The Adaptation of Botrytis cinerea Extracellular Vesicles Proteome to Surrounding Conditions: Revealing New Tools for Its Infection Process
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Optimizations of EVs Isolation
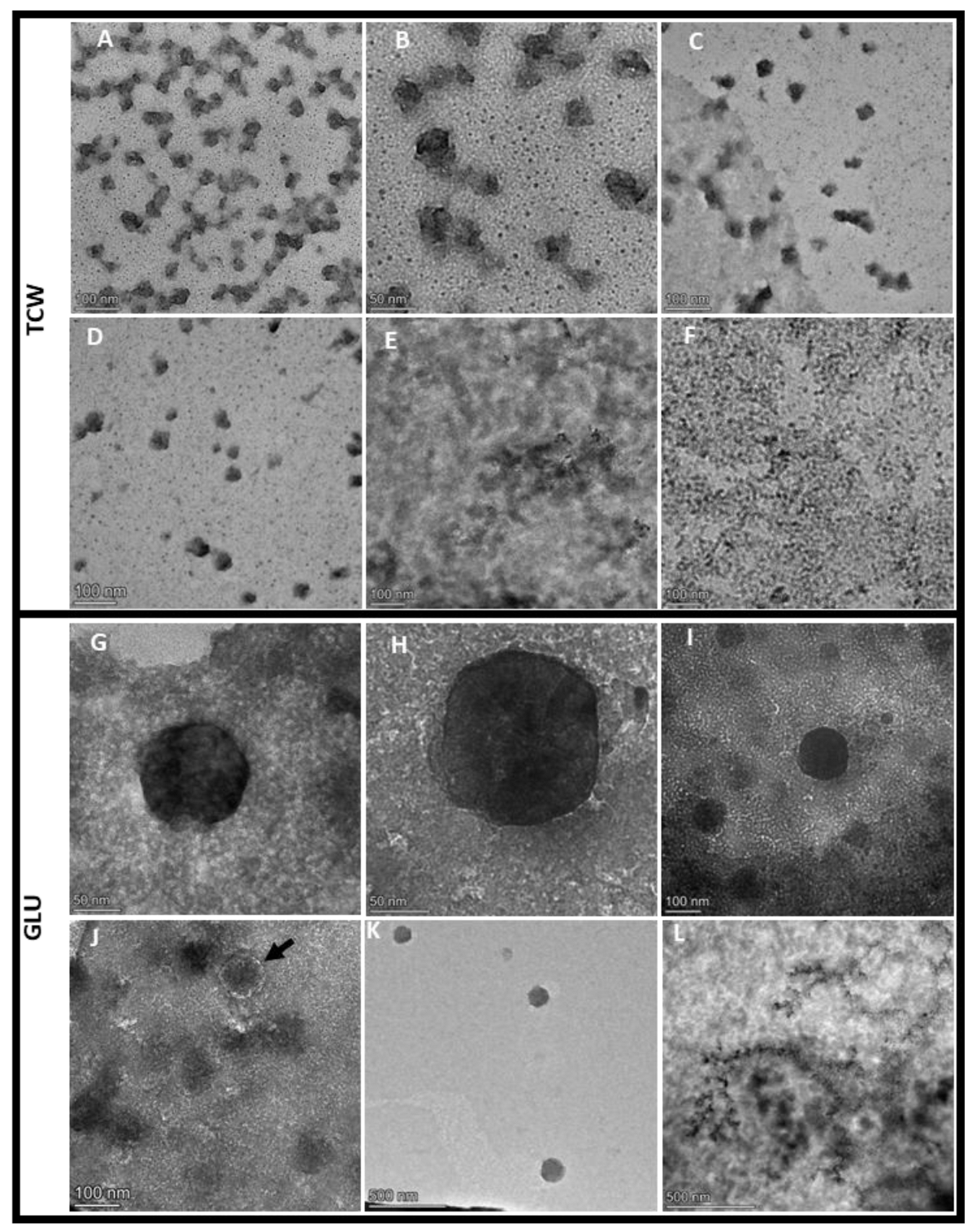
2.3. Transmission Electron Microscopy
2.4. Protein Extraction
2.5. Proteomic Analysis by LC-MS Analysis
2.6. Bioinformatic Analysis
3. Results and Discussion
3.1. EVs Isolated from B. cinerea Showed Different Morphologies Depending on Used Plant-Based Elicitor
3.2. Applied Methods Allowed the Characterization of EVs by Their Protein Composition
3.3. In Silico Analysis Corroborates the Isolation of High-Purity EVs Fraction
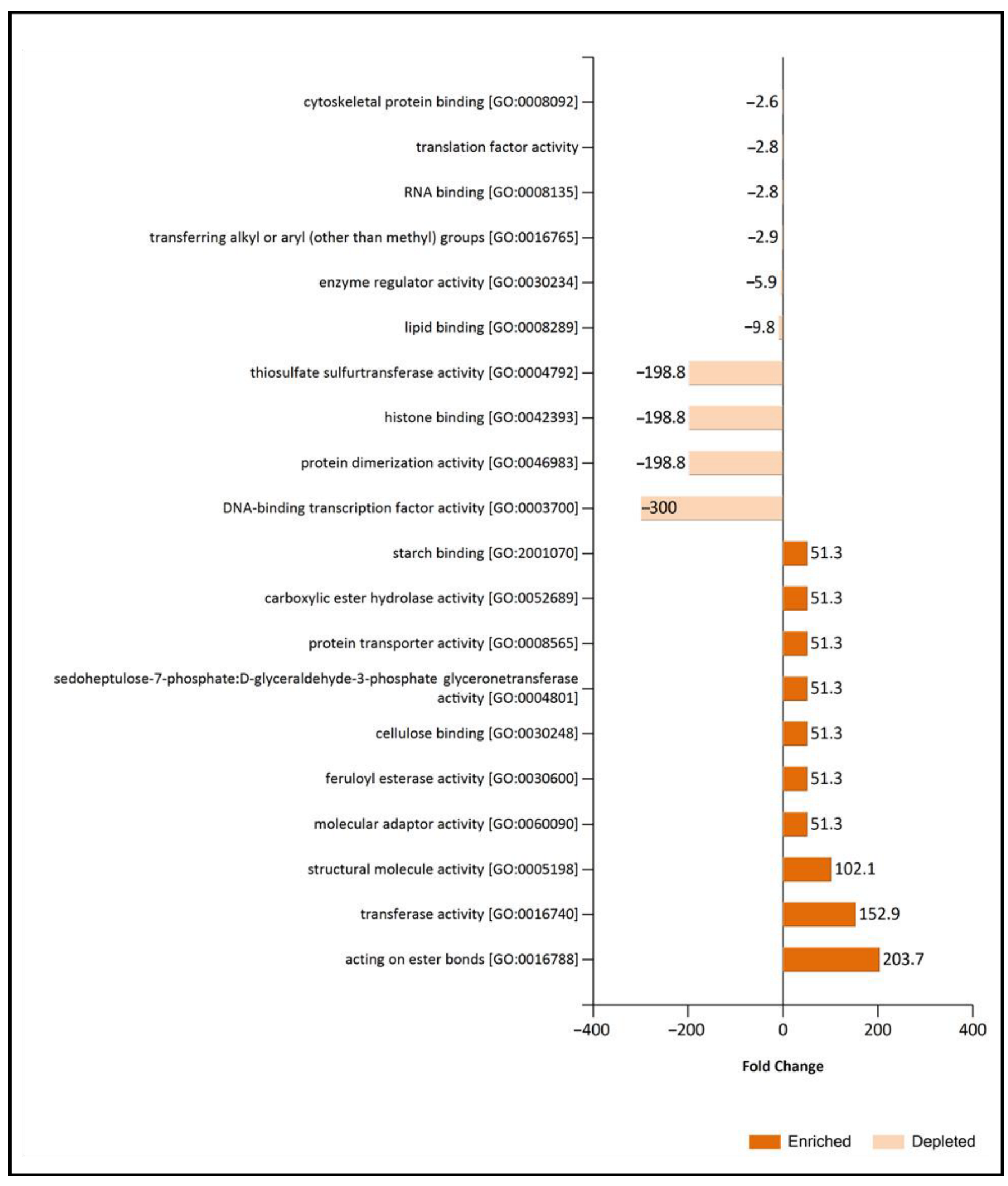
3.4. B. cinerea EVs Have a Functional Protein Profile Distinct from Supernatant Control, and It Is Also Adapted to the Environmental Condition
3.5. KEGG Analysis Reinforce the Involvement of B. cinerea EVs in the Infection Process
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rybak, K.; Robatzek, S. Functions of extracellular vesicles in immunity and Virulence. Plant Physiol. 2019, 179, 1236–1247. [Google Scholar] [CrossRef]
- Rizzo, J.; Taheraly, A.; Janbon, G. Structure, composition and biological properties of fungal extracellular vesicles. microLife 2021, 2, uqab009. [Google Scholar] [CrossRef]
- Takeo, K.; Uesaka, I.; Uehira, K.; Nishiura, M. Fine Structure of Cryptococcus neoformans Grown in vitro as Observed by Freeze-Etching. J. Bacteriol. 1973, 113, 1442–1448. [Google Scholar] [CrossRef]
- Liebana-Jordan, M.; Brotons, B.; Falcon-Perez, J.M.; Gonzalez, E. Extracellular vesicles in the fungik. Int. J. Mol. Sci. 2021, 22, 7221. [Google Scholar] [CrossRef]
- Garcia-Ceron, D.; Lowe, R.G.T.; McKenna, J.A.; Brain, L.M.; Dawson, C.S.; Clark, B.; Berkowitz, O.; Faou, P.; Whelan, J.; Bleackley, M.R.; et al. Extracellular Vesicles from Fusarium graminearum Contain Protein Effectors Expressed during Infection of Corn. J. Fungi 2021, 7, 977. [Google Scholar] [CrossRef]
- Rutter, B.D.; Chu, T.T.; Dallery, J.F.; Zajt, K.K.; O’Connell, R.J.; Innes, R.W. The development of extracellular vesicle markers for the fungal phytopathogen Colletotrichum higginsianum. J. Extracell. Vesicles 2022, 11, e12216. [Google Scholar] [CrossRef]
- De Vallee, A.; Dupuy, J.W.; Moriscot, C.; Gallet, B.; Vanderperre, S.; Guignard, G.; Rascle, C.; Calvar, G.; Malbert, B.; Gillet, F.X.; et al. Extracellular Vesicles of the Plant Pathogen Botrytis cinerea. J. Fungi 2023, 9, 495. [Google Scholar] [CrossRef]
- Fillinger, S.; Elad, Y. Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems; Springer International Publishing: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Staats, M.; van Kan, J.A. Genome update of Botrytis cinerea strains B05.10 and T4. Eukaryot. Cell 2012, 11, 1413–1414. [Google Scholar] [CrossRef]
- Atwell, S.; Corwin, J.A.; Soltis, N.E.; Subedy, A.; Denby, K.J.; Kliebenstein, D.J. Whole genome resequencing of Botrytis cinerea isolates identifies high levels of standing diversity. Front. Microbiol. 2015, 6, 996. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Rutherford, K.; Irvine, A.; Pedro, H.; Pant, R.; Sadanadan, V.; Khamari, L.; Billal, S.; Mohanty, S.; et al. PHI-base: A new interface and further additions for the multi-species pathogen-host interactions database. Nucleic Acids Res. 2017, 45, D604–D610. [Google Scholar] [CrossRef]
- Escobar-Niño, A.; Morano Bermejo, I.M.; Carrasco Reinado, R.; Fernandez-Acero, F.J. Deciphering the Dynamics of Signaling Cascades and Virulence Factors of B. cinerea during Tomato Cell Wall Degradation. Microorganisms 2021, 9, 1837. [Google Scholar] [CrossRef]
- Lineiro, E.; Macias-Sanchez, A.J.; Espinazo, M.; Cantoral, J.M.; Moraga, J.; Collado, I.G.; Fernandez-Acero, F.J. Phenotypic Effects and Inhibition of Botrydial Biosynthesis Induced by Different Plant-Based Elicitors in Botrytis cinerea. Curr. Microbiol. 2018, 75, 431–440. [Google Scholar] [CrossRef]
- Rizzo, J.; Rodrigues, M.L.; Janbon, G. Extracellular Vesicles in Fungi: Past, Present, and Future Perspectives. Front. Cell Infect. Microbiol. 2020, 10, 346. [Google Scholar] [CrossRef]
- Bleackley, M.R.; Dawson, C.S.; Anderson, M.A. Fungal Extracellular Vesicles with a Focus on Proteomic Analysis. Proteomics 2019, 19, e1800232. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Acero, F.J.; Colby, T.; Harzen, A.; Carbu, M.; Wieneke, U.; Cantoral, J.M.; Schmidt, J. 2-DE proteomic approach to the Botrytis cinerea secretome induced with different carbon sources and plant-based elicitors. Proteomics 2010, 10, 2270–2280. [Google Scholar] [CrossRef]
- Vallejo, I.; Carbú, M.; Muñoz, F.; Rebordinos, L.; Cantoral, J.M. Inheritance of chromosome-length polymorphisms in the phytopathogenic ascomycete Botryotinia fuckeliana (anam. Botrytis cinerea). Mycol. Res. 2002, 106, 1075–1085. [Google Scholar] [CrossRef]
- English, P.D.; Jurale, J.B.; Albersheim, P. Host-Pathogen Interactions: II. Parameters Affecting Polysaccharide-degrading Enzyme Secretion by Colletotrichum lindemuthianum Grown in Culture. Plant Physiol. 1971, 47, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hill, E.H.; Solomon, P.S. Extracellular vesicles from the apoplastic fungal wheat pathogen Zymoseptoria tritici. Fungal Biol. Biotechnol. 2020, 7, 13. [Google Scholar] [CrossRef]
- Bleackley, M.R.; Samuel, M.; Garcia-Ceron, D.; McKenna, J.A.; Lowe, R.G.T.; Pathan, M.; Zhao, K.; Ang, C.S.; Mathivanan, S.; Anderson, M.A. Extracellular Vesicles from the Cotton Pathogen Fusarium oxysporum f. sp. vasinfectum Induce a Phytotoxic Response in Plants. Front. Plant Sci. 2019, 10, 1610. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Rawat, S.; Arora, V.; Kottarath, S.K.; Dinda, A.K.; Vaishnav, P.K.; Nayak, B.; Mohanty, S. An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Stem. Cell Res. Ther. 2018, 9, 180. [Google Scholar] [CrossRef]
- Rappsilber, J.; Ishihama, Y.; Mann, M. Stop and Go Extraction Tips for Matrix-Assisted Laser Desorption/Ionization, Nanoelectrospray, and LC/MS Sample Pretreatment in Proteomics. Anal. Chem. 2003, 75, 663–670. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, F.M.; Wang, N.; Magee, G.B.; Nanduri, B.; Lawrence, M.L.; Camon, E.B.; Barrell, D.G.; Hill, D.P.; Dolan, M.E.; Williams, W.P.; et al. AgBase: A functional genomics resource for agriculture. BMC Genom. 2006, 7, 229. [Google Scholar] [CrossRef] [PubMed]
- Fonseka, P.; Pathan, M.; Chitti, S.V.; Kang, T.; Mathivanan, S. FunRich enables enrichment analysis of OMICs datasets. J. Mol. Biol. 2021, 433, 166747. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Gíslason, M.H.; Nielsen, H.; Almagro Armenteros, J.J.; Johansen, A.R. Prediction of GPI-anchored proteins with pointer neural networks. Curr. Res. Biotechnol. 2021, 3, 6–13. [Google Scholar] [CrossRef]
- Xie, Y.; Zheng, Y.; Li, H.; Luo, X.; He, Z.; Cao, S.; Shi, Y.; Zhao, Q.; Xue, Y.; Zuo, Z.; et al. GPS-Lipid: A robust tool for the prediction of multiple lipid modification sites. Sci. Rep. 2016, 6, 28249. [Google Scholar] [CrossRef]
- Zhao, L.; Poschmann, G.; Waldera-Lupa, D.; Rafiee, N.; Kollmann, M.; Stuhler, K. OutCyte: A novel tool for predicting unconventional protein secretion. Sci. Rep. 2019, 9, 19448. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N. EffectorP 3.0: Prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. Mol. Plant Microbe Interact. 2022, 35, 146–156. [Google Scholar] [CrossRef]
- Silva, B.M.; Prados-Rosales, R.; Espadas-Moreno, J.; Wolf, J.M.; Luque-Garcia, J.L.; Goncalves, T.; Casadevall, A. Characterization of Alternaria infectoria extracellular vesicles. Med. Mycol. 2014, 52, 202–210. [Google Scholar] [CrossRef]
- Reis, F.C.G.; Gimenez, B.; Jozefowicz, L.J.; Castelli, R.F.; Martins, S.T.; Alves, L.R.; Oliveira, H.C.d.; Rodrigues, M.L. Analysis of Cryptococcal Extracellular Vesicles: Experimental Approaches for Studying Their Diversity Among Multiple Isolates, Kinetics of Production, Methods of Separation, and Detection in Cultures of Titan Cells. Microbiol. Spectr. 2021, 9, e0012521. [Google Scholar] [CrossRef]
- Cleare, L.G.; Zamith, D.; Heyman, H.M.; Couvillion, S.P.; Nimrichter, L.; Rodrigues, M.L.; Nakayasu, E.S.; Nosanchuk, J.D. Media matters! Alterations in the loading and release of Histoplasma capsulatum extracellular vesicles in response to different nutritional milieus. Cell Microbiol. 2020, 22, e13217. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Dawson, C.S.; Garcia-Ceron, D.; Rajapaksha, H.; Faou, P.; Bleackley, M.R.; Anderson, M.A. Protein markers for Candida albicans EVs include claudin-like Sur7 family proteins. J. Extracell. Vesicles 2020, 9, 1750810. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ceron, D.; Dawson, C.S.; Faou, P.; Bleackley, M.R.; Anderson, M.A. Size-exclusion chromatography allows the isolation of EVs from the filamentous fungal plant pathogen Fusarium oxysporum f. sp. vasinfectum (Fov). Proteomics 2021, 21, e2000240. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.J.; Chirico, W.J.; Lipke, P.N. Through the back door: Unconventional protein secretion. Cell Surf. 2020, 6, 100045. [Google Scholar] [CrossRef]
- Parreira, V.d.S.C.; Santos, L.G.C.; Rodrigues, M.L.; Passetti, F. ExVe: The knowledge base of orthologous proteins identified in fungal extracellular vesicles. Comput. Struct. Biotechnol. J. 2021, 19, 2286–2296. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borras, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [PubMed]
- ten Have, A.; Espino, J.J.; Dekkers, E.; Van Sluyter, S.C.; Brito, N.; Kay, J.; Gonzalez, C.; van Kan, J.A. The Botrytis cinerea aspartic proteinase family. Fungal Genet. Biol. 2010, 47, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Gee, H.Y.; Kim, J.; Lee, M.G. Unconventional secretion of transmembrane proteins. Semin. Cell Dev. Biol. 2018, 83, 59–66. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Lenardon, M.D. Architecture of the dynamic fungal cell wall. Nat. Rev. Microbiol. 2022, 21, 248–259. [Google Scholar] [CrossRef]
- Choquer, M.; Rascle, C.; Gonçalves, I.R.; de Vallée, A.; Ribot, C.; Loisel, E.; Smilevski, P.; Ferria, J.; Savadogo, M.; Souibgui, E.; et al. The infection cushion of Botrytis cinerea: A fungal ‘weapon’ of plant-biomass destruction. Environ. Microbiol. 2021, 23, 2293–2314. [Google Scholar] [CrossRef]
- Frias, M.; Brito, N.; Gonzalez, M.; Gonzalez, C. The phytotoxic activity of the cerato-platanin BcSpl1 resides in a two-peptide motif on the protein surface. Mol. Plant Pathol. 2014, 15, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Frias, M.; Gonzalez, C.; Brito, N. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 2011, 192, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Bleackley, M.; Chisanga, D.; Gangoda, L.; Fonseka, P.; Liem, M.; Kalra, H.; Al Saffar, H.; Keerthikumar, S.; Ang, C.S.; et al. Extracellular vesicles secreted by Saccharomyces cerevisiae are involved in cell wall remodelling. Commun. Biol. 2019, 2, 305. [Google Scholar] [CrossRef] [PubMed]
- Mudholkar, K.; Fitzke, E.; Prinz, C.; Mayer, M.P.; Rospert, S. The Hsp70 homolog Ssb affects ribosome biogenesis via the TORC1-Sch9 signaling pathway. Nat. Commun. 2017, 8, 937. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Dickman, M.B.; Wang, Z. Cytoprotective Co-chaperone BcBAG1 Is a Component for Fungal Development, Virulence, and Unfolded Protein Response (UPR) of Botrytis cinerea. Front. Microbiol. 2019, 10, 685. [Google Scholar] [CrossRef]
- Du, W.; Zhai, P.; Liu, S.; Zhang, Y.; Lu, L. The Copper Chaperone CcsA, Coupled with Superoxide Dismutase SodA, Mediates the Oxidative Stress Response in Aspergillus fumigatus. Appl. Environ. Microbiol. 2021, 87, e0101321. [Google Scholar] [CrossRef]
- Gleason, J.E.; Li, C.X.; Odeh, H.M.; Culotta, V.C. Species-specific activation of Cu/Zn SOD by its CCS copper chaperone in the pathogenic yeast Candida albicans. J. Biol. Inorg. Chem. 2014, 19, 595–603. [Google Scholar] [CrossRef]
- Cotoras, M.; Castro, P.; Vivanco, H.; Melo, R.; Mendoza, L. Farnesol induces apoptosis-like phenotype in the phytopathogenic fungus Botrytis cinerea. Mycologia 2013, 105, 28–33. [Google Scholar] [CrossRef]
- Gioti, A.; Simon, A.; Le Pecheur, P.; Giraud, C.; Pradier, J.M.; Viaud, M.; Levis, C. Expression profiling of Botrytis cinerea genes identifies three patterns of up-regulation in planta and an FKBP12 protein affecting pathogenicity. J. Mol. Biol. 2006, 358, 372–386. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.; Zhang, C.; Wu, T.; Ma, Z.; Chen, Y. Phospholipid homeostasis plays an important role in fungal development, fungicide resistance and virulence in Fusarium graminearum. Phytopathol. Res. 2019, 1, 16. [Google Scholar] [CrossRef]
- Ma, L.; Salas, O.; Bowler, K.; Oren-Young, L.; Bar-Peled, M.; Sharon, A. Genetic alteration of UDP-rhamnose metabolism in Botrytis cinerea leads to the accumulation of UDP-KDG that adversely affects development and pathogenicity. Mol. Plant Pathol. 2017, 18, 263–275. [Google Scholar] [CrossRef]
- Santhanam, P.; Boshoven, J.C.; Salas, O.; Bowler, K.; Islam, M.T.; Saber, M.K.; van den Berg, G.C.; Bar-Peled, M.; Thomma, B.P. Rhamnose synthase activity is required for pathogenicity of the vascular wilt fungus Verticillium dahliae. Mol. Plant Pathol. 2017, 18, 347–362. [Google Scholar] [CrossRef]
- Kars, I.; Krooshof, G.H.; Wagemakers, L.; Joosten, R.; Benen, J.A.; van Kan, J.A. Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris. Plant J. 2005, 43, 213–225. [Google Scholar] [CrossRef]
- Zhang, L.; van Kan, J.A. Botrytis cinerea mutants deficient in D-galacturonic acid catabolism have a perturbed virulence on Nicotiana benthamiana and Arabidopsis, but not on tomato. Mol. Plant Pathol. 2013, 14, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hua, C.; Stassen, J.H.M.; Chatterjee, S.; Cornelissen, M.; van Kan, J.A.L. Genome-wide analysis of pectate-induced gene expression in Botrytis cinerea: Identification and functional analysis of putative d-galacturonate transporters. Fungal Genet. Biol. 2014, 72, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Thiewes, H.; van Kan, J.A. The D-galacturonic acid catabolic pathway in Botrytis cinerea. Fungal Genet. Biol. 2011, 48, 990–997. [Google Scholar] [CrossRef]
- Beccaccioli, M.; Reverberi, M.; Scala, V. Fungal lipids: Biosynthesis and signalling during plant-pathogen interaction. Front. Biosci. 2019, 24, 172–185. [Google Scholar] [CrossRef]
- Chen, D.; Shu, D.; Wei, Z.; Luo, D.; Yang, J.; Li, Z.; Tan, H. Combined transcriptome and proteome analysis of Bcfrp1 involved in regulating the biosynthesis of abscisic acid and growth in Botrytis cinerea TB-31. Front. Microbiol. 2022, 13, 1085000. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Que, Y.; Wang, H.; Wang, C.; Li, Y.; Yue, X.; Ma, Z.; Talbot, N.J.; Wang, Z. The MET13 methylenetetrahydrofolate reductase gene is essential for infection-related morphogenesis in the rice blast fungus Magnaporthe oryzae. PLoS ONE 2013, 8, e76914. [Google Scholar] [CrossRef]
- Sprenger, M.; Hartung, T.S.; Allert, S.; Wisgott, S.; Niemiec, M.J.; Graf, K.; Jacobsen, I.D.; Kasper, L.; Hube, B. Fungal biotin homeostasis is essential for immune evasion after macrophage phagocytosis and virulence. Cell Microbiol. 2020, 22, e13197. [Google Scholar] [CrossRef]
- Siegmund, U.; Viefhues, A. Reactive Oxygen Species in the Botrytis—Host Interaction. In Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems; Fillinger, S., Elad, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 269–289. [Google Scholar]
- Zou, X.; Wei, Y.; Jiang, S.; Xu, F.; Wang, H.; Zhan, P.; Shao, X. ROS Stress and Cell Membrane Disruption are the Main Antifungal Mechanisms of 2-Phenylethanol against Botrytis cinerea. J. Agric. Food Chem. 2022, 70, 14468–14479. [Google Scholar] [CrossRef] [PubMed]
- Viefhues, A.; Heller, J.; Temme, N.; Tudzynski, P. Redox systems in Botrytis cinerea: Impact on development and virulence. Mol. Plant Microbe Interact. 2014, 27, 858–874. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Cruz, J.; Oscar, C.S.; Emma, F.C.; Pilar, G.A.; Carmen, G.B. Absence of Cu-Zn superoxide dismutase BCSOD1 reduces Botrytis cinerea virulence in Arabidopsis and tomato plants, revealing interplay among reactive oxygen species, callose and signalling pathways. Mol. Plant Pathol. 2017, 18, 16–31. [Google Scholar] [CrossRef] [PubMed]
- Dautt-Castro, M.; Rosendo-Vargas, M.; Casas-Flores, S. The Small GTPases in Fungal Signaling Conservation and Function. Cells 2021, 10, 1039. [Google Scholar] [CrossRef]
- Zhang, X.; Jia, X.; Tian, S.; Zhang, C.; Lu, Z.; Chen, Y.; Chen, F.; Li, Z.; Su, X.; Han, X.; et al. Role of the small GTPase Rho1 in cell wall integrity, stress response, and pathogenesis of Aspergillus fumigatus. Fungal Genet. Biol. 2018, 120, 30–41. [Google Scholar] [CrossRef]
- An, B.; Li, B.; Qin, G.; Tian, S. Function of small GTPase Rho3 in regulating growth, conidiation and virulence of Botrytis cinerea. Fungal Genet. Biol. 2015, 75, 46–55. [Google Scholar] [CrossRef]
- Cox, R.; Mason-Gamer, R.J.; Jackson, C.L.; Segev, N. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol. Biol. Cell 2004, 15, 1487–1505. [Google Scholar] [CrossRef]
- Araiza-Cervantes, C.A.; Valle-Maldonado, M.I.; Patiño-Medina, J.A.; Alejandre-Castañeda, V.; Guzmán-Pérez, J.B.; Ramírez-Díaz, M.I.; Macias-Sánchez, K.L.; López-Berges, M.S.; Meza-Carmen, V. Transcript pattern analysis of Arf-family genes in the phytopathogen Fusarium oxysporum f. sp. lycopersici reveals the role of Arl3 in the virulence. Antonie Van Leeuwenhoek 2021, 114, 1619–1632. [Google Scholar] [CrossRef]






| TMHMM | SP | UPS | Intracellular | Localization | |
|---|---|---|---|---|---|
| EVs GLU exclusive and overrepresented proteins | 12.63% | 6.59% | 24.7% | 56.08% | 59.9% cytoplasm (53.21% lipidated) |
| 13.2% mitochondrion | |||||
| 7.7% nucleus | |||||
| 6% endoplasmic reticulum | |||||
| 4.4% lysosome | |||||
| 4.4% extracellular (50% lipidated) | |||||
| 2.2% cell membrane | |||||
| 1.64% peroxisome | |||||
| 0.54% golgi | |||||
| EVs TCW exclusive and overrepresented proteins | 15.38% | 2.4% | 28.4% | 53.85% | 63% cytoplasm (52.7% lipidated) |
| 12% mitochondrion | |||||
| 7.21% cell membrane | |||||
| 6.7% nucleus | |||||
| 4.8% endoplasmic reticulum | |||||
| 2.9% lysosome/vacuole | |||||
| 1.9% extracellular (50% lipidated) | |||||
| 0.96% peroxisome | |||||
| 0.5% golgi | |||||
| Supernatant GLU exclusive and overrepresented proteins overrepresented proteins | 3.9% | 60.6% | 6.3% | 29.1% | 74.8% extracellular (58.9% lipidated) |
| 16.5% cytoplasm (66.7% lipidated) | |||||
| 3.1% lysosome/vacuole | |||||
| 3.1% nucleus | |||||
| 1.6% endoplasmic reticulum | |||||
| 0.8% mitochondrion | |||||
| Supernatant TCW exclusive and overrepresented proteins | 6.89% | 27.58% | 24,13% | 41.38% | 51.7% cytoplasm (53.3% lipidated) |
| 20.7% extracellular (50% lipidated) | |||||
| 10.3% endoplasmic reticulum | |||||
| 6.9% mitochondrion | |||||
| 6.9% cell membrane | |||||
| 3.4% nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escobar-Niño, A.; Harzen, A.; Stolze, S.C.; Nakagami, H.; Fernández-Acero, F.J. The Adaptation of Botrytis cinerea Extracellular Vesicles Proteome to Surrounding Conditions: Revealing New Tools for Its Infection Process. J. Fungi 2023, 9, 872. https://doi.org/10.3390/jof9090872
Escobar-Niño A, Harzen A, Stolze SC, Nakagami H, Fernández-Acero FJ. The Adaptation of Botrytis cinerea Extracellular Vesicles Proteome to Surrounding Conditions: Revealing New Tools for Its Infection Process. Journal of Fungi. 2023; 9(9):872. https://doi.org/10.3390/jof9090872
Chicago/Turabian StyleEscobar-Niño, Almudena, Anne Harzen, Sara C. Stolze, Hirofumi Nakagami, and Francisco J. Fernández-Acero. 2023. "The Adaptation of Botrytis cinerea Extracellular Vesicles Proteome to Surrounding Conditions: Revealing New Tools for Its Infection Process" Journal of Fungi 9, no. 9: 872. https://doi.org/10.3390/jof9090872





