Typical Fluorescent Sensors Exploiting Molecularly Imprinted Hydrogels for Environmentally and Medicinally Important Analytes Detection
Abstract
:1. Introduction
2. Results and Discussion
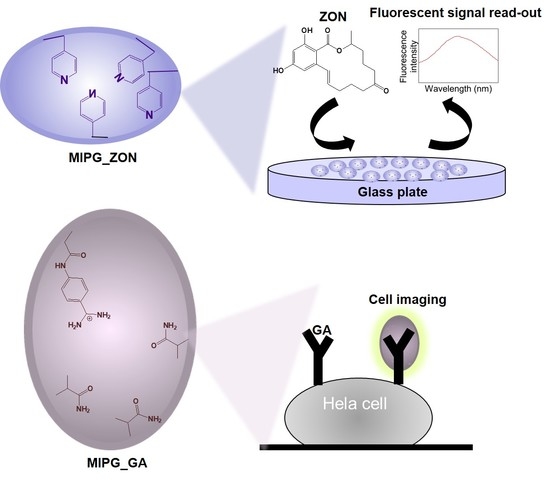
2.1. Characterization of Synthesized MIPGs_ZON
2.2. Equilibrium Binding Studies on MIPGs_ZON
2.3. Application of MIPGs_ZON-Based Fluorescent Sensor for Real Sample Tests
2.4. Synthesis and Binding Characterization of MIPG_GA
2.5. Application of MIPG_GA-Based Fluorescent Probe for Cell Imaging
3. Conclusions
4. Materials and Methods
4.1. Reagents
4.2. Synthesis of MIPGs_ZON and MIPG_GA
4.3. Equilibrium Binding Studies
4.4. Application of MIPG_ZON-Based Fluorescent Sensor for Real Sample Tests
4.5. Application of MIPG_GA-Based Fluorescent Probe for Cell Imaging
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Patel, B.R.; Noroozifar, M.; Kerman, K. Review-Nanocomposite-Based Sensors for Voltammetric Detection of Hazardous Phenolic Pollutants in Water. J. Electrochem. Soc. 2020, 167, 037568. [Google Scholar] [CrossRef]
- Sow, W.T.; Ye, F.; Zhang, C.; Li, H. Smart materials for point-of-care testing: From sample extraction to analyte sensing and readout signal generator. Biosens. Bioelectron. 2020, 170, 112682. [Google Scholar] [CrossRef] [PubMed]
- Beloglazova, N.; Lenain, P.; Tessier, M.; Goryacheva, I.; Hens, Z.; De Saeger, S. Bioimprinting for multiplex luminescent detection of deoxynivalenol and zearalenone. Talanta 2019, 192, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Yao, M.; Saeger, S.D.; Yan, L.; Song, S. Carbon Quantum Dots Encapsulated Molecularly Imprinted Fluorescence Quenching Particles for Sensitive Detection of Zearalenone in Corn Sample. Toxins 2018, 10, 438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lhotska, I.; Gajdosova, B.; Solich, P.; Satinsky, D. Molecularly imprinted vs. reversed-phase extraction for the determination of zearalenone: A method development and critical comparison of sample clean-up efficiency achieved in an on-line coupled SPE chromatography system. Anal. Bioanal. Chem. 2018, 410, 3265–3273. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhang, K.; Fizir, M.; Niu, M.; Sun, C.; Xi, S.; Hui, X.; Shi, J.; He, H. Rational design, preparation and adsorption study of a magnetic molecularly imprinted polymer using a dummy template and a bifunctional monomer. New J. Chem. 2017, 41, 7092–7101. [Google Scholar] [CrossRef]
- Li, Q.; Shinde, S.; Grasso, G.; Caroli, A.; Abouhany, R.; Lanzillotta, M.; Pan, G.; Wan, W.; Rurack, K.; Sellergren, B. Selective detection of phospholipids using molecularly imprinted fluorescent sensory core-shell particles. Sci. Rep. 2020, 10, 9924. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Feng, Y.; Pan, G.; Liu, L. Advances in Molecularly Imprinting Technology for Bioanalytical Applications. Sensors 2019, 19, 177. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Yin, Y.; Ni, L.; Miao, H.; Wang, Y.; Pan, C.; Tian, X.; Pan, J.; You, T.; Li, B.; et al. Thermo-responsive imprinted hydrogel with switchable sialic acid recognition for selective cancer cell isolation from blood. Bioact. Mater. 2021, 6, 1308–1317. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dai, J.; Xu, Y.; Dai, X.; Zhang, Y.; Shi, W.; Sellergren, B.; Pan, G. Molecularly Imprinted Fluorescent Test Strip for Direct, Rapid, and Visual Dopamine Detection in Tiny Amount of Biofluid. Small 2019, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Biosens. Technol. Detect. Biomol. 2016, 60, 1–8. [Google Scholar]
- Morales, M.A.; Halpern, J.M. Guide to Selecting a Biorecognition Element for Biosensors. Bioconjugate Chem. 2018, 29, 3231–3239. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, Y.; Soppera, O.; Haupt, K. Photopolymerization and photostructuring of molecularly imprinted polymers for sensor applications-A review. Anal. Chim. Acta 2012, 717, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Haupt, K.; Rangel, P.X.M.; Bui, B.T.S. Molecularly Imprinted Polymers: Antibody Mimics for Bioimaging and Therapy. Chem. Rev. 2020, 120, 9554–9582. [Google Scholar] [CrossRef]
- Xu, J.; Miao, H.; Wang, J.; Pan, G. Molecularly Imprinted Synthetic Antibodies: From Chemical Design to Biomedical Applications. Small 2020, 16. [Google Scholar] [CrossRef] [PubMed]
- Poma, A.; Turner, A.P.F.; Piletsky, S.A. Advances in the manufacture of MIP nanoparticles. Trends Biotechnol. 2010, 28, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ambrosini, S.; Tamahkar, E.; Rossi, C.; Haupt, K.; Bui, B.T.S. Toward a Universal Method for Preparing Molecularly Imprinted Polymer Nanoparticles with Antibody-like Affinity for Proteins. Biomacromolecules 2016, 17, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Wang, L.; Liu, Z. Molecularly Imprinted Polymer Nanoparticles: An Emerging Versatile Platform for Cancer Therapy. Angew. Chem. Int. Ed. 2021, 60, 3858–3869. [Google Scholar] [CrossRef] [PubMed]
- Cheong, W.J.; Yang, S.H.; Ali, F. Molecular imprinted polymers for separation science: A review of reviews. J. Sep. Sci. 2013, 36, 609–628. [Google Scholar] [CrossRef] [PubMed]
- Saylan, Y.; Yilmaz, F.; Ozgur, E.; Derazshamshir, A.; Yavuz, H.; Denizli, A. Molecular Imprinting of Macromolecules for Sensor Applications. Sensors 2017, 17, 898. [Google Scholar] [CrossRef]
- Vaneckova, T.; Bezdekova, J.; Han, G.; Adam, V.; Vaculovicova, M. Application of molecularly imprinted polymers as artificial receptors for imaging. Acta Biomater. 2020, 101, 444–458. [Google Scholar] [CrossRef]
- Korde, B.A.; Mankar, J.S.; Phule, S.; Krupadam, R.J. Nanoporous imprinted polymers (nanoMIPs) for controlled release of cancer drug. Mater. Sci. Eng. C 2019, 99, 222–230. [Google Scholar]
- Wackerlig, J.; Schirhagl, R. Applications of Molecularly Imprinted Polymer Nanoparticles and Their Advances toward Industrial Use: A Review. Anal. Chem. 2016, 88, 250–261. [Google Scholar] [CrossRef]
- Lowdon, J.W.; Dilien, H.; Singla, P.; Peeters, M.; Cleij, T.J.; van Grinsven, B.; Eersels, K. MIPs for commercial application in low-cost sensors and assays—An overview of the current status quo. Sens. Actuators B 2020, 325, 128973. [Google Scholar]
- Urraca, J.L.; Marazuela, M.D.; Merino, E.R.; Orellana, G.; Moreno-Bondi, M.C. Molecularly imprinted polymers with a streamlined mimic for zearalenone analysis. J. Chromatogr. A 2006, 1116, 127–134. [Google Scholar] [CrossRef]
- Lucci, P.; Derrien, D.; Alix, F.; Perollier, C.; Bayoudh, S. Molecularly imprinted polymer solid-phase extraction for detection of zearalenone in cereal sample extracts. Anal. Chim. Acta 2010, 672, 15–19. [Google Scholar] [CrossRef]
- Xuan-Anh, T.; Acha, V.; Haupt, K.; Bernadette Tse Sum, B. Direct fluorimetric sensing of UV-excited analytes in biological and environmental samples using molecularly imprinted polymer nanoparticles and fluorescence polarization. Biosens. Bioelectron. 2012, 36, 22–28. [Google Scholar]
- Demir, B.; Lemberger, M.M.; Panagiotopoulou, M.; Rangel, P.X.M.; Timur, S.; Hirsch, T.; Bui, B.T.S.; Wegener, J.; Haupt, K. Tracking Hyaluronan: Molecularly Imprinted Polymer Coated Carbon Dots for Cancer Cell Targeting and Imaging. ACS Appl. Mater. Interfaces 2018, 10, 3305–3313. [Google Scholar] [CrossRef]
- Panagiotopoulou, M.; Salinas, Y.; Beyazit, S.; Kunath, S.; Duma, L.; Prost, E.; Mayes, A.G.; Resmini, M.; Bui, B.T.S.; Haupt, K. Molecularly Imprinted Polymer Coated Quantum Dots for Multiplexed Cell Targeting and Imaging. Angew. Chem. Int. Ed. 2016, 55, 8244–8248. [Google Scholar]
- Thongrussamee, T.; Kuzmina, N.S.; Shim, W.B.; Jiratpong, T.; Eremin, S.A.; Intrasook, J.; Chung, D.H. Monoclonal-based enzyme-linked immunosorbent assay for the detection of zearalenone in cereals. Food Addit. Contam. Part A 2008, 25, 997–1006. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Z.; Zhang, Q.; Wang, Z.; Zhang, W.; Yu, L.; Li, H.; Jiang, J.; Li, P. Rapid and sensitive double-label based immunochromatographic assay for zearalenone detection in cereals. Electrophoresis 2018, 39, 2125–2130. [Google Scholar] [CrossRef] [PubMed]
- Caglayan, M.O.; Ustundag, Z. Detection of zearalenone in an aptamer assay using attenuated internal reflection ellipsometry and it’s cereal sample applications. Food Chem. Toxicol. 2020, 136, 111081. [Google Scholar] [CrossRef]
- Sun, S.; Xie, Y. An enhanced enzyme-linked aptamer assay for the detection of zearalenone based on gold nanoparticles. Anal. Methods 2021, 13, 1255–1260. [Google Scholar] [CrossRef]
- Nestora, S.; Merlier, F.; Beyazit, S.; Prost, E.; Duma, L.; Baril, B.; Greaves, A.; Haupt, K.; Bui, B.T.S. Plastic Antibodies for Cosmetics: Molecularly Imprinted Polymers Scavenge Precursors of Malodors. Angew. Chem. Int. Ed. 2016, 55, 6252–6256. [Google Scholar] [CrossRef]
- Kolapalli, S.P.; Kumaraswamy, S.B.; Mortha, K.K.; Thomas, A.; Das Banerjee, S. UNIVmAb reactive albumin associated hyaladherin as a potential biomarker for colorectal cancer. Cancer Biomark. 2021, 30, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Guo, L.; Li, J.W.; Liu, N.; Qi, R.; Liu, J. Expression of hyaluronan receptors CD44 and RHAMM in stomach cancers: Relevance with tumor progression. Int. J. Oncol. 2000, 17, 927–932. [Google Scholar] [CrossRef]
- Rangel, P.X.M.; Lacief, S.; Xu, J.; Panagiotopoulou, M.; Kovensky, J.; Bui, B.T.S.; Haupt, K. Solid-phase synthesis of molecularly imprinted polymer nanolabels: Affinity tools for cellular bioimaging of glycans. Sci. Rep. 2019, 9, 3923. [Google Scholar] [CrossRef] [Green Version]





| Polymer | ZAN (mmol) | 4-VPY (mmol) | EGDMA (mmol) | ABDV (mg) | Size (nm) | PDI |
|---|---|---|---|---|---|---|
| MIPG1 | 0.03 | 0.06 | 0.6 | 3.1 | 345 ± 5 | 0.163 |
| NIPG1 | - | 0.06 | 0.6 | 3.1 | 455 ± 16 | 0.160 |
| MIPG2 | 0.03 | 0.12 | 0.6 | 3.2 | 443 ± 11 | 0.137 |
| NIPG2 | - | 0.12 | 0.6 | 3.2 | 483 ± 6 | 0.199 |
| MIPG3 | 0.03 | 0.18 | 0.6 | 3.4 | 468 ± 12 | 0.156 |
| NIPG3 | - | 0.18 | 0.6 | 3.4 | 503 ± 9 | 0.085 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, L.; Ding, R.; Li, X.; Miao, H.; Xu, J.; Pan, G. Typical Fluorescent Sensors Exploiting Molecularly Imprinted Hydrogels for Environmentally and Medicinally Important Analytes Detection. Gels 2021, 7, 67. https://doi.org/10.3390/gels7020067
Zou L, Ding R, Li X, Miao H, Xu J, Pan G. Typical Fluorescent Sensors Exploiting Molecularly Imprinted Hydrogels for Environmentally and Medicinally Important Analytes Detection. Gels. 2021; 7(2):67. https://doi.org/10.3390/gels7020067
Chicago/Turabian StyleZou, Lihua, Rong Ding, Xiaolei Li, Haohan Miao, Jingjing Xu, and Guoqing Pan. 2021. "Typical Fluorescent Sensors Exploiting Molecularly Imprinted Hydrogels for Environmentally and Medicinally Important Analytes Detection" Gels 7, no. 2: 67. https://doi.org/10.3390/gels7020067