Surface Mesh Reconstruction from Cardiac MRI Contours †
Abstract
:1. Introduction
- Development of a surface reconstruction method capable of dealing with non-parallel, cross-sectional, sparse, heterogeneous, and non-coincidental contours,
- Use of our technique on the surfaces of the left ventricle as well as on the right ventricle,
- Evaluation of our method on several realistic shapes generated from a statistical shape model, as well as on real data data consisting of normal and abnormal pathologies.
State of the Art
2. Materials and Methods
2.1. Synthetic and Real Data
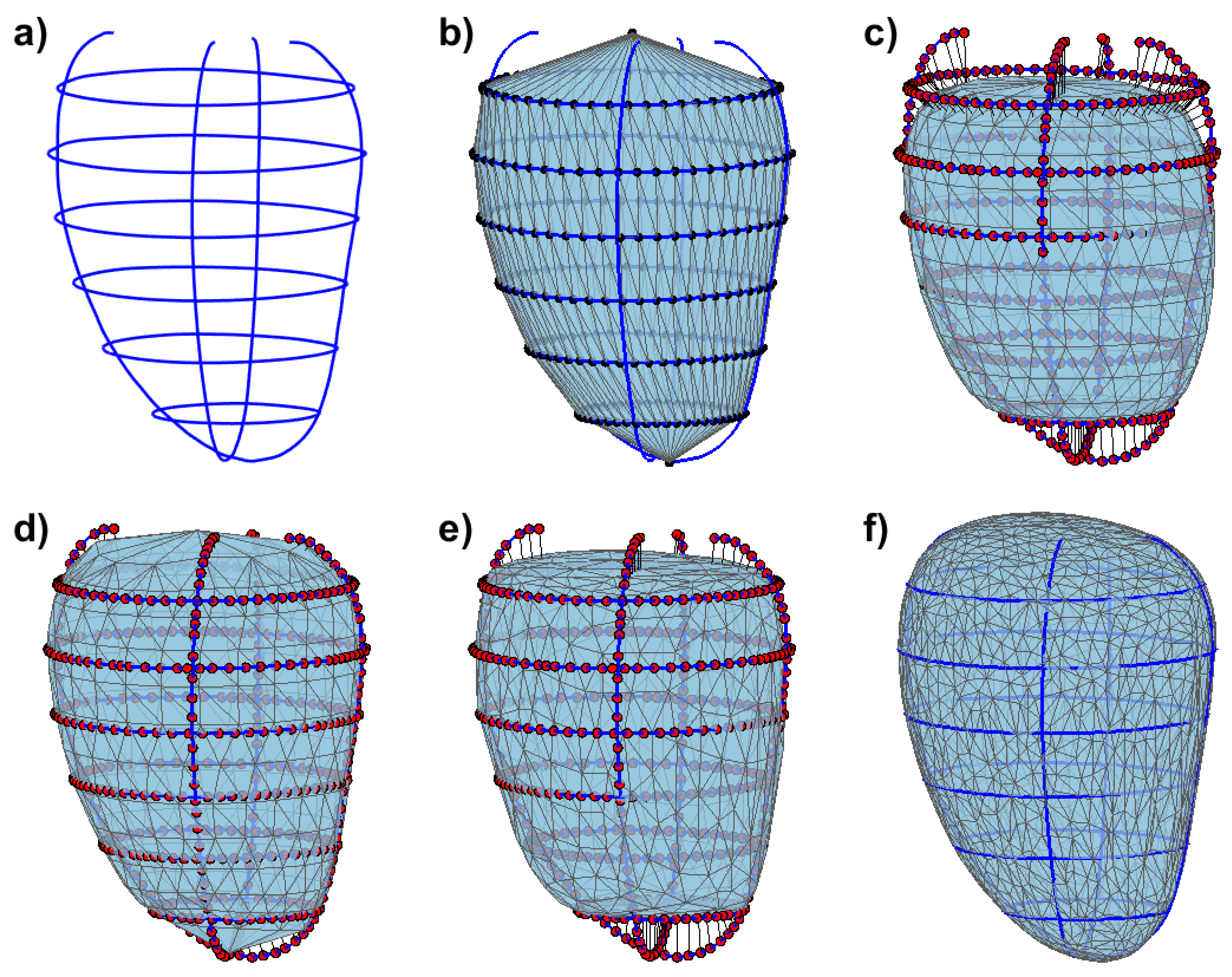
2.2. Initial Meshes
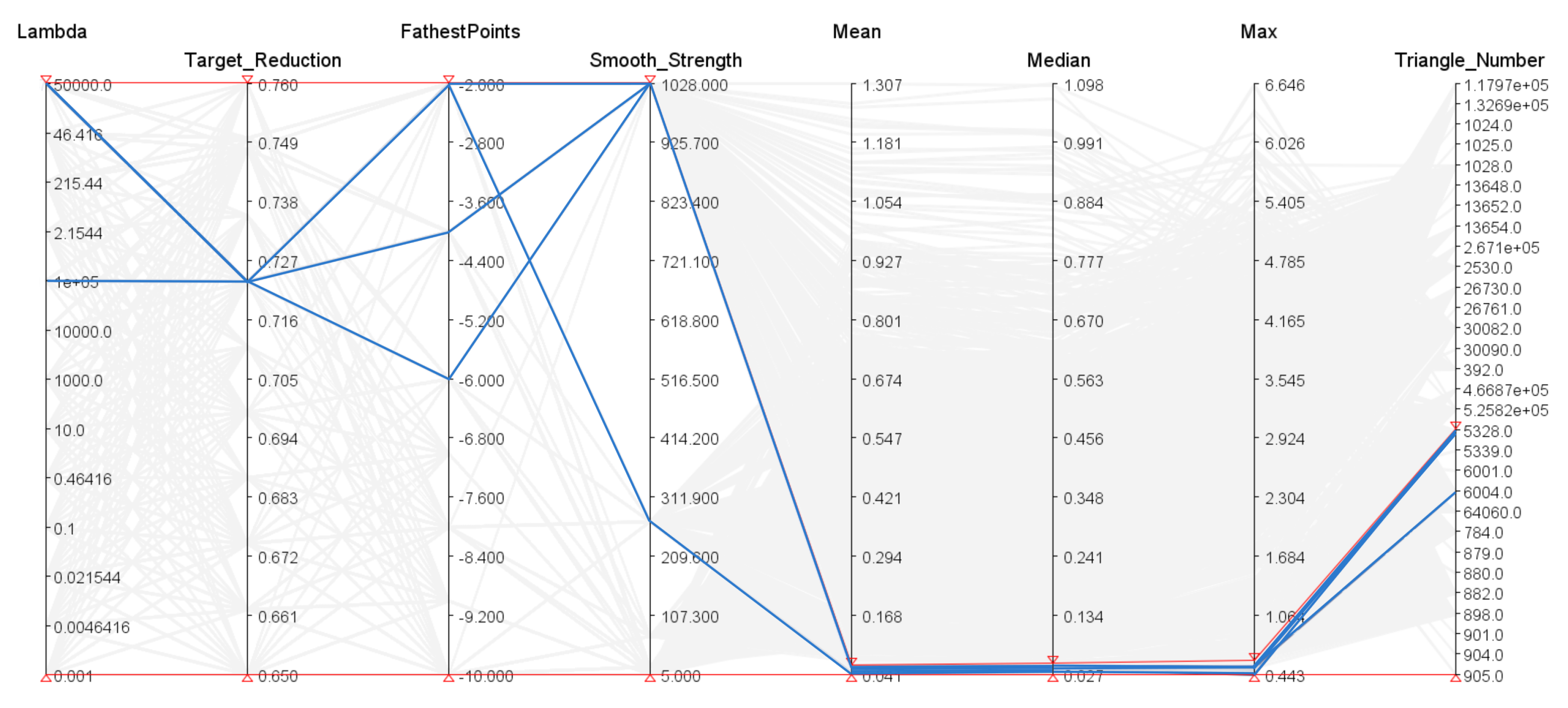
2.3. Mesh Deformation
3. Results
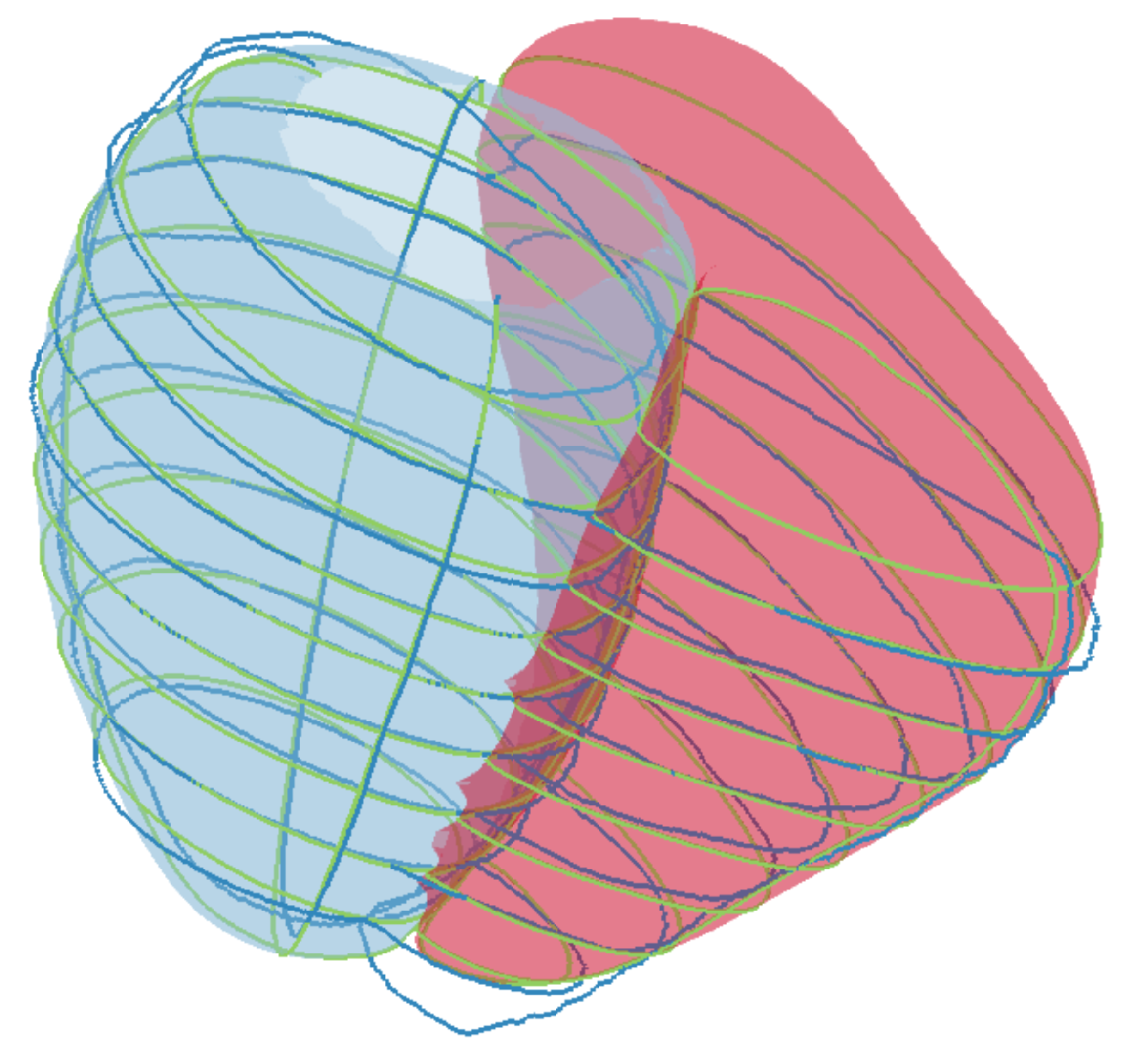
3.1. Simulation of CMR Acquisitions
3.2. Synthetic Data
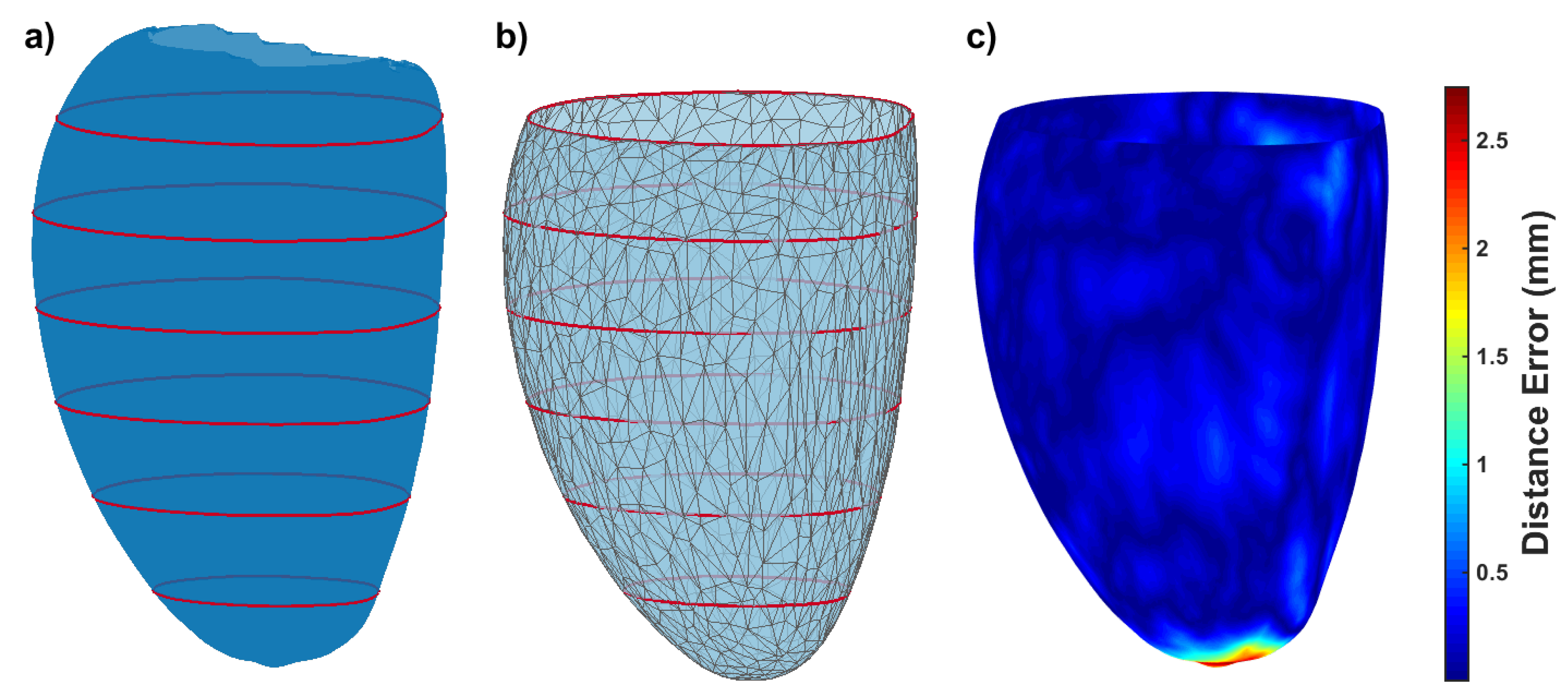
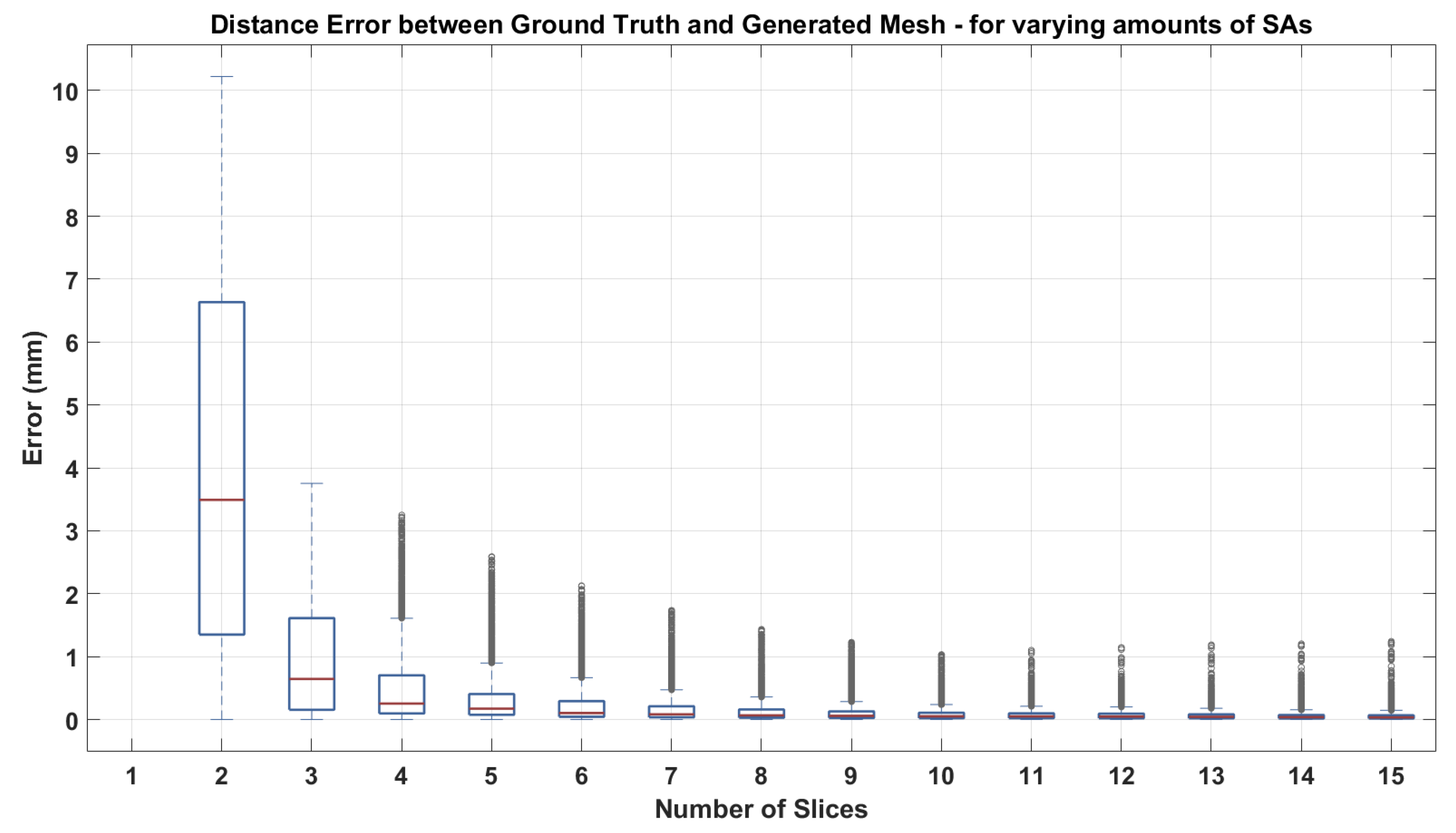
3.2.1. Short Axis Contours
3.2.2. Short Axis and Long Axis Contours
3.2.3. Non-Parallel, Non-Coincidental Contours
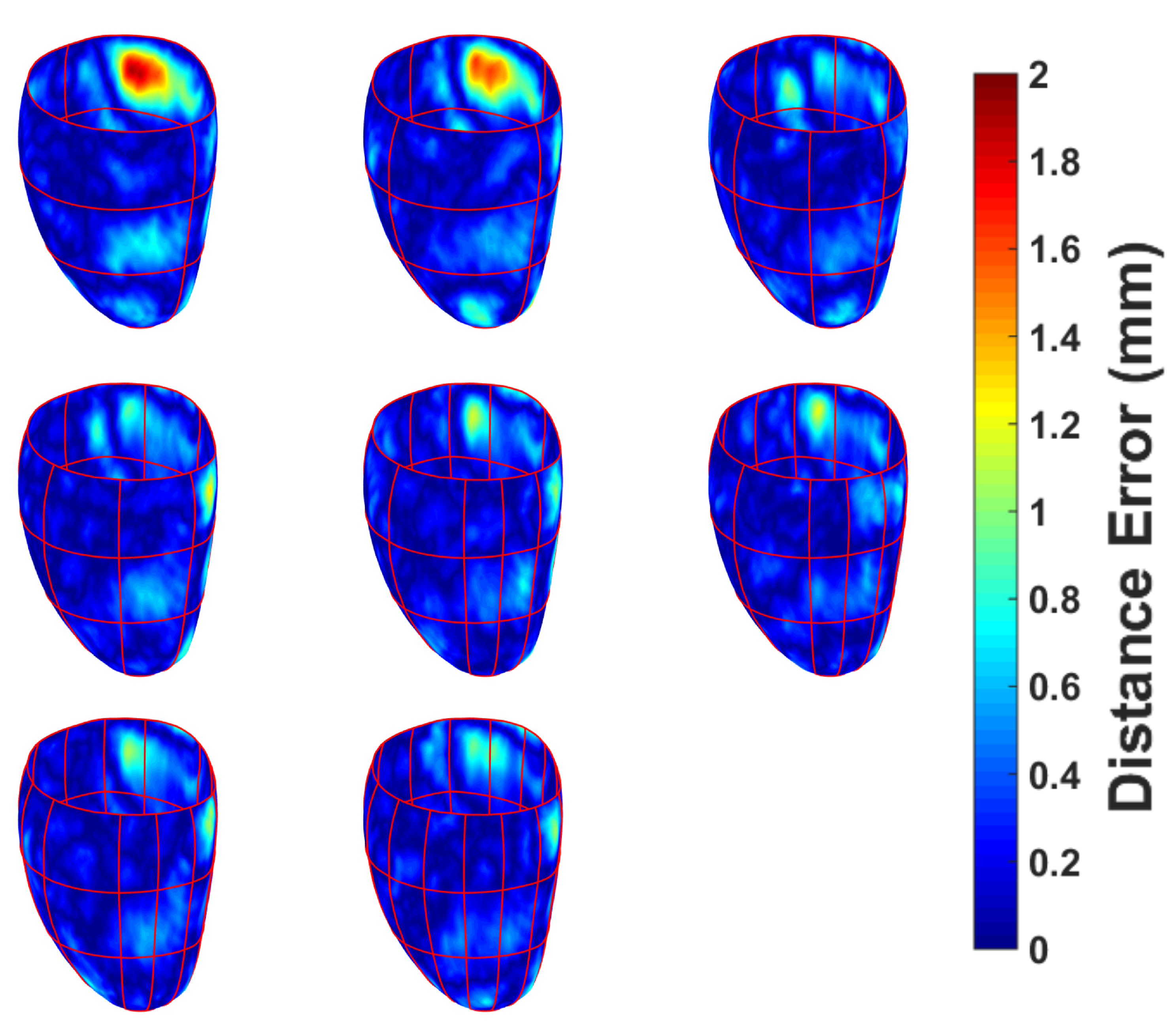
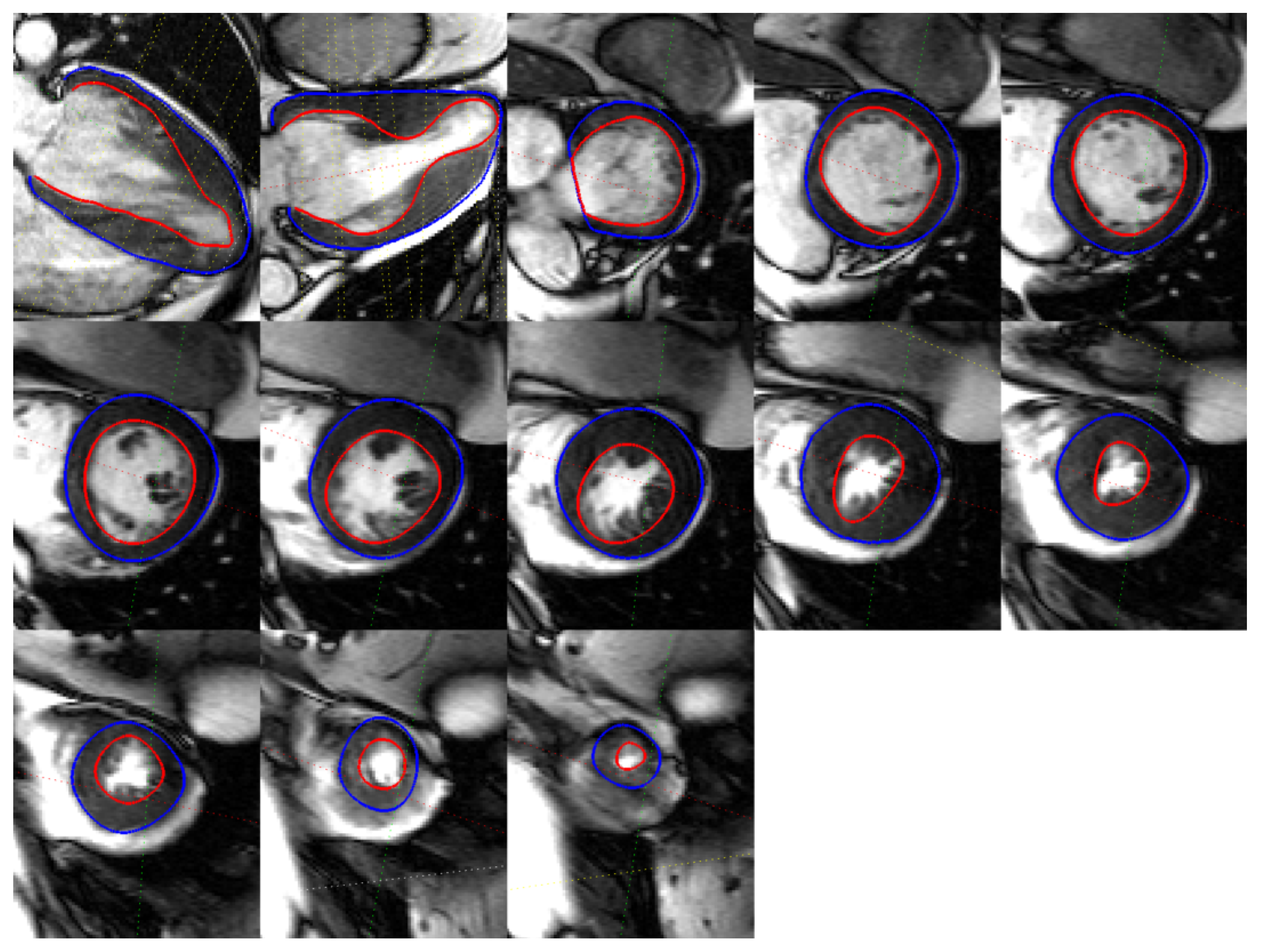
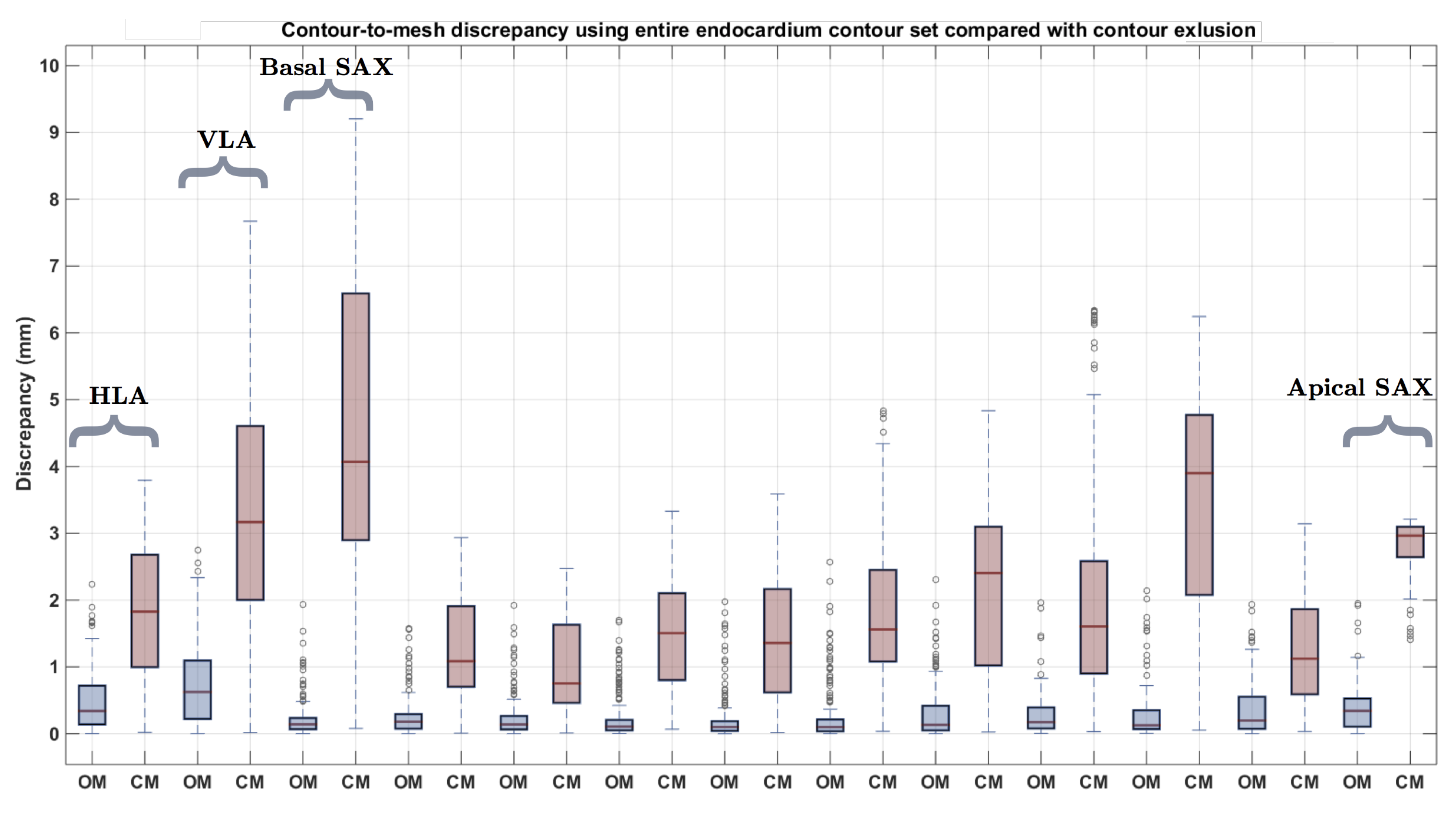
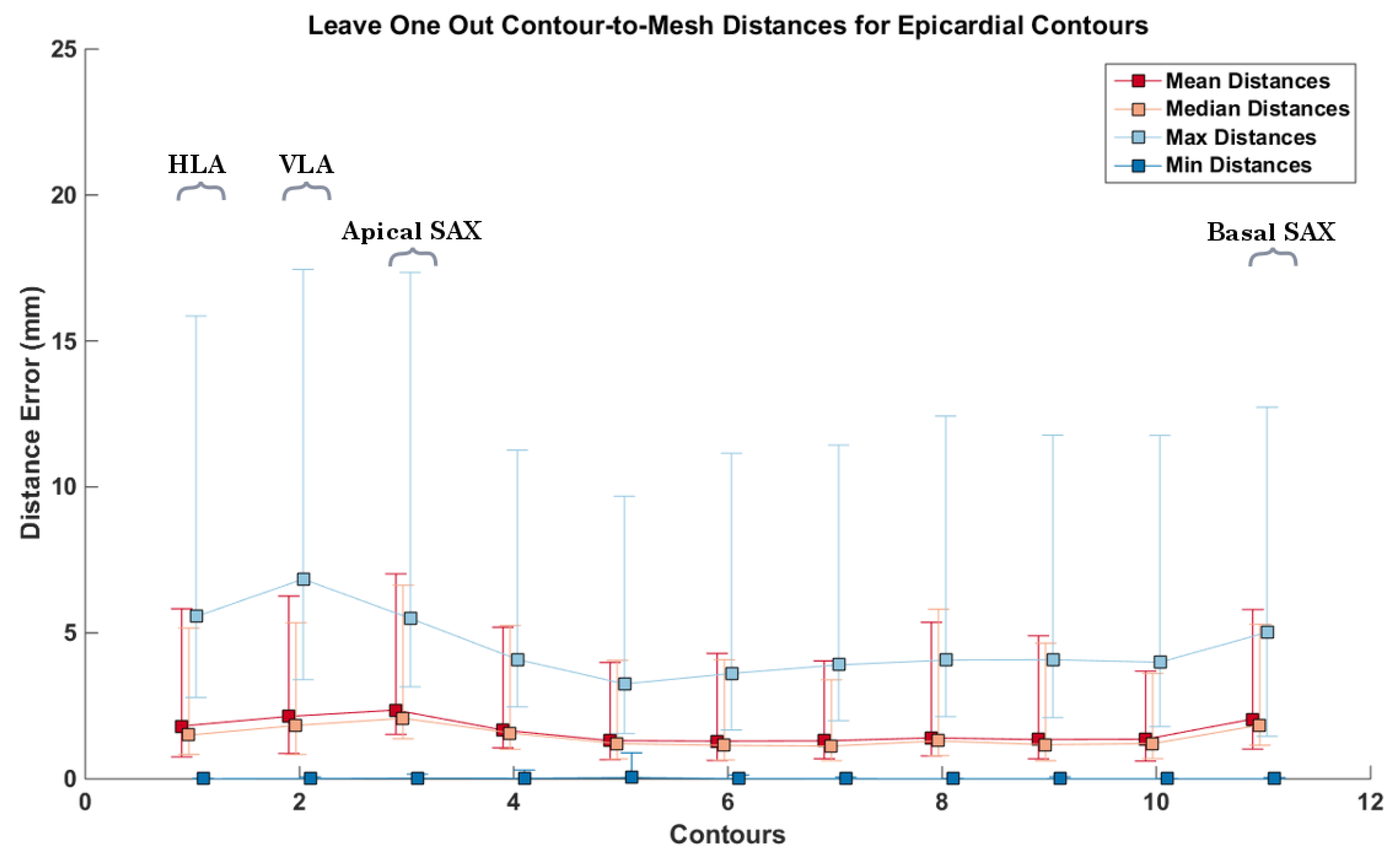
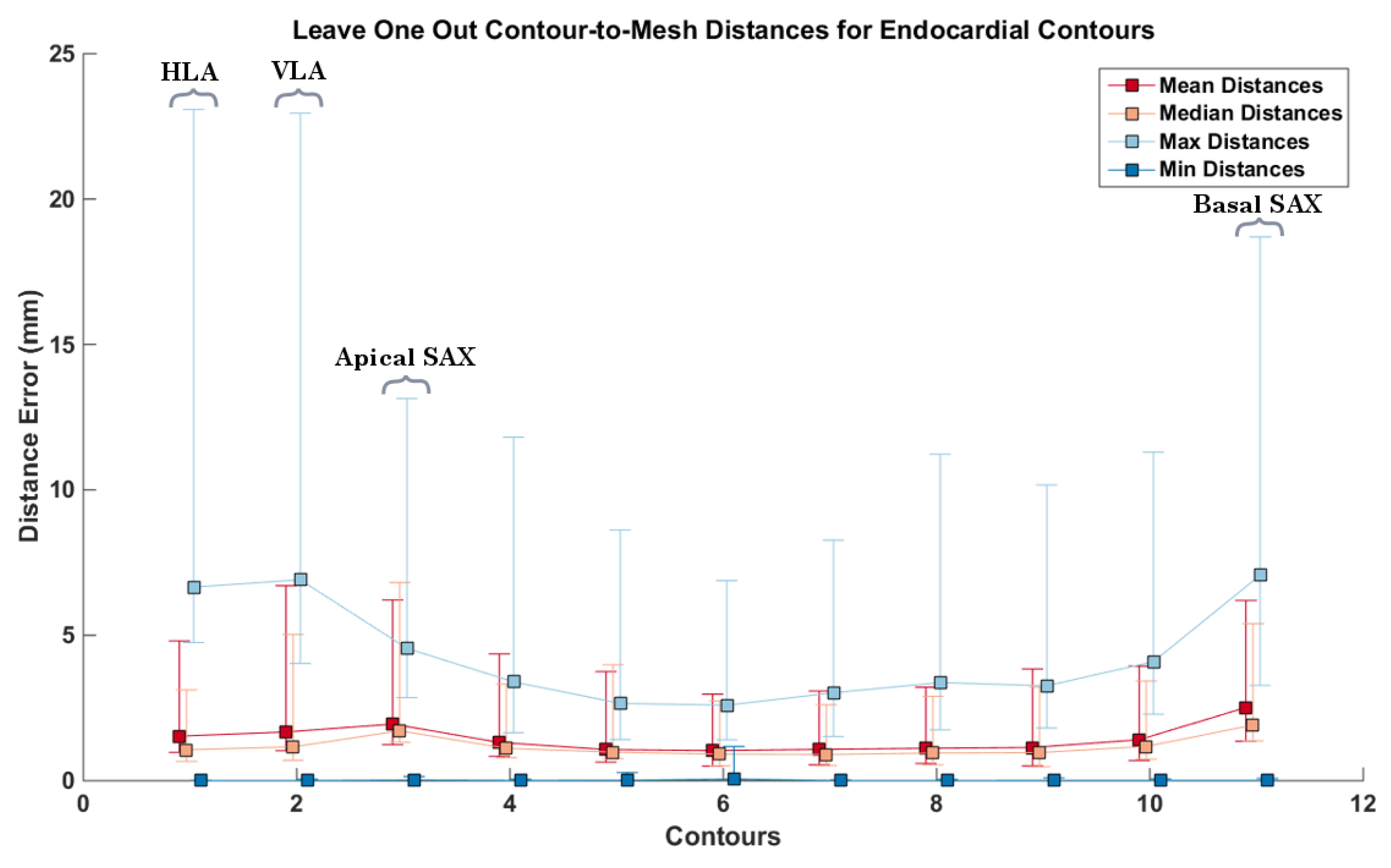
3.3. CMR Acquired Data
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MRI | Magnetic resonance imaging |
| CMR | Cardiac magnetic resonance |
| SAX | Short axis slice |
| LAX | Long axis slice |
| TPS | Thin plate spline |
| SSM | Statistical shape model |
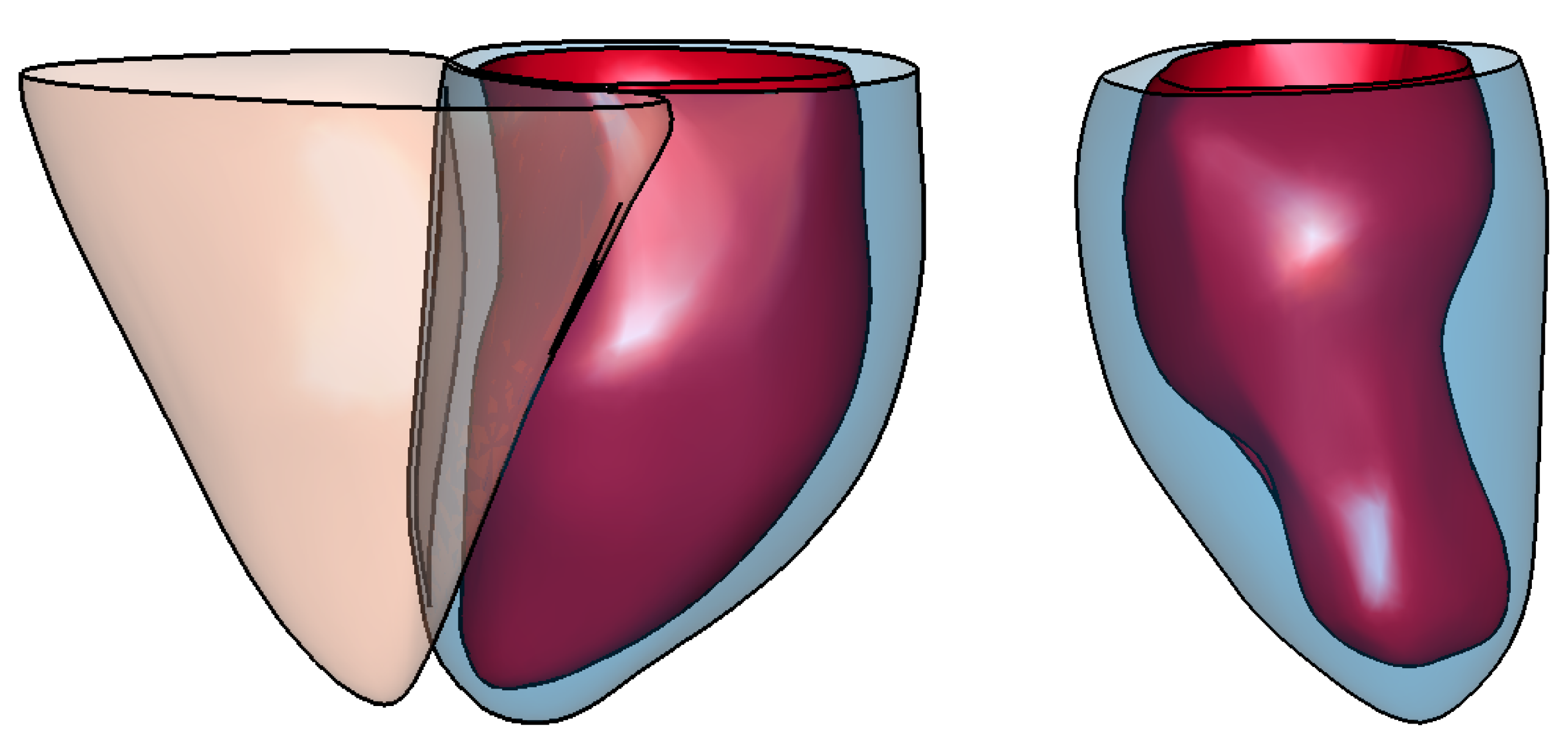
Appendix A. Illustration of Extreme Cases
Appendix B. Simulation of CMR Acquisitions

References
- Arevalo, H.; Vadakkumpadan, F.; Guallar, E.; Jebb, A.; Malamas, P.; Wu, K.; Trayanova, N. Arrhythmia risk stratification of patients after myocardial infarction using personalized heart models. Nat. Commun. 2016, 7, 11437. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Holloway, M.; Carr, N.; Ju, T. Topology-Constrained Surface Reconstruction From Cross-sections. ACM Trans. Graph. 2015, 34, 128. [Google Scholar] [CrossRef]
- Sunderland, K.; Boyeong, W.; Pinter, C.; Fichtinger, G. Reconstruction of surfaces from planar contours through Contour interpolations. In Proceedings of the Image-Guided Procedures, Robotic Interventions, and Modeling, Orlando, FL, USA, 22–24 February 2015. [Google Scholar]
- Smith, N.; de Vecchi, A.; McCormick, M.; Nordsletten, D.; Camara, O.; Frangi, A.; Delingette, H.; Sermesant, M.; Relan, J.; Ayache, N.; et al. euHeart: Personalized and integrated cardiac care using patient-specific cardiovascular modelling. Interface Focus 2017, 1, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Zhang, J.; Xia, L. Three-Dimensional Mesh Generation for Human Heart Model. Life Syst. Model. Intell. Comput. 2010, 98, 157–162. [Google Scholar]
- Vukicevic, M.; Mosadegh, B.; Min, J.; Little, S. Cardiac 3D Printing and its Future Directions. JACC Cardiovasc. Imaging 2017, 10, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, H.; Kneser, R.; Neizel, M.; Peters, J.; Ecabert, O.; Kühl, H.; Kelm, M.; Weese, J. Integrating Viability Information into a Cardiac Model for Interventional Guidance. In Proceedings of the 5th International Conference Functional Imaging and Modeling of the Heart (FIMH), Nice, France, 3–5 June 2009; pp. 312–320. [Google Scholar]
- Young, A.; Cowan, B.; Thrupp, S.; Hedley, W.; DellItalia, L. Left Ventricular Mass and Volume: Fast Calculation with Guide-Point Modelling on MR Images. Radiology 2000, 2, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Lamata, P.; Niederer, S.; Nordsletten, D.; Barber, D.; Roy, I.; Hose, D.; Smith, N. An accurate, fast and robust method to generate patient-specific cubic Hermite meshes. Med. Image Anal. 2011, 15, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Albà, X.; Pereañez, M.; Hoogendoorn, C.; Swift, A.; Wild, J.; Frangi, A.; Lekadir, K. An algorithm for the segmentation of highly abnormal hearts using a generic statistical shape model. IEEE Trans. Med. Imaging 2016, 35, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Villard, B.; Zacur, E.; Dall’Armellina, E.; Grau, V. Correction of Slice Misalignment in Multi-breath-hold Cardiac MRI Scans. In International Workshop on Statistical Atlases and Computational Models of the Heart. (Imaging and Modelling Challenges. STACOM 2016); Lecture Notes in Computer Science (LNCS); Springer: Cham, Switzerland, 2016; Volume 10124. [Google Scholar]
- Villard, B.; Carapella, V.; Ariga, R.; Grau, V.; Zacur, E. Cardiac Mesh Reconstruction from Sparse, Heterogeneous Contours. In Medical Image Understanding and Analysis. (MIUA 2017); Communications in Computer and Information Science; Springer: Cham, Germany, 2017; Volume 723. [Google Scholar]
- Lopez-Perez, A.; Sebastian, R.; Ferrero, J. Three-dimensional cardiac computational modelling: Methods, features and applications. BioMed. Eng. OnLine 2015, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Schall, O.; Samozino, M. Surface from scattered points. In Proceedings of the International Workshop on Semantic Virtual Environments, Villars, Switzerland, 16–18 March 2005; pp. 138–147. [Google Scholar]
- Berger, M.; Tagliasacchi, A.; Saversky, L.; Alliez, P.; Guennebaud, G.; Levine, J.; Sharf, A.; Silva, C. A survey of surface reconstruction from Points clouds. Comput. Graph. Forum 2017, 36, 301–329. [Google Scholar] [CrossRef]
- Khatamian, A.; Arabnia, H. Survey on 3D Surface Reconstruction. J. Inf. Process. Syst. 2016, 12, 338–357. [Google Scholar]
- Hoppe, H.; DeRose, T.; Duchamp, T.; McDonald, J.; Stuetzle, W. Surface Reconstruction from Unorganized Points. SIGGRAPH Comput. Graph. 1992, 26, 71–78. [Google Scholar] [CrossRef]
- Bernardini, F.; Mittleman, J.; Rushmeier, H.; Silva, C.; Taubin, G. The Ball-pivoting algorithm for surface reconstruction. IEEE Trans. Vis. Comput. Graph. 1999, 5, 349–359. [Google Scholar] [CrossRef]
- Lim, C.; Su, Y.; Yeo, S.; Ng, G.; Nguyen, V.; Zhong, L.; Tan, R.S.; Poh, K.K.; Chai, P. Automatic 4D Reconstruction of Patient-Specific Cardiac Mesh with 1-to-1 Vertex Correspondence from Segmented Contours Lines. PLoS ONE 2014, 9, e93747. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Bajaj, C.; Deasy, J.; Ju, T. Surface Reconstruction from Non-parallel Curve Networks. Eurographics 2008, 27, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Konno, K.; Tokuyama, K. 3D Terrain Reconstruction Based on Contours. In Proceedings of the Ninth International Conference on Computer Aided Design and Computer Graphics, Hong Kong, China, 7–10 December 2005; pp. 325–330. [Google Scholar]
- Wang, Z.; Geng, N.; Zhang, Z. Surface Mesh Reconstruction Based on Contours. In Proceedings of the International Conference on Computational Intelligence and Software Engineering, Wuhan, China, 11–13 December 2009; pp. 325–330. [Google Scholar]
- Kazhdan, M.; Bolitho, M.; Hoppe, H. Poisson Surface Reconstruction. In Proceedings of the Eurographics Symposium on Geometry Processing, Cagliari, Sardinia, 26–28 June 2006. [Google Scholar]
- De Marvao, A.; Dawes, T.; Shi, W.; Minas, C.; Keenan, N.; Diamond, T.; Durighel, G.; Montana, G.; Rueckert, D.; Cook, S.; et al. Population-based studies of myocardial hypertrophy: High resolution cardiovascular magnetic resonance atlases improve statistical power. J. Cardiovasc. Magn. Reson. 2015, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.; Shi, W.; de Marvao, A.; Dawes, T.; O’Regan, D.; Cook, S.; Rueckert, D. A bi-ventricular cardiac atlas built from 1000+ high resolution MR images of healthy subjects and an analysis of shape and motion. Med. Image Anal. 2015, 26, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, J.G.; Arce, G.R. Optimality of the myriad filter in practical impulsive-noise environments. IEEE Trans. on Signal Process. 2001, 49, 438–441. [Google Scholar] [CrossRef]
- Moenning, C.; Dodgson, N.A. Fast marching farthest point sampling for implicit surfaces and point clouds. Comput. Lab. Tech. Rep. 2003, 565, 1–11. [Google Scholar]
- Rohr, K.; Stiehl, H.; Sprengel, R.; Buzug, T.; Weese, J.; Kuhn, M. Landmark-based elastic registration using approximating thin-plate splines. IEEE Trans. Med. Imaging 2001, 20, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Sprengel, R.; Rohr, K.; Stiehl, H. Thin-Plate Sline Approximation for Image Registration. Eng. Med. Biol. Soc. 1996, 18, 1190–1191. [Google Scholar]
- Dyn, N.; Levine, D.; Gregory, J. A Butterfly Subdivision Scheme for Surface Interpolation with Tension Control. ACM Trans. Graph. 1990, 9, 160–169. [Google Scholar] [CrossRef]
- Nealen, A.; Igarashi, T.; Sorkine, O.; Alexa, M. Laplacian Mesh Optimization. In Proceedings of the 4th International Conference on Computer Graphics and Interactive Techniques in Australasia and Southeast Asia, Kuala Lumpur, Malaysia, 29 November–2 December 2006. [Google Scholar]
- Taubin, G. A signal processing approach to fair surface design. In Proceedings of the 22nd annual conference on Computer Graphics and Interactive Techniques, Los Angeles, CA, USA, 6–11 August 1995; pp. 351–358. [Google Scholar]
- Garland, M.; Heckbert, P. Surface simplification using quadric error metrics. In Proceedings of the 24th Annual Conference on Computer Graphics and Interactive Techniques, Los Angeles, CA, USA, 3–8 August 1997; pp. 209–216. [Google Scholar]
- McLeish, K.; Hill, D.L.G.; Atkinson, D.; Blackall, J.M.; Razavi, R. A study of the motion and deformation of the heart due to respiration. IEEE Trans. Med. Imaging 2002, 21, 1142–1150. [Google Scholar] [CrossRef] [PubMed]
- Shechter, G.; Ozturk, C.; Resar, J.R.; McVeigh, E.R. Respiratory motion of the heart from free breathing coronary angiograms. IEEE Trans. Med. Imaging 2004, 23, 1046–1056. [Google Scholar] [CrossRef] [PubMed]

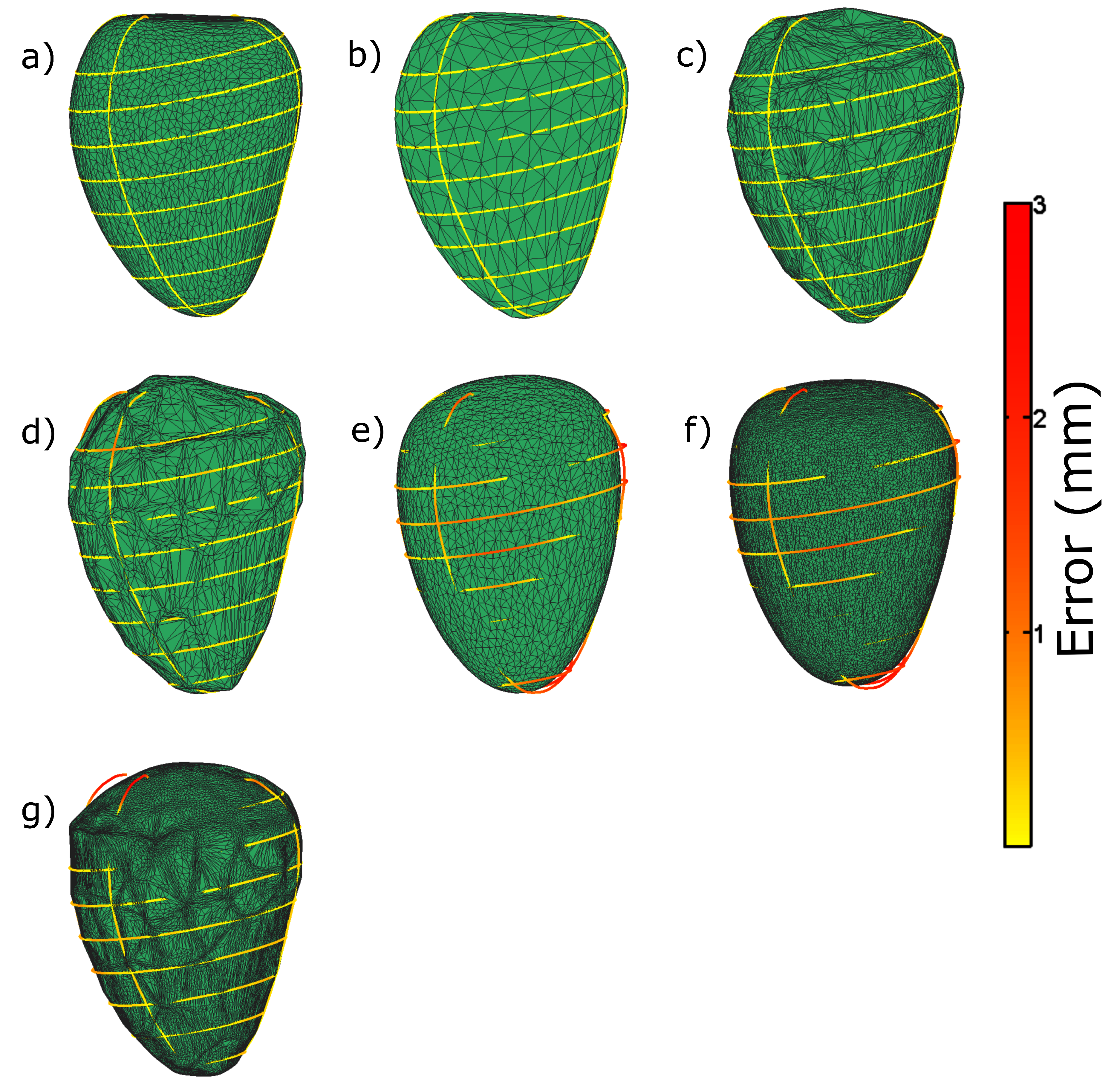
| Measure | Epicardium (mm) | Endocardium (mm) | Right Ventricle (mm) |
|---|---|---|---|
| Mean | 0.12 | 0.14 | 0.29 |
| Median | 0.09 | 0.09 | 0.22 |
| Max | 1.11 | 1.29 | 3.34 |
| Min | 1.35 | 2.04 | 7.49 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villard, B.; Grau, V.; Zacur, E. Surface Mesh Reconstruction from Cardiac MRI Contours. J. Imaging 2018, 4, 16. https://doi.org/10.3390/jimaging4010016
Villard B, Grau V, Zacur E. Surface Mesh Reconstruction from Cardiac MRI Contours. Journal of Imaging. 2018; 4(1):16. https://doi.org/10.3390/jimaging4010016
Chicago/Turabian StyleVillard, Benjamin, Vicente Grau, and Ernesto Zacur. 2018. "Surface Mesh Reconstruction from Cardiac MRI Contours" Journal of Imaging 4, no. 1: 16. https://doi.org/10.3390/jimaging4010016
APA StyleVillard, B., Grau, V., & Zacur, E. (2018). Surface Mesh Reconstruction from Cardiac MRI Contours. Journal of Imaging, 4(1), 16. https://doi.org/10.3390/jimaging4010016





