Pohl, M.; Bortfeldt, R.H.; Grützmann, K.; Schuster, S. Alternative splicing of mutually exclusive exons—A review. Biosystems 2013, 114, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Koren, E.; Lev-Maor, G.; Ast, G. The Emergence of Alternative 3′ and 5′ Splice Site Exons from Constitutive Exons. PLoS Comput. Biol. 2007, 3, e95. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.-T.; Lin, C.-X.; Fang, Z.-Y.; Li, H.-D. Intron Retention as a Mode for RNA-Seq Data Analysis. Front. Genet. 2020, 11, 586. [Google Scholar] [CrossRef] [PubMed]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef] [Green Version]
- Turunen, J.J.; Verma, B.; Nyman, T.A.; Frilander, M.J. HnRNPH1/H2, U1 snRNP, and U11 snRNP cooperate to regulate the stability of the U11-48K pre-mRNA. RNA 2013, 19, 380–389. [Google Scholar] [CrossRef] [Green Version]
- Wachutka, L.; Caizzi, L.; Gagneur, J.; Cramer, P. Global donor and acceptor splicing site kinetics in human cells. Elife 2019, 8, e45056. [Google Scholar] [CrossRef]
- Wickramasinghe, V.O.; Gonzàlez-Porta, M.; Perera, D.; Bartolozzi, A.R.; Sibley, C.R.; Hallegger, M.; Ule, J.; Marioni, J.C.; Venkitaraman, A.R. Regulation of constitutive and alternative mRNA splicing across the human transcriptome by PRPF8 is determined by 5′ splice site strength. Genome Biol. 2015, 16, 201. [Google Scholar] [CrossRef] [Green Version]
- Kondo, Y.; Oubridge, C.; Van Roon, A.-M.M.; Nagai, K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife 2015, 4, e04986. [Google Scholar] [CrossRef] [PubMed]
- Perriman, R.; Ares, M. Invariant U2 snRNA Nucleotides Form a Stem Loop to Recognize the Intron Early in Splicing. Mol. Cell 2010, 38, 416–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertram, K.; Agafonov, D.E.; Liu, W.-T.; Dybkov, O.; Will, C.L.; Hartmuth, K.; Urlaub, H.; Kastner, B.; Stark, H.; Lührmann, R. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nat. Cell Biol. 2017, 542, 318–323. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Yan, C.; Zhan, X.; Li, L.; Lei, J.; Shi, Y. Structure of the human activated spliceosome in three conformational states. Cell Res. 2018, 28, 307–322. [Google Scholar] [CrossRef]
- Zheng, C.L.; Fu, X.-D.; Gribskov, M. Characteristics and regulatory elements defining constitutive splicing and different modes of alternative splicing in human and mouse. RNA 2005, 11, 1777–1787. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, M.J.; de la Mata, M.; Kornblihtt, A.R. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem. Sci. 2010, 35, 497–504. [Google Scholar] [CrossRef]
- Shabalina, S.A.; Spiridonov, A.N.; Spiridonov, N.A.; Koonin, E.V. Connections between Alternative Transcription and Alternative Splicing in Mammals. Genome Biol. Evol. 2010, 2, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Oren, Y.S.; Pranke, I.M.; Kerem, B.; Sermet-Gaudelus, I. The suppression of premature termination codons and the repair of splicing mutations in CFTR. Curr. Opin. Pharmacol. 2017, 34, 125–131. [Google Scholar] [CrossRef]
- Jimeno-González, S.; Reyes, J.C. Chromatin structure and pre-mRNA processing work together. Transcription 2016, 7, 63–68. [Google Scholar] [CrossRef] [Green Version]
- Chabot, B.; Shkreta, L. Defective control of pre–messenger RNA splicing in human disease. J. Cell Biol. 2016, 212, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Sterne-Weiler, T.; Sanford, J.R. Exon identity crisis: Disease-causing mutations that disrupt the splicing code. Genome Biol. 2014, 15, 201–208. [Google Scholar] [CrossRef] [Green Version]
- Krawczak, M.; Thomas, N.S.; Hundrieser, B.; Mort, M.; Wittig, M.; Hampe, J.; Cooper, D.N. Single base-pair substitutions in exon-intron junctions of human genes: Nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007, 28, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Habara, Y.; Takeshima, Y.; Awano, H.; Okizuka, Y.; Zhang, Z.; Saiki, K.; Yagi, M.; Matsuo, M. In vitro splicing analysis showed that availability of a cryptic splice site is not a determinant for alternative splicing patterns caused by +1G->A mutations in introns of the dystrophin gene. J. Med. Genet. 2008, 46, 542–547. [Google Scholar] [CrossRef] [Green Version]
- Cariola, F.; Disciglio, V.; Valentini, A.M.; Lotesoriere, C.; Fasano, C.; Forte, G.; Russo, L.; Di Carlo, A.; Guglielmi, F.; Manghisi, A.; et al. Characterization of a rare variant (c.2635-2A>G) of the MSH2 gene in a family with Lynch syndrome. Int. J. Biol. Markers 2018, 33, 534–539. [Google Scholar] [CrossRef] [Green Version]
- Anna, A.; Monika, G. Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 2018, 59, 253–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufner-Almeida, L.G.; Carmo, R.T.D.; Masotti, C.; Haddad, L.A. Understanding human DNA variants affecting pre-mRNA splicing in the NGS era. Adv. Genet. 2019, 103, 39–90. [Google Scholar] [CrossRef]
- Caminsky, N.G.; Mucaki, E.J.; Rogan, P.K. Interpretation of mRNA splicing mutations in genetic disease: Review of the literature and guidelines for information-theoretical analysis. F1000Research 2014, 3, 282. [Google Scholar] [CrossRef]
- Ward, A.J.; Cooper, T.A. The pathobiology of splicing. J. Pathol. 2009, 220, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, K.; Roca, X.; Beiglböck, H.; Callens, T.; Etzler, J.; Rao, A.R.; Krainer, A.R.; Fonatsch, C.; Messiaen, L. Extensive in silico analysis of NF1 splicing defects uncovers determinants for splicing outcome upon 5′ splice-site disruption. Hum. Mutat. 2007, 28, 599–612. [Google Scholar] [CrossRef]
- Vaz-Drago, R.; Custódio, N.; Carmo-Fonseca, M. Deep intronic mutations and human disease. Hum. Genet. 2017, 136, 1093–1111. [Google Scholar] [CrossRef] [PubMed]
- Popp, M.W.-L.; Maquat, L.E. Organizing Principles of Mammalian Nonsense-Mediated mRNA Decay. Annu. Rev. Genet. 2013, 47, 139–165. [Google Scholar] [CrossRef] [Green Version]
- Diederichs, S.; Bartsch, L.; Berkmann, J.C.; Fröse, K.; Heitmann, J.; Hoppe, C.; Iggena, D.; Jazmati, D.; Karschnia, P.; Linsenmeier, M.; et al. The dark matter of the cancer genome: Aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol. Med. 2016, 8, 442–457. [Google Scholar] [CrossRef]
- Conboy, J.G. Unannotated splicing regulatory elements in deep intron space. Wiley Interdiscip. Rev. RNA 2021, 12, e1656. [Google Scholar] [CrossRef] [PubMed]
- Rowlands, C.F.; Baralle, D.; Ellingford, J.M. Machine Learning Approaches for the Prioritization of Genomic Variants Impacting Pre-mRNA Splicing. Cells 2019, 8, 1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frebourg, T. The Challenge for the Next Generation of Medical Geneticists. Hum. Mutat. 2014, 35, 909–911. [Google Scholar] [CrossRef]
- Baralle, D.; Lucassen, A.; Buratti, E. Missed Threads. The Impact of Pre-mRNA Splicing Defects on Clinical Practice. EMBO Rep. 2009, 10, 810–816. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.-S.; Cooper, T.A. Splicing in disease: Disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007, 8, 749–761. [Google Scholar] [CrossRef]
- Lim, K.H.; Ferraris, L.; Filloux, M.E.; Raphael, B.J.; Fairbrother, W.G. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc. Natl. Acad. Sci. USA 2011, 108, 11093–11098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Lin, H.; Zhao, H.; Hao, Y.; Mort, M.; Cooper, D.N.; Zhou, Y.; Liu, Y. Impact of human pathogenic micro-insertions and micro-deletions on post-transcriptional regulation. Hum. Mol. Genet. 2014, 23, 3024–3034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogozin, I.B.; Milanesi, L. Analysis of donor splice sites in different eukaryotic organisms. J. Mol. Evol. 1997, 45, 50–59. [Google Scholar] [CrossRef]
- Pertea, M.; Lin, X.; Salzberg, S.L. GeneSplicer: A new computational method for splice site prediction. Nucleic Acids Res. 2001, 29, 1185–1190. [Google Scholar] [CrossRef] [Green Version]
- Dogan, R.I.; Getoor, L.; Wilbur, W.J.; Mount, S. SplicePort--An interactive splice-site analysis tool. Nucleic Acids Res. 2007, 35, W285–W291. [Google Scholar] [CrossRef]
- Burge, C.B.; Karlina, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997, 268, 78–94. [Google Scholar] [CrossRef] [Green Version]
- Hebsgaard, S.M.; Korning, P.G.; Tolstrup, N.; Engelbrecht, J.; Rouzé, P.; Brunak, S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996, 24, 3439–3452. [Google Scholar] [CrossRef] [Green Version]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved Splice Site Detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Yeo, E.; Burge, C.B. Maximum Entropy Modeling of Short Sequence Motifs with Applications to RNA Splicing Signals. J. Comput. Biol. 2004, 11, 377–394. [Google Scholar] [CrossRef]
- Corvelo, A.; Hallegger, M.; Smith, C.W.J.; Eyras, E. Genome-Wide Association between Branch Point Properties and Alternative Splicing. PLoS Comput. Biol. 2010, 6, e1001016. [Google Scholar] [CrossRef] [Green Version]
- Shibata, A.; Okuno, T.; Rahman, M.A.; Azuma, Y.; Takeda, J.-I.; Masuda, A.; Selcen, D.; Engel, A.G.; Ohno, K. IntSplice: Prediction of the splicing consequences of intronic single-nucleotide variations in the human genome. J. Hum. Genet. 2016, 61, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Divina, P.; Kvitkovicova, A.; Buratti, E.; Vorechovsky, I. Ab initio prediction of mutation-induced cryptic splice-site activation and exon skipping. Eur. J. Hum. Genet. 2009, 17, 759–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, K.H.; Fairbrother, W.G. Spliceman—A computational web server that predicts sequence variations in pre-mRNA splicing. Bioinformatics 2012, 28, 1031–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cartegni, L.; Wang, J.; Zhu, Z.; Zhang, M.Q.; Krainer, A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003, 31, 3568–3571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, P.J.; Zhang, C.; Wang, J.; Chew, S.L.; Zhang, M.Q.; Krainer, A.R. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006, 15, 2490–2508. [Google Scholar] [CrossRef] [Green Version]
- Ke, S.; Shang, S.; Kalachikov, S.M.; Morozova, I.; Yu, L.; Russo, J.J.; Ju, J.; Chasin, L.A. Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res. 2011, 21, 1360–1374. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Rolish, M.E.; Yeo, E.; Tung, V.; Mawson, M.; Burge, C.B. Systematic Identification and Analysis of Exonic Splicing Silencers. Cell 2004, 119, 831–845. [Google Scholar] [CrossRef] [Green Version]
- Erkelenz, S.; Theiss, S.; Otte, M.; Widera, M.; Peter, J.O.; Schaal, H. Genomic HEXploring allows landscaping of novel potential splicing regulatory elements. Nucleic Acids Res. 2014, 42, 10681–10697. [Google Scholar] [CrossRef] [Green Version]
- Fairbrother, W.G.; Yeh, R.-F.; Sharp, P.A.; Burge, C.B. Predictive Identification of Exonic Splicing Enhancers. Science 2002, 297, 1007–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piva, F.; Giulietti, M.; Nocchi, L.; Principato, G. SpliceAid: A database of experimental RNA target motifs bound by splicing proteins in humans. Bioinformatics 2009, 25, 1211–1213. [Google Scholar] [CrossRef]
- Paz, I.; Kosti, I.; Ares, M., Jr.; Cline, M.; Mandel-Gutfreund, Y. RBPmap: A web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014, 42, W361–W367. [Google Scholar] [CrossRef]
- Akerman, M.; David-Eden, H.; Pinter, R.Y.; Mandel-Gutfreund, Y. A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol. 2009, 10, R30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knudsen, B.; Hein, J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003, 31, 3423–3428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markham, N.R.; Zuker, M. UNAFold: Software for Nucleic Acid Folding and Hybridization. Methods Mol. Biol. 2008, 453, 3–31. [Google Scholar] [CrossRef]
- Shapiro, M.B.; Senapathy, P. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987, 15, 7155–7174. [Google Scholar] [CrossRef] [Green Version]
- Desmet, F.-O.; Hamroun, D.; Lalande, M.; Collod-Beroud, G.; Claustres, M.; Béroud, C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009, 37, e67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, S.; Hall, E.; Ast, G. SROOGLE: Webserver for integrative, user-friendly visualization of splicing signals. Nucleic Acids Res. 2009, 37, W189–W192. [Google Scholar] [CrossRef] [Green Version]
- Faber, K.; Glatting, K.-H.; Mueller, P.J.; Risch, A.; Hotz-Wagenblatt, A. Genome-wide prediction of splice-modifying SNPs in human genes using a new analysis pipeline called AASsites. BMC Bioinform. 2011, 12 (Suppl 4), S2. [Google Scholar] [CrossRef] [Green Version]
- Raponi, M.; Kralovicova, J.; Copson, E.; Divina, P.; Eccles, D.M.; Johnson, P.; Baralle, D.; Vorechovsky, I. Prediction of single-nucleotide substitutions that result in exon skipping: Identification of a splicing silencer inBRCA1exon 6. Hum. Mutat. 2011, 32, 436–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samuel, A.L. Some Studies in Machine Learning Using the Game of Checkers. IBM J. Res. Dev. 1959, 3, 210–229. [Google Scholar] [CrossRef]
- Bishop, C.M. Pattern Recognition and Machine Learning; Information Science and Statistics; Springer: New York, YN, USA, 2006. [Google Scholar]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public archive of rela-tionships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef] [Green Version]
- Libbrecht, M.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [Green Version]
- Camacho, D.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-Generation Machine Learning for Biological Networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- Kirke, J.; Jin, X.-L.; Zhang, X.-H. Expression of a Tardigrade Dsup Gene Enhances Genome Protection in Plants. Mol. Biotechnol. 2020, 62, 563–571. [Google Scholar] [CrossRef]
- Gelfman, S.; Wang, Q.; McSweeney, K.M.; Ren, Z.; La Carpia, F.; Halvorsen, M.; Schoch, K.; Ratzon, F.; Heinzen, E.L.; Boland, M.J.; et al. Annotating pathogenic non-coding variants in genic regions. Nat. Commun. 2017, 8, 236. [Google Scholar] [CrossRef]
- Davydov, E.V.; Goode, D.; Sirota, M.; Cooper, G.M.; Sidow, A.; Batzoglou, S. Identifying a High Fraction of the Human Genome to be under Selective Constraint Using GERP++. PLoS Comput. Biol. 2010, 6, e1001025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.; Roos, P.; Sharma, N.; Atalar, M.; Evans, T.A.; Pellicore, M.J.; Davis, E.; Lam, A.-T.N.; Stanley, S.E.; Khalil, S.E.; et al. Systematic Computational Identification of Variants That Activate Exonic and Intronic Cryptic Splice Sites. Am. J. Hum. Genet. 2017, 100, 751–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagadeesh, K.A.; Paggi, J.M.; Ye, J.S.; Stenson, P.D.; Cooper, D.N.; Bernstein, J.A.; Bejerano, G. S-CAP extends pathogenicity prediction to genetic variants that affect RNA splicing. Nat. Genet. 2019, 51, 755–763. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Y.; Hu, Z. Using secondary structure to predict the effects of genetic variants on alternative splicing. Hum. Mutat. 2019, 40, 1270–1279. [Google Scholar] [CrossRef] [Green Version]
- Jaganathan, K.; Panagiotopoulou, S.K.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Pan, Z.; Ying, Y.; Xie, Z.; Adhikari, S.; Phillips, J.; Carstens, R.P.; Black, D.L.; Wu, Y.; Xing, Y. Deep-learning augmented RNA-seq analysis of transcript splicing. Nat. Methods 2019, 16, 307–310. [Google Scholar] [CrossRef]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Nguyen TY, D.; Cygan, K.J.; Çelik, M.H.; Fairbrother, W.G.; Avsec, Ž.; Gagneur, J. MMSplice: Modular modeling improves the predictions of genetic variant effects on splicing. Genome Biol. 2019, 20, 48. [Google Scholar] [CrossRef]
- The 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mort, M.; Sterne-Weiler, T.; Li, B.; Ball, E.V.; Cooper, D.N.; Radivojac, P.; Sanford, J.R.; Mooney, S.D. MutPred Splice: Machine learning-based prediction of exonic variants that disrupt splicing. Genome Biol. 2014, 15, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, H.Y.; Alipanahi, B.; Lee, L.J.; Bretschneider, H.; Merico, D.; Yuen, R.; Hua, Y.; Gueroussov, S.; Najafabadi, H.; Hughes, T.R.; et al. The human splicing code reveals new insights into the genetic determinants of disease. Science 2015, 347, 1254806. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Wang, Z.; Wang, J.; Li, S. SpliceFinder: Ab initio prediction of splice sites using convolutional neural network. BMC Bioinform. 2019, 20 (Suppl. 23), 652. [Google Scholar] [CrossRef] [PubMed]
- Jian, X.; Boerwinkle, E.; Liu, X. In silico tools for splicing defect prediction: A survey from the viewpoint of end users. Genet. Med. 2014, 16, 497–503. [Google Scholar] [CrossRef] [Green Version]
- Coolidge, C.J.; Seely, R.J.; Patton, J.G. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 1997, 25, 888–896. [Google Scholar] [CrossRef] [Green Version]
- Bryen, S.; Joshi, H.; Evesson, F.J.; Girard, C.; Ghaoui, R.; Waddell, L.B.; Testa, A.C.; Cummings, B.; Arbuckle, S.; Graf, N.; et al. Pathogenic Abnormal Splicing Due to Intronic Deletions that Induce Biophysical Space Constraint for Spliceosome Assembly. Am. J. Hum. Genet. 2019, 105, 573–587. [Google Scholar] [CrossRef] [Green Version]
- De Conti, L.; Baralle, M.; Buratti, E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip. Rev. RNA 2013, 4, 49–60. [Google Scholar] [CrossRef]
- Ke, S.; Chasin, L.A. Intronic motif pairs cooperate across exons to promote pre-mRNA splicing. Genome Biol. 2010, 11, R84. [Google Scholar] [CrossRef] [Green Version]
- Dunnen, J.T.D. RNA-Based Variant Detection. In Molecular Diagnostics; Elsevier: Amsterdam, The Netherlands, 2010; pp. 293–298. [Google Scholar]
- Frayling, I.M. Methods of molecular analysis: Mutation detection in solid tumours. Mol. Pathol. 2002, 55, 73–79. [Google Scholar] [CrossRef] [Green Version]
- Baralle, D. Splicing in action: Assessing disease causing sequence changes. J. Med. Genet. 2005, 42, 737–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietz, H.C.; Kendzior, R.J. Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat. Genet. 1994, 8, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Vossen, R.; Dunnen, J.T.D. Protein Truncation Test. Curr. Protoc. Hum. Genet. 2004, 42, 9.11.1–9.11.23. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Wan, R.; Zhang, Q. Application of Reverse Transcription-PCR and Real-Time PCR in Nanotoxicity Research. In Nanotoxicity; Reineke, J., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; Volume 926, pp. 99–112. [Google Scholar]
- He, S.L.; Green, R. Northern Blotting. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2013; Volume 530, pp. 75–87. [Google Scholar]
- Harvey, S.E.; Cheng, C. Methods for Characterization of Alternative RNA Splicing. In Long Non-Coding RNAs; Feng, Y., Zhang, L., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2016; Volume 1402, pp. 229–241. [Google Scholar]
- Freeman, W.M.; Walker, S.J.; Vrana, K.E. Quantitative RT-PCR: Pitfalls and Potential. Biotechniques 1999, 26, 112–125. [Google Scholar] [CrossRef]
- Cooper, T.A. Use of minigene systems to dissect alternative splicing elements. Methods 2005, 37, 331–340. [Google Scholar] [CrossRef]
- Singh, G.; Cooper, T.A. Minigene reporter for identification and analysis of cis elements and trans factors affecting pre-mRNA splicing. Biotechniques 2006, 41, 177–181. [Google Scholar] [CrossRef] [Green Version]
- Qian, X.; Ba, Y.; Zhuang, Q.; Zhong, G. RNA-Seq Technology and Its Application in Fish Transcriptomics. OMICS A J. Integr. Biol. 2014, 18, 98–110. [Google Scholar] [CrossRef] [Green Version]
- Knudsen, S.; Knudsen, S. Guide to Analysis of DNA Microarray Data, 2nd ed.; Wiley-Liss: Hoboken, NJ, USA, 2004. [Google Scholar]
- Al-Haggar, M. Evolving Molecular Methods for Detection of Mutations. Gene Technol. 2013, 2, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Tarca, A.L.; Romero, R.; Draghici, S. Analysis of microarray experiments of gene expression profiling. Am. J. Obstet. Gynecol. 2006, 195, 373–388. [Google Scholar] [CrossRef] [Green Version]
- Jaksik, R.; Iwanaszko, M.; Rzeszowska-Wolny, J.; Kimmel, M. Microarray experiments and factors which affect their reliability. Biol. Direct 2015, 10, 46. [Google Scholar] [CrossRef]
- Haddad, R.; Tromp, G.; Kuivaniemi, H.; Chaiworapongsa, T.; Kim, Y.M.; Mazor, M.; Romero, R. Human spontaneous labor without histologic chorioamnionitis is characterized by an acute inflammation gene expression signature. Am. J. Obstet. Gynecol. 2006, 195, 394–405.e12. [Google Scholar] [CrossRef] [Green Version]
- Maier, T.; Güell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yazaki, J.; Gregory, B.D.; Ecker, J.R. Mapping the genome landscape using tiling array technology. Curr. Opin. Plant Biol. 2007, 10, 534–542. [Google Scholar] [CrossRef] [Green Version]
- Eklund, A.C.; Turner, L.R.; Chen, P.; Jensen, R.V.; Defeo, G.; Kopf-Sill, A.R.; Szallasi, Z. Replacing cRNA targets with cDNA reduces microarray cross-hybridization. Nat. Biotechnol. 2006, 24, 1071–1073. [Google Scholar] [CrossRef] [PubMed]
- Okoniewski, M.J.; Miller, C.J. Hybridization interactions between probesets in short oligo microarrays lead to spurious correlations. BMC Bioinform. 2006, 7, 276. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.A.; Dehler, C.E.; Krol, E. Transcriptomic responses in the fish intestine. Dev. Comp. Immunol. 2016, 64, 103–117. [Google Scholar] [CrossRef]
- Kukurba, K.R.; Montgomery, S. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, pdb.top084970. [Google Scholar] [CrossRef] [Green Version]
- Chmel, N.; Danescu, S.; Gruler, A.; Kiritsi, D.; Bruckner-Tuderman, L.; Kreuter, A.; Kohlhase, J.; Has, C. A Deep-Intronic FERMT1 Mutation Causes Kindler Syndrome: An Explanation for Genetically Unsolved Cases. J. Investig. Dermatol. 2015, 135, 2876–2879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeidian, A.H.; Youssefian, L.; Vahidnezhad, H.; Uitto, J. Research Techniques Made Simple: Whole-Transcriptome Sequencing by RNA-Seq for Diagnosis of Monogenic Disorders. J. Investig. Dermatol. 2020, 140, 1117–1126.e1. [Google Scholar] [CrossRef]
- Zeng, W.; Mortazavi, A. Technical considerations for functional sequencing assays. Nat. Immunol. 2012, 13, 802–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Whitley, S.K.; Horne, W.T.; Kolls, J.K. Research Techniques Made Simple: Methodology and Clinical Applications of RNA Sequencing. J. Investig. Dermatol. 2016, 136, e77–e82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, M.S.; Van Vleet, T.R.; Ciurlionis, R.; Buck, W.R.; Mittelstadt, S.W.; Blomme, E.A.G.; Liguori, M.J. Comparison of RNA-Seq and Microarray Gene Expression Platforms for the Toxicogenomic Evaluation of Liver from Short-Term Rat Toxicity Studies. Front. Genet. 2019, 9, 636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eilertsen, I.A.; Moosavi, S.H.; Strømme, J.M.; Nesbakken, A.; Johannessen, B.; Lothe, R.A.; Sveen, A. Technical differences between sequencing and microarray platforms impact transcriptomic subtyping of colorectal cancer. Cancer Lett. 2019, 469, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Roest, P.A.; Roberts, R.; van der Tuijn, A.C.; Heikoop, J.C.; van Ommen, G.-J.B.; Dunnen, J.T.D. Protein truncation test (PTT) to rapidly screen the DMD gene for translation terminating mutations. Neuromuscul. Disord. 1993, 3, 391–394. [Google Scholar] [CrossRef]
- Hauss, O.; Müller, O. The Protein Truncation Test in Mutation Detection and Molecular Diagnosis. In In Vitro Transcription and Translation Protocols; Grandi, G., Ed.; Humana Press: Totowa, NJ, USA, 2007; pp. 151–164. [Google Scholar]
- Gite, S.; Lim, M.; Carlson, R.; Olejnik, J.; Zehnbauer, B.; Rothschild, K. A high-throughput nonisotopic protein truncation test. Nat. Biotechnol. 2003, 21, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Denier, C.; Labauge, P.; Brunereau, L.; Cavé-Riant, F.; Marchelli, F.; Arnoult, M.; Cecillon, M.; Maciazek, J.; Joutel, A.; Tournier-Lasserve, E.; et al. Clinical features of cerebral cavernous malformations patients withKRIT1mutations. Ann. Neurol. 2003, 55, 213–220. [Google Scholar] [CrossRef]
- Canson, D.; Glubb, D.; Spurdle, A.B. Variant effect on splicing regulatory elements, branchpoint usage, and pseudoexonization: Strategies to enhance bioinformatic prediction using hereditary cancer genes as exemplars. Hum. Mutat. 2020, 41, 1705–1721. [Google Scholar] [CrossRef]
- Agiannitopoulos, K.; Pepe, G.; Papadopoulou, E.; Tsaousis, G.N.; Kampouri, S.; Maravelaki, S.; Fassas, A.; Christodoulou, C.; Iosifidou, R.; Karageorgopoulou, S.; et al. Clinical Utility of Functional RNA Analysis for the Reclassification of Splicing Gene Variants in Hereditary Cancer. Cancer Genom. Proteom. 2021, 18, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Soukarieh, O.; Gaildrat, P.; Hamieh, M.; Drouet, A.; Baert-Desurmont, S.; Frébourg, T.; Tosi, M.; Martins, A. Exonic Splicing Mutations Are More Prevalent than Currently Estimated and Can Be Predicted by Using In Silico Tools. PLoS Genet. 2016, 12, e1005756. [Google Scholar] [CrossRef] [Green Version]
- Ahlborn, L.; Dandanell, M.; Steffensen, A.Y.; Jønson, L.; Nielsen, M.D.; Hansen, T.V.O. Splicing analysis of 14 BRCA1 missense variants classifies nine variants as pathogenic. Breast Cancer Res. Treat. 2015, 150, 289–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci, C.; Riolo, G.; Battistini, S. Molecular genetic analysis of cerebral cavernous malformations: An update. Vessel. Plus 2021, 5. [Google Scholar] [CrossRef]






| Splice Sites | Nucleotides |
|---|---|
| 5’ splice site | CAG/GUAAGU |
| Branch point sequence | YUNAY |
| Polypyrimidine tract | Yn |
| 3’ splice site | NYAG/G |
| Method | Main Characteristics |
|---|---|
| Regression |
|
| Classification |
|
| Support Vector Machine (SVM) |
|
| Decision Tree |
|
| Random Forest |
|
| Neural Network (NN) |
|
| Deep Neural Networks (DNNs) |
|
| Convolutional Neural Networks (CNN) |
|
| Microarray | Rna-Seq | |
|---|---|---|
| Benefits |
|
|
| Drawbacks |
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riolo, G.; Cantara, S.; Ricci, C. What’s Wrong in a Jump? Prediction and Validation of Splice Site Variants. Methods Protoc. 2021, 4, 62. https://doi.org/10.3390/mps4030062
Riolo G, Cantara S, Ricci C. What’s Wrong in a Jump? Prediction and Validation of Splice Site Variants. Methods and Protocols. 2021; 4(3):62. https://doi.org/10.3390/mps4030062
Chicago/Turabian StyleRiolo, Giulia, Silvia Cantara, and Claudia Ricci. 2021. "What’s Wrong in a Jump? Prediction and Validation of Splice Site Variants" Methods and Protocols 4, no. 3: 62. https://doi.org/10.3390/mps4030062