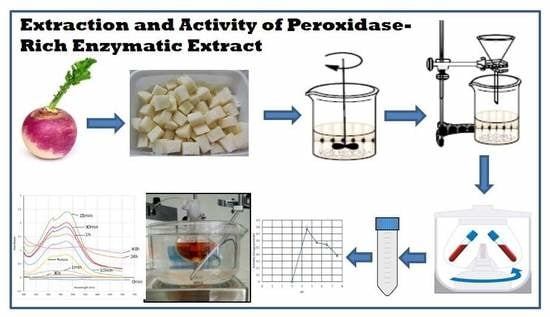
A Green and Simple Protocol for Extraction and Application of a Peroxidase-Rich Enzymatic Extract
Abstract
:1. Introduction
2. Experimental Design
2.1. Materials
2.1.1. Preparation of Enzymatic Extract
- Fresh turnip (Brassica rapa var. rapa Barkant) roots.
- Phosphate buffer 100 mM pH 6.5.
- Cotton wool.
2.1.2. Protein Quantification
- Bradford Reagent
- Bovine Serum Albumin (BSA) (Sigma, Darmstadt, Germany)
2.1.3. Biotransformation Reaction
- 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) (Sigma, Darmstadt, Germany).
- Phosphate buffer 300 mM pH 5.5, 6.5 and 7.5.
- Acetate buffer 300 mM pH 3.5 and 4.5.
- Hydrogen peroxide (H2O2) (Fluka, Charlotte, NC, USA).
- Dimethyl sulfoxide (DMSO) (Sigma, Darmstadt, Germany).
- Acetone (Sigma, Darmstadt, Germany).
- Guaiacol (2-methoxyphenol) (Sigma, Darmstadt, Germany).
2.2. Equipment
- Eppendorf 5810R centrifuge (Eppendorf—Hamburg, Germany).
- EVOLUTION 160 UV–Vis Thermo Scientific thermostated spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
3. Procedure
3.1. Preparation of Enzymatic Extract. Time for Completion: 00:15 h
- Wash and peel the turnip roots.
- Dice 20 g of the peeled turnip roots.
- Add 100 mL of cold phosphate buffer 100 mM, pH 6.5 to the diced roots.
- Homogenize the mixture, immersed in ice using a commercial blender, until no lumps are observed.
- Filter the homogenized mixture with cotton wool to remove suspended fibrous solid particles.
- Centrifuge the filtrate at 18,514× g for 5 min at 4 °C.
- Collect the supernatant and store it in ice.
PAUSE STEP If the extract prepared is not going to be readily used, it can be stored at −80 °C without loss of activity.
3.2. Protein Quantification—Bradford Assay. Time for Completion: 00:30 h
- Prepare BSA dilutions for standards (from 100–1000 µg/mL).
- Use phosphate buffer 100 mM, pH 6.5 as a blank (0 µg/mL).
- Mix 2500 µL of Bradford protein-dye solution with standards or enzymatic extract samples.
- Incubate for 10 min.
- Measure the absorbance at 595 nm.
- Plot a concentration/absorbance curve with the values obtained for standards.
- Substitute the absorbance values of the extract samples on the equation of the curve to obtain the total protein concentration.
3.3. Biotransformation Reaction
3.3.1. Assessment of Extract Activity. Time for Completion: 00:05 h
- On two 3 mL cuvettes, add 2150 μL of acetate buffer 300 mM (pH 4.5), each.
CRITICAL STEP All reagents and samples need to be brought to 35 °C in advance, to prevent temperature fluctuations during the test.
- Proceed to add 100 μL ABTS (0.7 mM in acetate buffer 300 mM pH 4.5) to both cuvettes.OPTIONAL STEP Other colored substrates can be used, as long as they react with the enzymatic extract, resulting in a change in absorbance.
- Add 50 μL of H2O2 0.3% (v/v) in acetate buffer 300 mM (pH 4.5) to both cuvettes and mix them thoroughly.
- Put the cuvette in the thermostated spectrophotometer, with the temperature set to 35 °C, and let it remain there for 2–3 min, to reach temperature equilibrium.
- To the first cuvette, add 50 μL of acetate buffer 300 mM (pH 4.5), and set the blank in the spectrophotometer, at 414 nm, with the temperature set to 35 °C.
- To the remaining cuvette, add 50 μL of the enzymatic extract and shake the mixture.
- Immediately after the addition of the extract, start registering the absorbance in the thermostated spectrophotometer, always with the temperature set to 35 °C.
CRITICAL STEP To obtain a more correct assessment of the enzymatic activity, the absorbance has to be registered from the moment the reaction starts, so step 6 must be done in the fastest way possible.
- Follow the absorbance of the sample at 414 nm, for 90 s.
- With the data obtained, plot a curve which will allows calculating the enzymatic activity of the extract.OPTIONAL STEP All the reaction parameters (temperature, pH, etc.) described are optimal for this enzymatic extract activity to the substrate ABTS. However, if there is the need to test the effect of changing these parameters in the activity of the enzymatic extract or, if a different substrate is used, the working parameters could be reassessed using the same methodology used with ABTS.
3.3.2. Biotransformation. Time for Completion: 48:00 h
- Set a water bath to 35 °C.
- Prepare 20 mL of substrate at a concentration of 7.25 mM in acetate buffer 300 mM (pH 4.5). Other volumes can be used, if needed.OPTIONAL STEP If the substrate is not soluble under these conditions, prepare the substrate with an appropriate solvent and add the buffer to obtain a mixture where the enzymatic extract retains if not all, at least a % of its activity. This proportion can be assessed by the procedure described in Section 3.3.1.
- Gradually add 0.4 mL of H2O2 (9.43 mM) to the reaction mixture every 8 h, to a final volume of 2.4 mL.
CRITICAL STEP The concentration of H2O2 is higher than substrate concentration to prevent H2O2 from becoming a limiting step. The addition of this reactive should be performed in small portions during the reaction, rather than adding all in the beginning. This is due to the fact that adding a large amount at once may cause the decomposition of the prosthetic heme and also modification of the higher structure of peroxidase, both essential for enzyme activity [31].
OPTION Other time intervals for adding H2O2 can be chosen, depending on the laboratory equipment to ensure the gradual and fractionated addition of the reactive, like peristaltic pumps. - Start the reaction by adding 750 μL of turnip enzymatic extract and repeat every 12 h, to ensure that there are always active molecules of the enzyme in the reaction medium.OPTION The addition of enzymatic extract and H2O2 can be done simultaneously, but it also depends on laboratory equipment. In fact, in this work the conditions chosen were limited by only one peristaltic pump with a timer, used to the hydrogen peroxide addition, while enzyme addition was performed manually. Considering that in the blank reaction, performed with peroxide but not enzymatic extract, there were no changes in the reactional medium, the conditions chosen did not affect the development of the catalytic reaction.
- Stop the reaction after 48 h.
4. Expected Results
4.1. Properties of Enzymatic Extract
4.2. Assessment of Extract Activity in Different Conditions
4.3. Biotransformation Reaction
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dunn, P.J. The importance of green chemistry in process research and development. Chem. Soc. Rev. 2012, 41, 1452–1461. [Google Scholar] [CrossRef] [PubMed]
- Anastas, P.; Warner, J. Green Chemistry: Theory and Practice; Oxford University Press: New York, NY, USA, 1998; ISBN 9780198506980. [Google Scholar]
- Mulvihill, M.J.; Beach, E.S.; Zimmerman, J.B.; Anastas, P.T. Green chemistry and green engineering: A framework for sustainable technology development. Annu. Rev. Environ. Resour. 2011, 36, 271–293. [Google Scholar] [CrossRef] [Green Version]
- Ivanković, A.; Dronjić, A.; Bevanda, A.M.; Talić, S. Review of 12 principles of green chemistry in practice. Int. J. Green Energy 2017, 6, 39–48. [Google Scholar] [CrossRef] [Green Version]
- Chaudhari, R.V. Fundamentals of homogeneous catalysis. In Industrial Catalytic Processes for Fine and Specialty Chemicals; Joshi, S.S., Ranade, V.V., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 17–39. [Google Scholar] [CrossRef]
- Unnikrishnan, P.; Srinivas, D. Heterogeneous catalysis. In Industrial Catalytic Processes for Fine and Specialty Chemicals; Joshi, S.S., Ranade, V.V., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 41–111. [Google Scholar] [CrossRef]
- Faber, K. Biotransformations in Organic Chemistry: A Textbook; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar] [CrossRef]
- Robinson, P.K. Enzymes: Principles and biotechnological applications. Essays Biochem. 2015, 59, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Divakkar, S. Enzymatic Transformation; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar] [CrossRef]
- Mohamad, R.; Marzuki, H.C.; Buang, A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Tosa, T.; Mori, T.; Fuse, N.; Chibata, I. Studies on continuous enzyme reactions. I. Screening of carriers for preparation of water-insoluble aminoacylase. Enzymologia 1966, 31, 214–224. [Google Scholar] [PubMed]
- Hernandez, K.; Fernandez-Lafuente, R. Control of protein immobilization: Coupling immobilization and side-directed mutagenesis to improve biocatalyst or biosensor performance. Enzyme Microb. Technol. 2011, 48, 107–122. [Google Scholar] [CrossRef]
- Jesionowski, T.; Zdarta, J.; Krajewska, B. Enzyme immobilization by adsorption: A review. Adsorption 2014, 20, 801–821. [Google Scholar] [CrossRef] [Green Version]
- Basso, A.; Serban, S. Industrial applications of immobilized enzymes—A review. Mol. Catal. 2019, 479, 110607. [Google Scholar] [CrossRef]
- Singh, R.S.; Singh, T.; Pandey, A. Microbial enzymes—An overview. In Biomass, Biofuels and Biochemical: Advances in Enzyme Technology; Singh, R.S., Singhania, R.R., Pandey, A., Larroche, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–40. [Google Scholar] [CrossRef]
- Yadav, M.; Yadav, H.S. Applications of ligninolytic enzymes to pollutants, wastewater, dyes, soil, coal, paper and polymers. Environ. Chem. Lett. 2015, 13, 309–318. [Google Scholar] [CrossRef]
- Osuji, A.C.; Eze, S.O.O.; Osayi, E.E.; Chilaka, F.C. Biobleaching of industrial important dyes with peroxidase partially purified from garlic. Sci. World J. 2014, 2014, 183163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duarte-Vázquez, M.A.; Ortega-Tovar, M.A.; García-Almendarez, B.E.; Regalado, C. Removal of aqueous phenolic compounds from a model system by oxidative polymerization with turnip (Brassica napus L var. purple top white globe) peroxidase. J. Chem. Technol. Biotechnol. 2003, 78, 42–47. [Google Scholar] [CrossRef]
- Quintanilla-Guerrero, F.; Duarte-Vázquez, M.A.; García-Almendarez, B.E.; Tinoco, R.; Vazquez-Duhalt, R.; Regalado, C. Polyethylene glycol improves phenol removal by immobilized turnip peroxidase. Bioresour. Technol. 2008, 99, 8605–8611. [Google Scholar] [CrossRef] [PubMed]
- Matto, M.; Husain, Q. Decolorization of direct dyes by immobilized turnip peroxidase in batch and continuous processes. Ecotoxicol. Environ. Saf. 2009, 72, 965–971. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Corrêa, A.D.; Amorim, M.T.S.P.; Parpot, P.; Torres, J.A.; Chagas, P.M.B. Decolorization of the phthalocyanine dye reactive blue 21 by turnip peroxidase and assessment of its oxidation products. J. Mol. Catal. B Enzym. 2012, 77, 9–14. [Google Scholar] [CrossRef] [Green Version]
- Pandey, V.P.; Awasthi, M.; Singh, S.; Tiwari, S.; Dwivedi, U.N. A comprehensive review on function and application of plant peroxidases. Biochem. Anal. Biochem. 2017, 6, 308. [Google Scholar] [CrossRef]
- Krainer, F.W.; Glieder, A. An updated view on horseradish peroxidases: Recombinant production and biotechnological applications. Appl. Microbiol. Biotechnol. 2015, 99, 1611–1625. [Google Scholar] [CrossRef] [Green Version]
- Eggenreich, B.; Willim, M.; Wurm, D.J.; Herwig, C.; Spadiut, O. Production strategies for active heme-containing peroxidases from E. coli inclusion bodies—A review. Biotechnol. Rep. 2016, 10, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, S.F.; da Luz, J.M.R.; Kasuya, M.C.M.; Ladeira, L.O.; Junior, A.C. Enzymatic extract containing lignin peroxidase immobilized on carbon nanotubes: Potential biocatalyst in dye decolourization. Saudi J. Biol. Sci. 2018, 25, 651–659. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Penner, M.H. Selective oxidation of enzyme extracts for improved quantification of peroxidase activity. Anal. Biochem. 2015, 476, 20–25. [Google Scholar] [CrossRef]
- Enachi, E.; Grigore-Gurgu, L.; Aprodu, I.; Stănciuc, N.; Dalmadi, I.; Bahrim, G.; Râpeanu, G.; Croitoru, C. Extraction, purification and processing stability of peroxidase from plums (Prunus domestica). Int. J. Food Prop. 2018, 21, 2744–2757. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Xi, Y.; Luo, X.-Y.; Ni, H.; Li, H.-H. Preparation of peroxidase and phenolics using discarded sweet potato old stems. Sci. Rep. 2019, 9, 3769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkesman, J.; Castro, D.; Contreras, L.M.; Kurz, L. Guaiacol peroxidase zymography for the undergraduate laboratory. Biochem. Mol. Biol. Educ. 2014, 42, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Akita, M.; Tsutsumi, D.; Kobayashi, M.; Kise, H. Structural change and catalytic activity of horseradish peroxidase in oxidative polymerization of phenol. Biosci. Biotechnol. Biochem. 2001, 65, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Motamed, S.; Ghaemmaghami, F.; Alemzadeh, I. Turnip (Brassica rapa) peroxidase: Purification and characterization. Ind. Eng. Chem. Res. 2009, 48, 10614–10618. [Google Scholar] [CrossRef]
- Rathnamsamy, S.; Singh, R.; Auxilia, R.; Vedhahari, B.N. Extraction of peroxidase from various plant sources and its biodegradation studies on phenolic compounds. BTAIJ 2014, 9, 160–165, ISSN 0974-7435. [Google Scholar]
- Saboora, A.; Parsiavash, L.; Moosavi-Nejad, Z. Purification and kinetic properties of guaiacol peroxidase in turnip (Brassica napus var. okapi) root during different growth stages. Prog. Biol. Sci. 2012, 2, 76–86. [Google Scholar] [CrossRef]
- Doerge, D.R.; Divi, R.L.; Churchwell, M.I. Identification of the colored guaiacol oxidation product produced by peroxidases. Anal. Biochem. 1997, 250, 10–17. [Google Scholar] [CrossRef]
- Lopes, L.C.; Brandão, I.V.; Sánchez, O.C.; Franceschi, E.; Borges, G.; Dariva, C.; Fricks, A.T. Horseradish peroxidase biocatalytic reaction monitoring using Near-Infrared (NIR) Spectroscopy. Process Biochem. 2018, 71, 127–133. [Google Scholar] [CrossRef]
- Diederix, R.E.M.; Ubbink, M.; Canters, G.W. The peroxidase activity of cytochrome c-550 from Paracoccus versutus. Eur. J. Biochem. 2001, 268, 4207–4216. [Google Scholar] [CrossRef] [PubMed]








| Extraction | Season | Total Protein (μg/mL Extract) | Total Activity (U/μg Protein) |
|---|---|---|---|
| 1 | Autumn | 349 ± 11 | 0.77 ± 0.05 |
| 2 | Autumn | 358 ± 15 | 0.81 ± 0.02 |
| 3 | Autumn | 353 ± 26 | 0.79 ± 0.06 |
| 4 | Autumn | 351 ± 18 | 0.80 ± 0.03 |
| 5 | Winter | 201 ± 6 | 0.54 ± 0.01 |
| 6 | Winter | 205 ± 6 | 0.57 ± 0.05 |
| 7 | Winter | 211 ± 13 | 0.60 ± 0.02 |
| 8 | Winter | 210 ± 4 | 0.59 ± 0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosa, G.P.; Barreto, M.d.C.; Pinto, D.C.G.A.; Seca, A.M.L. A Green and Simple Protocol for Extraction and Application of a Peroxidase-Rich Enzymatic Extract. Methods Protoc. 2020, 3, 25. https://doi.org/10.3390/mps3020025
Rosa GP, Barreto MdC, Pinto DCGA, Seca AML. A Green and Simple Protocol for Extraction and Application of a Peroxidase-Rich Enzymatic Extract. Methods and Protocols. 2020; 3(2):25. https://doi.org/10.3390/mps3020025
Chicago/Turabian StyleRosa, Gonçalo P., Maria do Carmo Barreto, Diana C. G. A. Pinto, and Ana M. L. Seca. 2020. "A Green and Simple Protocol for Extraction and Application of a Peroxidase-Rich Enzymatic Extract" Methods and Protocols 3, no. 2: 25. https://doi.org/10.3390/mps3020025