Osmotic Stress Responses, Cell Wall Integrity, and Conidiation Are Regulated by a Histidine Kinase Sensor in Trichoderma atroviride
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Construction of ∆nik1 Mutants
2.3. Cellular Stress Assays
2.4. Sensitivity to the Cell Wall Lytic Enzyme Assay
2.5. Gene Expression Induced by Osmotic Stress
2.6. Radial Growth Measurement
2.7. Conidial Production Induced by Light
2.8. Expression Assays of Light-Regulated Genes
2.9. Statistical Analysis
3. Results
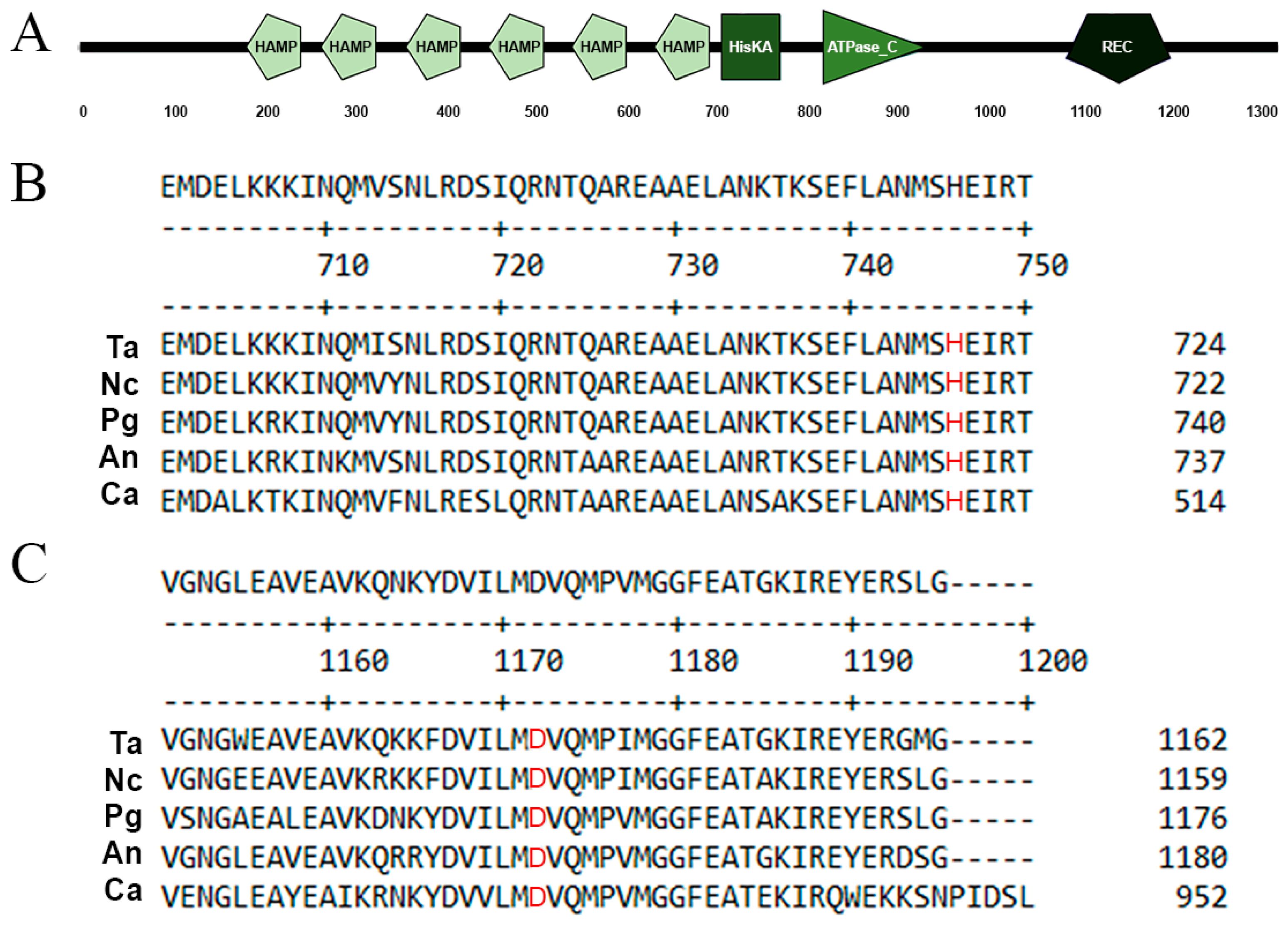
3.1. Identification and Deletion of the T. atroviride nik1 Gene
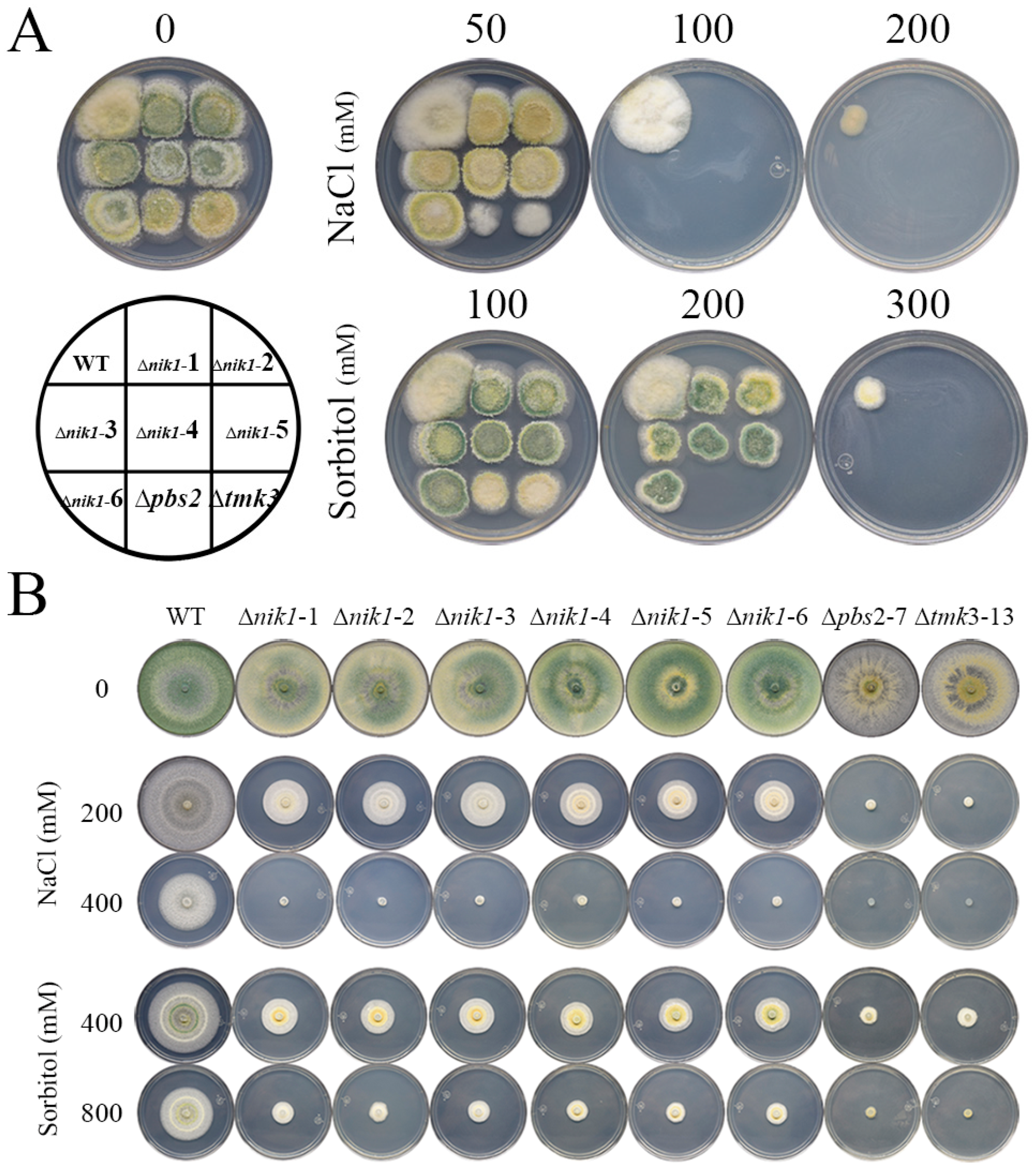
3.2. Nik1 Is Involved in Osmotic Stress Resistance
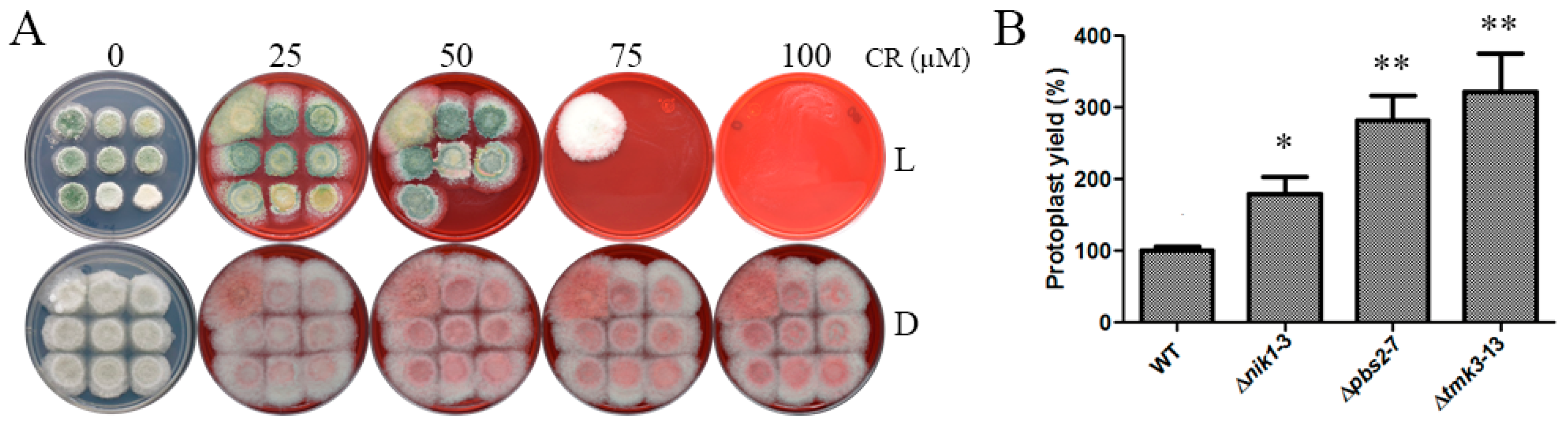
3.3. Nik1 Regulates the Cell Wall Integrity Maintenance of T. atroviride
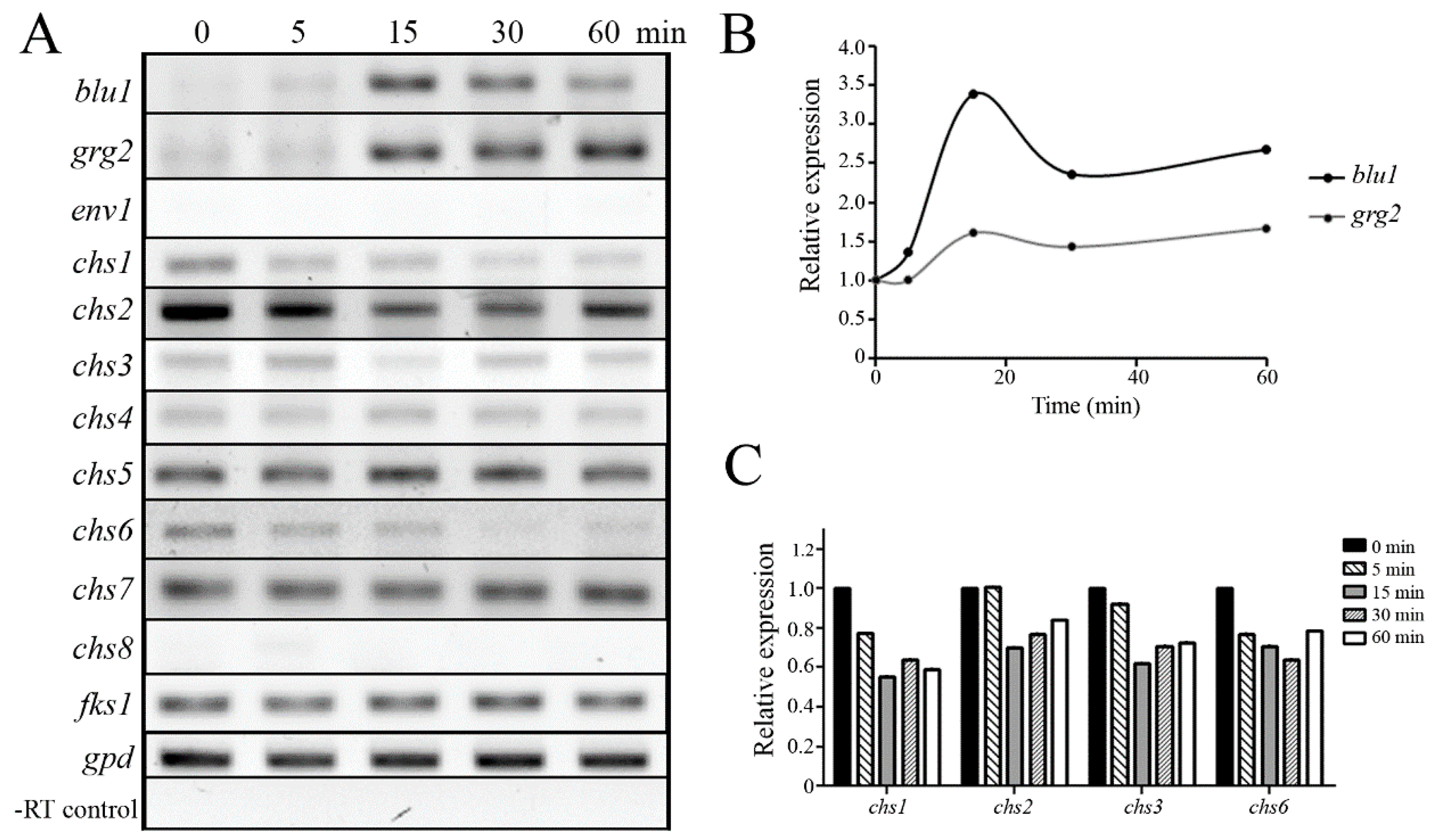
3.4. Osmotic Stress Provokes Changes in Gene Expression through the Tmk3 MAPK Pathway
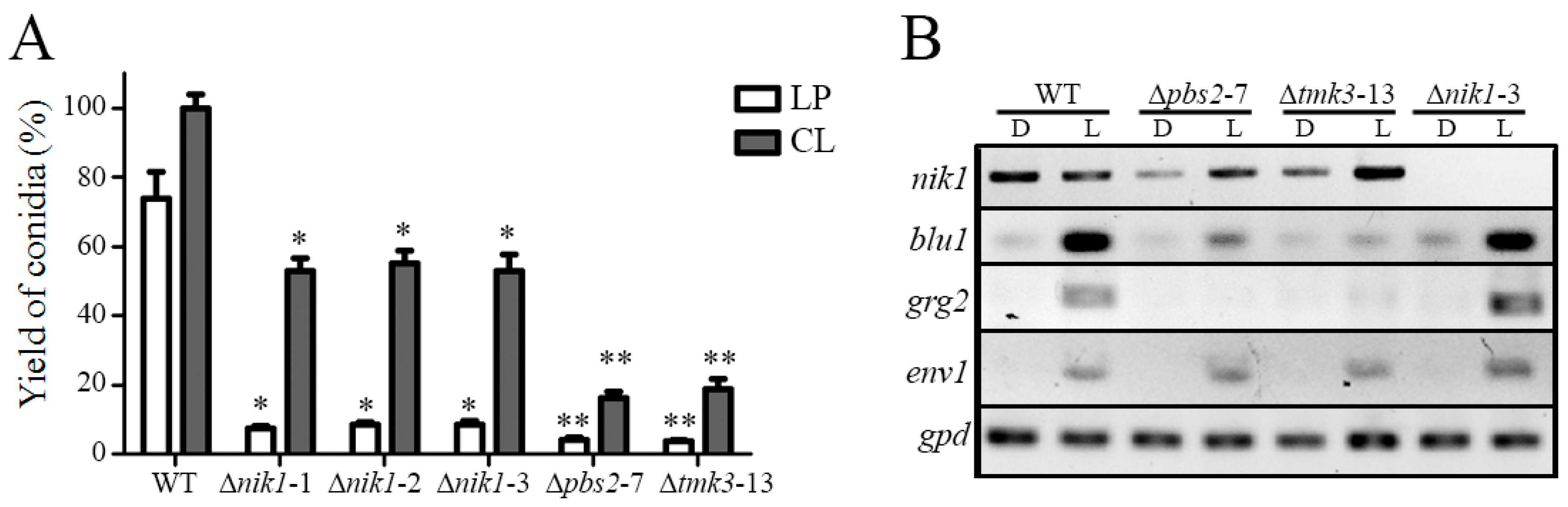
3.5. The HK Nik1 Is Involved in Mycelial Growth and Asexual Reproduction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Medina-Castellanos, E.; Esquivel-Naranjo, E.U.; Heil, M.; Herrera-Estrella, A. Extracellular ATP activates MAPK and ROS signaling during injury response in the fungus Trichoderma atroviride. Front. Plant Sci. 2004, 5, 659. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Horwitz, B.A.; Herrera-Estrella, A.; Schmoll, M.; Kenerley, C.M. Trichoderma Research in the Genome Era. Annu. Rev. Phytopathol. 2013, 51, 105–129. [Google Scholar] [CrossRef] [PubMed]
- Steyaert, J.M.; Weld, R.J.; Mendoza-Mendoza, A.; Stewart, A. Reproduction without sex: Conidiation in the filamentous fungus Trichoderma. Microbiology 2010, 156, 2887–2900. [Google Scholar] [CrossRef]
- Brewster, J.; de Valoir, T.; Dwyer, N.; Winter, E.; Gustin, M. An osmosensing signal transduction pathway in yeast. Science 1993, 259, 1760–1763. [Google Scholar] [CrossRef]
- Singh, K.K. The Saccharomyces cerevisiae sln1p-ssk1p two-component system mediates response to oxidative stress and in an oxidant-specific fashion. Free Radic. Biol. Med. 2000, 29, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Winkler, A.; Arkind, C.; Mattison, C.P.; Burkholder, A.; Knoche, K.; Ota, K. Heat Stress Activates the Yeast High-Osmolarity Glycerol Mitogen-Activated Protein Kinase Pathway, and Protein Tyrosine Phosphatases Are Essential under Heat Stress. Eukaryote Cell 2002, 1, 163–173. [Google Scholar] [CrossRef] [PubMed]
- García-Rodriguez, L.J.; Durán, A.; Roncero, C. Calcofluor Antifungal Action Depends on Chitin and a Functional High-Osmolarity Glycerol Response (HOG) Pathway: Evidence for a Physiological Role of the Saccharomyces cerevisiae HOG Pathway under Noninducing Conditions. J. Bacteriol. 2000, 182, 2428–2437. [Google Scholar] [CrossRef]
- Jiang, L.; Cao, C.; Zhang, L.; Lin, W.; Xia, J.; Xu, H.; Zhang, Y. Cadmium-induced activation of high osmolarity glycerol pathway through its Sln1 branch is dependent on the MAP kinase kinase kinase Ssk2, but not its paralog Ssk22, in budding yeast. FEMS Yeast Res. 2014, 14, 1263–1272. [Google Scholar] [CrossRef]
- Saito, H.; Posas, F. Response to Hyperosmotic Stress. Genetics 2012, 192, 289–318. [Google Scholar] [CrossRef]
- Esquivel-Naranjo, E.U.; García-Esquivel, M.; Medina-Castellanos, E.; Correa-Pérez, V.A.; Parra-Arriaga, J.L.; Landeros-Jaime, F.; Cervantes-Chávez, J.A.; Herrera-Estrella, A. A Trichoderma atroviride stress-activated MAPK pathway integrates stress and light signals. Mol. Microbiol. 2016, 100, 860–876. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, Q.; Yang, J.; Jiang, B.; Wang, F.; Liu, K.; Fang, X. A Mitogen-Activated Protein Kinase Tmk3 Participates in High Osmolarity Resistance, Cell Wall Integrity Maintenance and Cellulase Production Regulation in Trichoderma reesei. PLoS ONE 2013, 8, e72189. [Google Scholar] [CrossRef]
- Delgado-Jarana, J.; Sousa, S.; González, F.; Rey, M.; Llobell, A. ThHog1 controls the hyperosmotic stress response in Trichoderma harzianum. Microbiology 2006, 152, 1687–1700. [Google Scholar] [CrossRef]
- Horie, T.; Tatebayashi, K.; Yamada, R.; Saito, H. Phosphorylated Ssk1 Prevents Unphosphorylated Ssk1 from Activating the Ssk2 Mitogen-Activated Protein Kinase Kinase Kinase in the Yeast High-Osmolarity Glycerol Osmoregulatory Pathway. Mol. Cell. Biol. 2008, 28, 5172–5183. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Tatebayashi, K.; Nishimura, A.; Yamamoto, K.; Yang, H.Y.; Saito, H. Yeast Osmosensors Hkr1 and Msb2 Activate the Hog1 MAPK Cascade by Different Mechanisms. Sci. Signal. 2014, 7, ra21. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Kwok, S.; Bleecker, A.; Meyerowitz, E. Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science 1993, 262, 539–544. [Google Scholar] [CrossRef]
- Ota, I.; Varshavsky, A. A yeast protein similar to bacterial two-component regulators. Science 1993, 262, 566–569. [Google Scholar] [CrossRef]
- Schmoll, M.; Dattenböck, C.; Carreras-Villaseñor, N.; Mendoza-Mendoza, A.; Tisch, D.; Alemán, M.I.; Baker, S.E.; Brown, C.; Cervantes-Badillo, M.G.; Cetz-Chel, J.; et al. The Genomes of Three Uneven Siblings: Footprints of the Lifestyles of Three Trichoderma Species. Microbiol. Mol. Biol. Rev. 2016, 80, 205–327. [Google Scholar] [CrossRef]
- Catlett, N.L.; Yoder, O.C.; Turgeon, B.G. Whole-Genome Analysis of Two-Component Signal Transduction Genes in Fungal Pathogens. Eukaryote Cell 2003, 2, 1151–1161. [Google Scholar] [CrossRef]
- Schumacher, M.M.; Enderlin, C.S.; Selitrennikoff, C.P. The Osmotic-1 Locus of Neurospora crassa Encodes a Putative Histidine Kinase Similar to Osmosensors of Bacteria and Yeast. Curr. Microbiol. 1997, 34, 340–347. [Google Scholar] [CrossRef]
- Alex, L.A.; Borkovich, K.A.; Simon, M.I. Hyphal development in Neurospora crassa: Involvement of a two-component histidine kinase. Proc. Natl. Acad. Sci. USA 1996, 93, 3416–3421. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, R.; Banno, S.; Ichikawa, R.; Fukumori, F.; Ichiishi, A.; Kimura, M.; Yamaguchi, I.; Fujimura, M. Identification of OS-2 MAP kinase-dependent genes induced in response to osmotic stress, antifungal agent fludioxonil, and heat shock in Neurospora crassa. Fungal Genet. Biol. 2007, 44, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, D.; Takahashi-Nakaguchi, A.; Toyotome, T.; Yoshimi, A.; Abe, K.; Kamei, K.; Gonoi, T.; Kawamoto, S. NikA/TcsC Histidine Kinase Is Involved in Conidiation, Hyphal Morphology, and Responses to Osmotic Stress and Antifungal Chemicals in Aspergillus fumigatus. PLoS ONE 2013, 8, e80881. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.H.; Hamari, Z.; Han, K.H.; Seo, J.A.; Reyes-Domínguez, Y.; Scazzocchio, C. Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef]
- Carroll, A.M.; Sweigard, J.A.; Valent, B. Improved Vectors for Selecting Resistance to Hygromycin. Fungal Genet. Rep. 1994, 41, 22. [Google Scholar] [CrossRef]
- Calcáneo-Hernández, G.; Rojas-Espinosa, E.; Landeros-Jaime, F.; Cervantes-Chávez, J.A.; Esquivel-Naranjo, E.U. An efficient transformation system for Trichoderma atroviride using the pyr4 gene as a selectable marker. Braz. J. Microbiol. 2020, 51, 1631–1643. [Google Scholar] [CrossRef] [PubMed]
- Raeder, U.; Broda, P. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1985, 1, 17–20. [Google Scholar] [CrossRef]
- Schultz, J.; Copley, R.R.; Doerks, T.; Ponting, C.P.; Bork, P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000, 28, 231–234. [Google Scholar] [CrossRef]
- Ene, I.; Walker, L.A.; Schiavone, M.; Lee, K.K.; Martin-Yken, H.; Dague, E.; Gow, N.A.R.; Munro, C.A.; Brown, A.J.P. Cell Wall Remodeling Enzymes Modulate Fungal Cell Wall Elasticity and Osmotic Stress Resistance. mBio 2015, 6, e00986-15. [Google Scholar] [CrossRef]
- Rosales-Saavedra, T.; Esquivel-Naranjo, E.U.; Casas-Flores, S.; Martínez-Hernández, P.; Ibarra-Laclette, E.; Cortes-Penagos, C.; Herrera-Estrella, A. Novel light-regulated genes in Trichoderma atroviride: A dissection by cDNA microarrays. Microbiology 2006, 152, 3305–3317. [Google Scholar] [CrossRef]
- Schmoll, M.; Franchi, L.; Kubicek, C.P. Envoy, a PAS/LOV Domain Protein of Hypocrea jecorina (Anamorph Trichoderma reesei), Modulates Cellulase Gene Transcription in Response to Light. Eukaryote Cell 2005, 4, 1998–2007. [Google Scholar] [CrossRef]
- Levin, D.E. Regulation of Cell Wall Biogenesis in Saccharomyces cerevisiae: The Cell Wall Integrity Signaling Pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef]
- Maeda, T.; Wurgler-Murphy, S.M.; Saito, H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 1994, 369, 242–245. [Google Scholar] [CrossRef]
- Wang, Z.; An, N.; Xu, W.; Zhang, W.; Meng, X.; Chen, G.; Liu, W. Functional characterization of the upstream components of the Hog1-like kinase cascade in hyperosmotic and carbon sensing in Trichoderma reesei. Biotechnol. Biofuels 2018, 11, 97. [Google Scholar] [CrossRef]
- Ke, R.; Ingram, P.J.; Haynes, K. An Integrative Model of Ion Regulation in Yeast. PLoS Comput. Biol. 2013, 9, e1002879. [Google Scholar] [CrossRef]
- Garcia, M.J.; Rios, G.; Ali, R.; Belles, J.M.; Serrano, R. Comparative physiology of salt tolerance in Candida tropicalis and Saccharomyces cerevisiae. Microbiology 1997, 143, 1125–1131. [Google Scholar] [CrossRef]
- Motoyama, T.; Kadokura, K.; Ohira, T.; Ichiishi, A.; Fujimura, M.; Yamaguchi, I.; Kudo, T. A two-component histidine kinase of the rice blast fungus is involved in osmotic stress response and fungicide action. Fungal Genet. Biol. 2005, 42, 200–212. [Google Scholar] [CrossRef]
- Jacob, S.; Foster, A.J.; Yemelin, A.; Thines, E. Histidine kinases mediate differentiation, stress response, and pathogenicity in Magnaporthe oryzae. MicrobiologyOpen 2014, 3, 668–687. [Google Scholar] [CrossRef]
- Liu, J.; Tong, S.M.; Qiu, L.; Ying, S.H.; Feng, M.G. Two histidine kinases can sense different stress cues for activation of the MAPK Hog1 in a fungal insect pathogen: Two sensors of fungal Hog1 cascade. Environ. Microbiol. 2017, 19, 4091–4102. [Google Scholar] [CrossRef] [PubMed]
- Brewster, J.L.; Gustin, M.C. Hog1: 20 years of discovery and impact. Sci. Signal. 2014, 7, re7. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Tao, J.; Qian, Z.; You, S.; Dong, H.; Shen, H.; Chen, X.; Tang, S.; Ren, S. A histidine kinase PmHHK1 regulates polar growth, sporulation and cell wall composition in the dimorphic fungus Penicillium marneffei. Mycol. Res. 2009, 113, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Williams, D.; Lowman, D.; Monteiro, M.A.; Tan, X.; Kruppa, M.; Fonzi, W.; Roman, E.; Pla, J.; Calderone, R. The Candida albicans histidine kinase Chk1p: Signaling and cell wall mannan. Fungal Genet. Biol. 2009, 46, 731–741. [Google Scholar] [CrossRef]
- Li, M.; Jiang, C.; Wang, Q.; Zhao, Z.; Jin, Q.; Xu, J.R.; Liu, H. Evolution and Functional Insights of Different Ancestral Orthologous Clades of Chitin Synthase Genes in the Fungal Tree of Life. Front. Plant Sci. 2016, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Kappel, L.; Münsterkötter, M.; Sipos, G.; Escobar-Rodriguez, C.; Gruber, S. Chitin and chitosan remodeling defines vegetative development and Trichoderma biocontrol. PLOS Pathog. 2020, 16, e1008320. [Google Scholar] [CrossRef] [PubMed]
- Robledo-Briones, M.; Ruiz-Herrera, J. Transcriptional Regulation of the Genes Encoding Chitin and β-1,3-Glucan Synthases from Ustilago maydis. Curr. Microbiol. 2012, 65, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Gandía, M.; Harries, E.; Marcos, J.F. Identification and characterization of chitin synthase genes in the postharvest citrus fruit pathogen Penicillium digitatum. Fungal Biol. 2012, 116, 654–664. [Google Scholar] [CrossRef]
- Munro, C.A.; Selvaggini, S.; Bruijn, I.; Walker, L.; Lenardon, M.D.; Gerssen, B.; Milne, S.; Brown, A.J.P.; Gow, N.A.R. The PKC, HOG and Ca 2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol. Microbiol. 2007, 63, 1399. [Google Scholar] [CrossRef]
- García-Marqués, S.; Randez-Gil, F.; Prieto, J.A. Nuclear versus cytosolic activity of the yeast Hog1 MAP kinase in response to osmotic and tunicamycin-induced ER stress. FEBS Lett. 2015, 589, 2163–2168. [Google Scholar] [CrossRef]
- Yu, Z.; Armant, O.; Fischer, R. Fungi use the SakA (HogA) pathway for phytochrome-dependent light signalling. Nat. Microbiol. 2016, 1, 16019. [Google Scholar] [CrossRef]
- Mohanan, V.C.; Chandarana, P.M.; Chattoo, B.B.; Patkar, R.N.; Manjrekar, J. Fungal Histidine Phosphotransferase Plays a Crucial Role in Photomorphogenesis and Pathogenesis in Magnaporthe oryzae. Front. Chem. 2017, 5, 31. [Google Scholar] [CrossRef]

| Gene | Primer | Sequence (5′ → 3′) | Target Region |
|---|---|---|---|
| nik1 (EHK40885.1) | Pnik1-F | CTT GCA GGC ACA TCC TTG ACG | 5′ flanking region |
| PQnik1-R | TGC TCC TTC AAT ATC AGT TAA CGT CGA TCA CGC TCG GCT CGG GTA AGC | ||
| TQnik1-F | CCC AGC ACT CGT CCG AGG GCA AAG GAA TAG ACA GAA CCA GCT CAT CCA GAC C | 3′ flanking region | |
| Tnik1-R | CTC TTA TCC ACC TTC CGT CCG | ||
| N5nik1-F | TCG CCT GAG ACT TCC AAG ACG | Nested nik1 primers | |
| N3nik1-R | TCA AGC CTG CAG CTC TCT CTC | ||
| hph (AEJ60084.1) | Hyg-F | GAT CGA CGT TAA CTG ATA TTG AAG GAG CA | hph marker |
| Hyg-R | CTA TTC CTT TGC CCT CGG ACG AGT GCT GGG |
| Gene | Primer | Sequence (5′ → 3′) | Number of PCR Cycles |
|---|---|---|---|
| nik1 (EHK40885.1) | 1nik1-F | CGA ATG TCG AGG GCA AGT GG | 35 |
| 3nik1-R | GTG AAG TCG CCG TCT GTA GC | ||
| tmk3 (EHK43400.1) | tmk3-F | GTT TGG TCT TGT CTG CTC TGC G | 35 |
| tmk3-R | GCA GGT CGG TTC CGA GAA GC | ||
| gpd (EHK49005.1) | gpd-F | GCC GAT GGT GAG CTC AAG GG | 26 |
| gpd-R | GGT CGA GGA CAC GGC GGG A | ||
| blu1 (EHK44319.1) | qblu1-F | CGT TGG CTC TCG CCT GAC C | 27 |
| blu1-R | GAA CGC CAT TGA AGG CCT CG | ||
| grg2 (EHK50625.1) | grg2-F | GAT TCC ATC AAG CAG GGT GCC | 27 |
| grg2-R | GTT TAG ATA GCC TGC TTG TGG G | ||
| env1 (EHK44161.1) | env1-F | GCC AAA ATG GTT CCT TCA GGG TC | 27 |
| env1-R | GTT TGG TCG AGA CAC AAG TCG G | ||
| chs1 (EHK39721.1) | chs1-F | CTG ACG TTC CCG ACA CTG TTC C | 40 |
| chs1-R | TGC CAG TCC ACC AGC GAC G | ||
| chs2 (EHK46279.1) | chs2-F | GAT TCG CGC CAA CCA TGT CG | 32 |
| chs3-R | GTA GGA TAA AGC ATC AAC CGA GG | ||
| chs3 (EHK39554.1) | chs3-F | CCT CAG GCA GTA GCT ACC AC | 40 |
| chs3-R | CGT GGA CAG TGG AGG CAG G | ||
| chs4 (EHK40657.1) | chs4-F | GTC CGC GAT CTC TGT GGC AC | 32 |
| chs4-R | ACC AAG AGT GTG CGG TGA CG | ||
| chs5 (EHK48324.1) | chs5-F | CCG TGC ATG GCT AAG ACT TGG | 35 |
| chs5-R | GAG TCG GGT GTG TAG ATG CAG | ||
| chs6 (EHK48325.1) | chs6-F | GTT CGA CTG GGT CAG AAT GGC | 40 |
| chs6-R | CCA TTG GAG AAC TGA GAC GAC G | ||
| chs7 (EHK48360.1) | chs7-F | TCA TCA CAG CCG CAC CAG C | 35 |
| chs7-R | GAG TCG ATT GAT GCA GAG AAC C | ||
| chs8 (EHK41601.1) | chs8-F | TCT TCG GAA ATG TCT CGC ACC | 40 |
| chs8-R | CGG AGC CTT GCC TCT TCC | ||
| fks1 (EHK39881.1) | fks1-F | CTC TTC TGG TTA TTG CCC AGT C | 35 |
| fks1-R | GTT GCT TGG TTG TAA CAG TCG G |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calcáneo-Hernández, G.; Landeros-Jaime, F.; Cervantes-Chávez, J.A.; Mendoza-Mendoza, A.; Esquivel-Naranjo, E.U. Osmotic Stress Responses, Cell Wall Integrity, and Conidiation Are Regulated by a Histidine Kinase Sensor in Trichoderma atroviride. J. Fungi 2023, 9, 939. https://doi.org/10.3390/jof9090939
Calcáneo-Hernández G, Landeros-Jaime F, Cervantes-Chávez JA, Mendoza-Mendoza A, Esquivel-Naranjo EU. Osmotic Stress Responses, Cell Wall Integrity, and Conidiation Are Regulated by a Histidine Kinase Sensor in Trichoderma atroviride. Journal of Fungi. 2023; 9(9):939. https://doi.org/10.3390/jof9090939
Chicago/Turabian StyleCalcáneo-Hernández, Gabriela, Fidel Landeros-Jaime, José Antonio Cervantes-Chávez, Artemio Mendoza-Mendoza, and Edgardo Ulises Esquivel-Naranjo. 2023. "Osmotic Stress Responses, Cell Wall Integrity, and Conidiation Are Regulated by a Histidine Kinase Sensor in Trichoderma atroviride" Journal of Fungi 9, no. 9: 939. https://doi.org/10.3390/jof9090939





