Alkaliphilic/Alkali-Tolerant Fungi: Molecular, Biochemical, and Biotechnological Aspects
Abstract
:1. Introduction
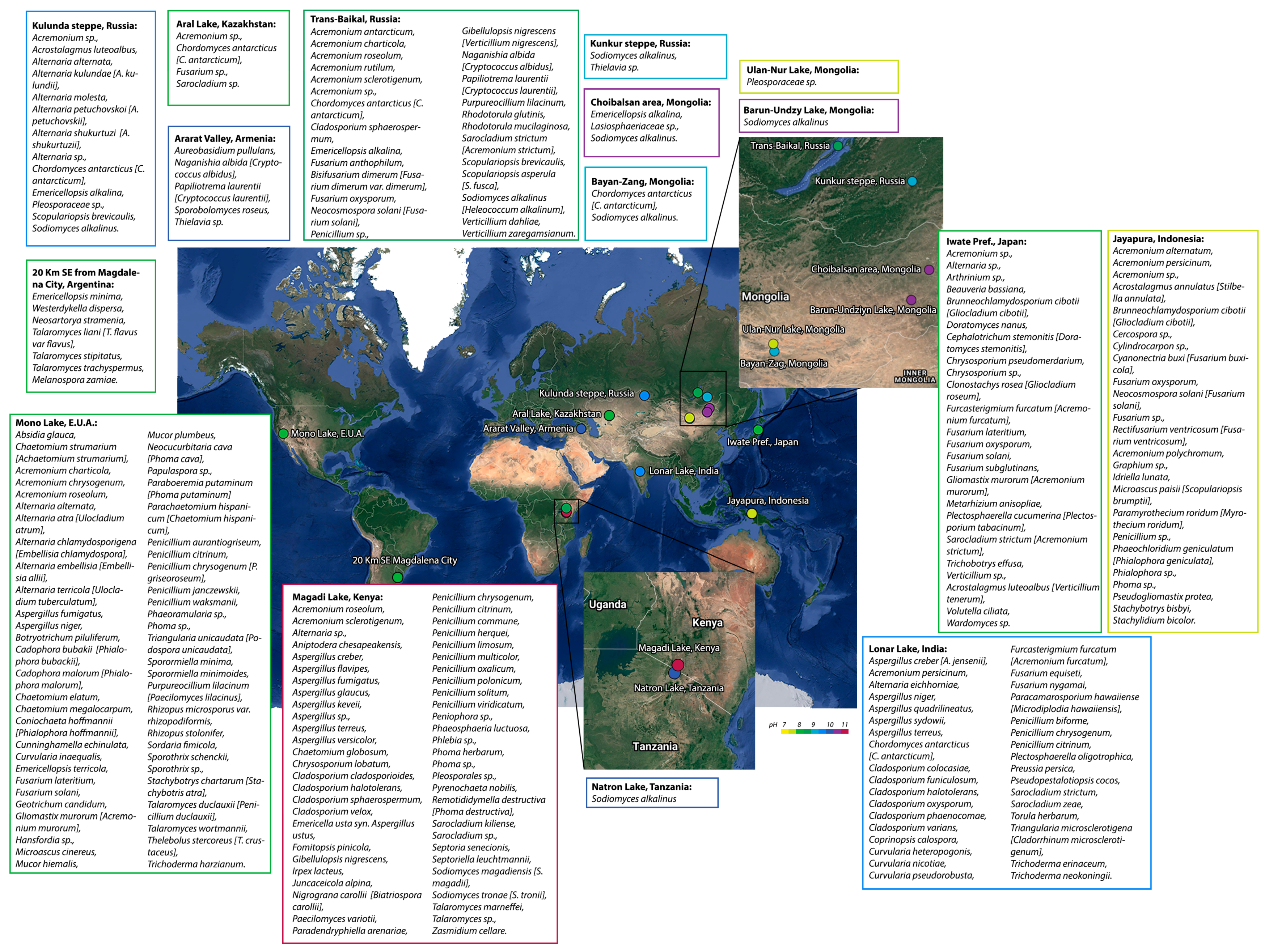
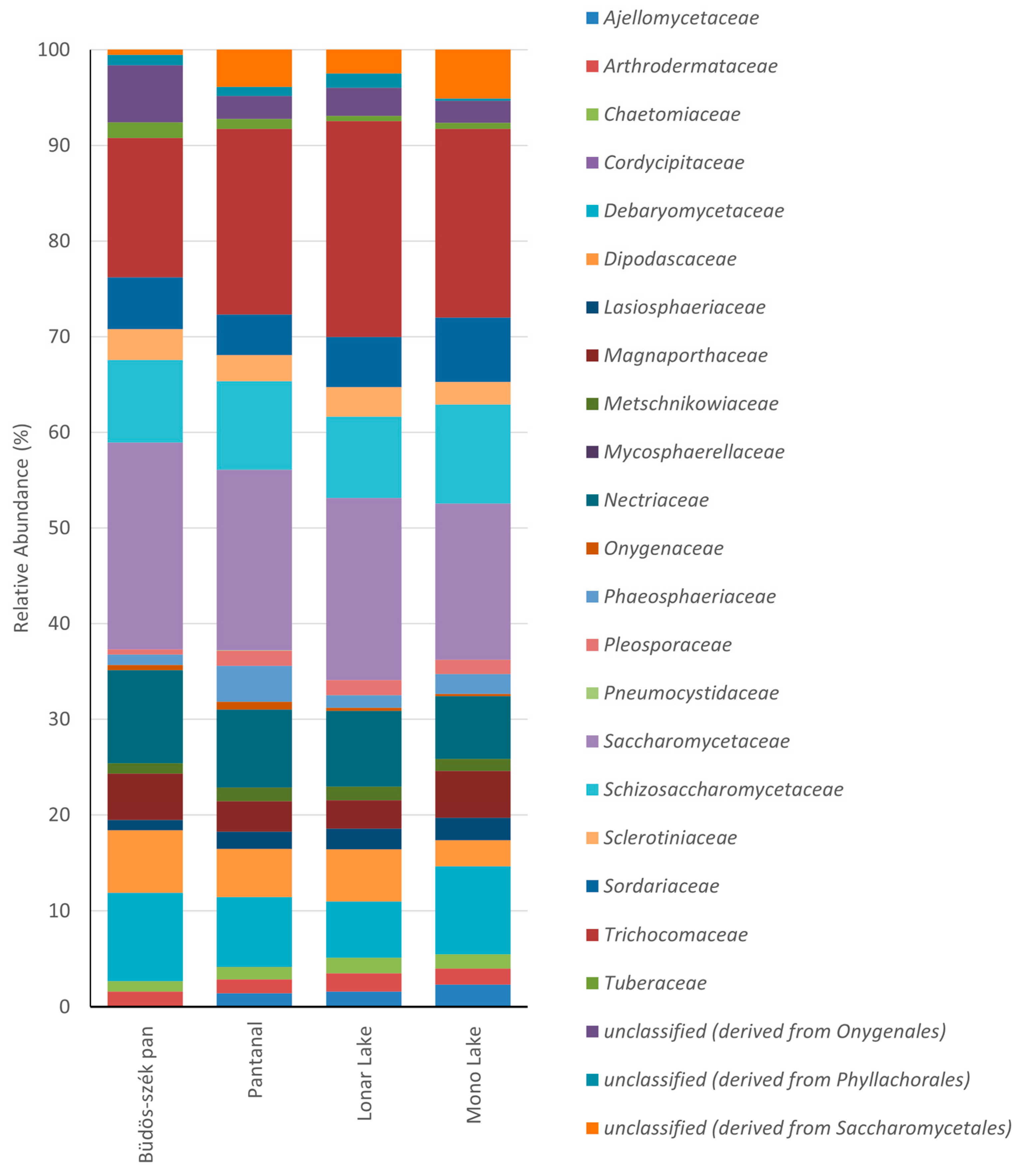
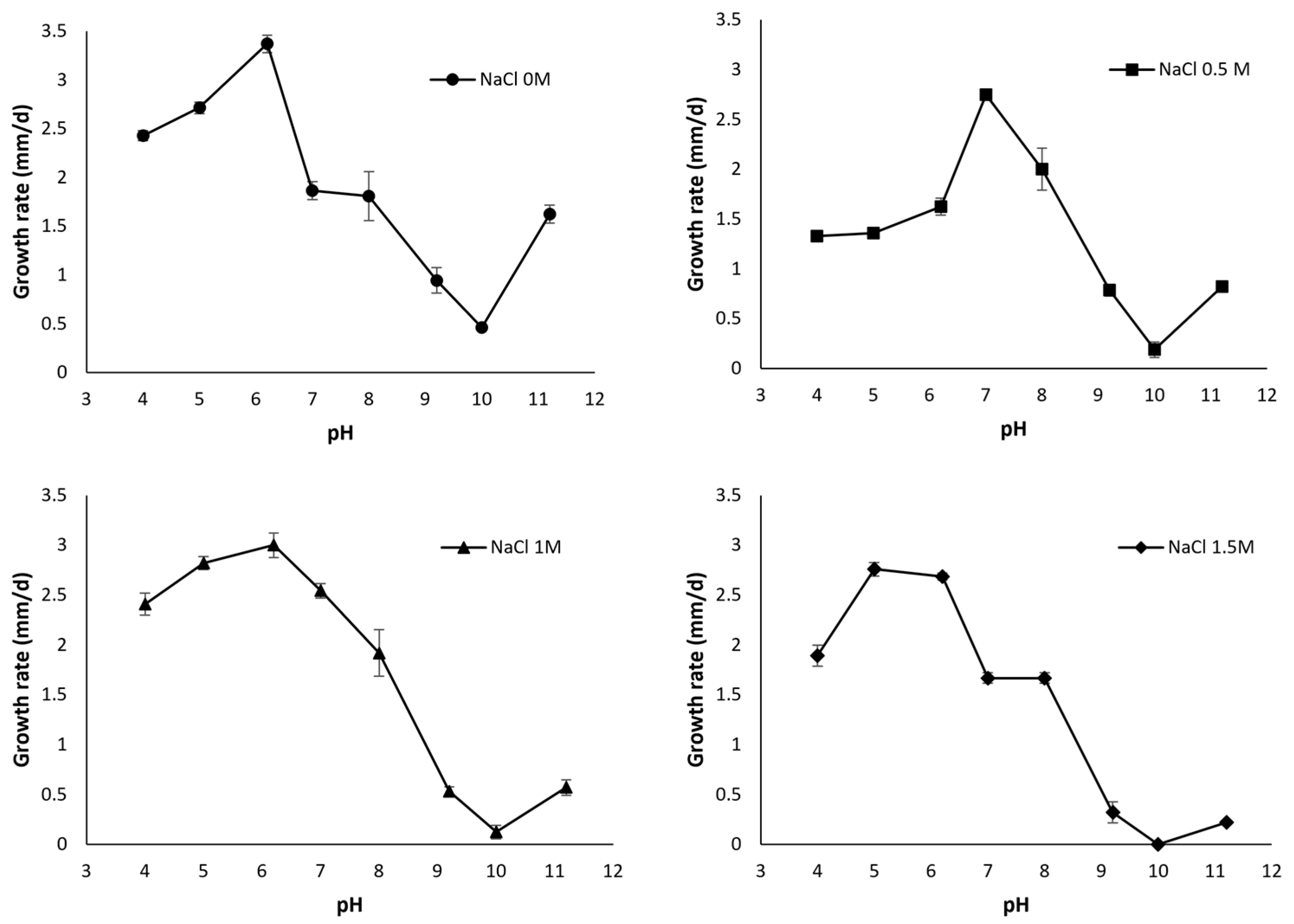
2. Fungal Diversity in Alkaline Environments
3. Physiology
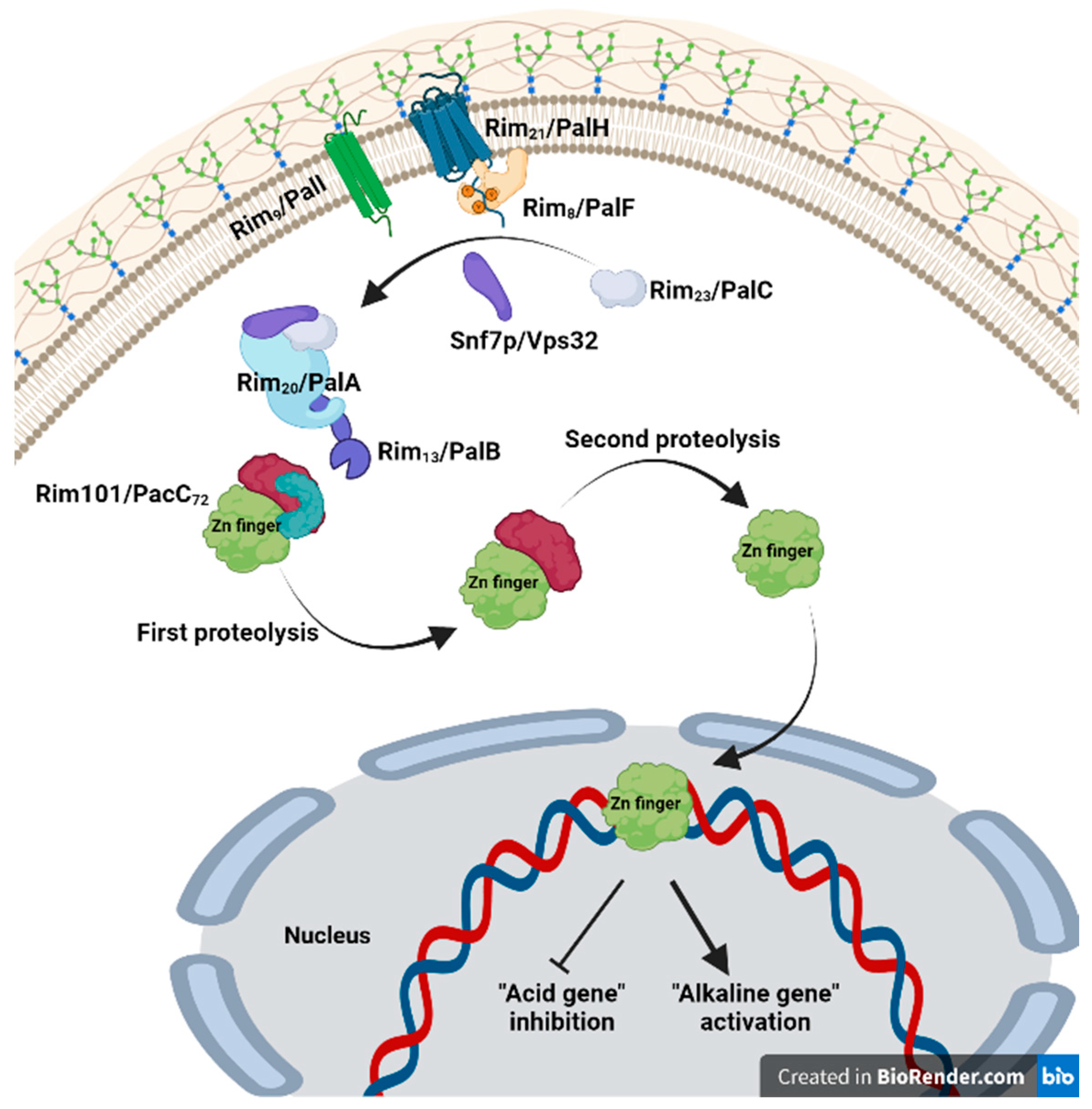
3.1. Genes, Transcription Factors, and Signaling Mechanism
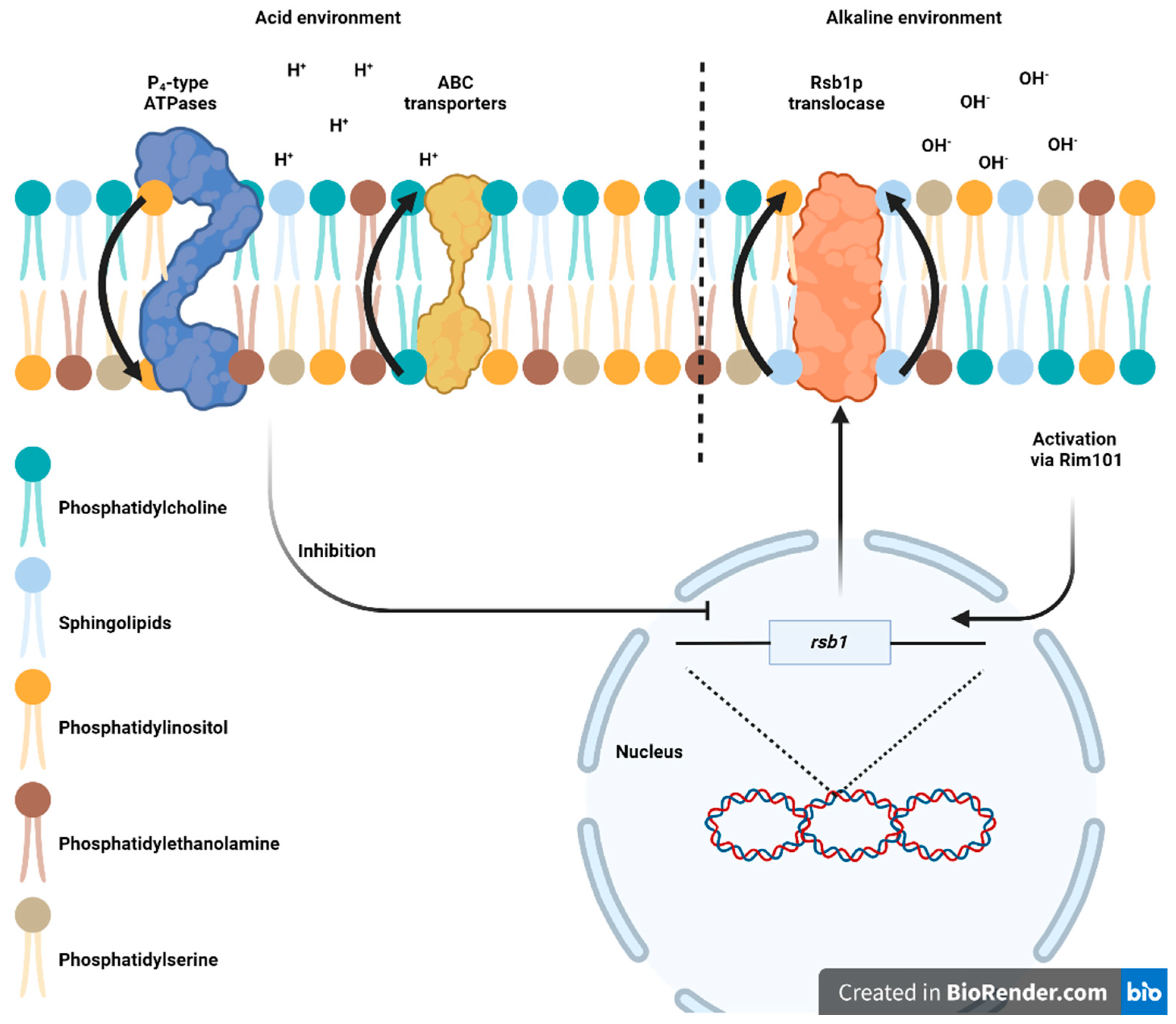
3.2. Membrane Adaptations
3.3. Protein Adaptations
4. Biotechnological Impact of Alkaliphilic and Alkali-Tolerant Fungi
| Enzyme | Microorganism | pH Enzyme Production | Optimum pH for Activity | Optimum T (°C) for Activity | References |
|---|---|---|---|---|---|
| Alkaline protease | Aspergillus flavus | 7 | Optimum activity in the pH range of 7.5–9.5. | Optimum activity at 42 °C. | [107] |
| Neosartorya clavata (syn. Aspergillus clavatus) | 8 | Optimum activity at pH 8.5. It is active in the pH range of 6.0–11.0. The enzyme is stable after an hour of incubation at pH 8.0–9.0. | The enzyme was active between 40 and 70 °C with an optimum around 50 °C. Stable activity after 1 h treatment at 30 °C. | [108,109] | |
| Sodiomyces alkalinus | 10 | Optimum activity at pH 8. | Optimum activity at 30 °C. | [64] | |
| Emericella usta (syn. Aspergillus ustus) | 9 | Enzyme active in a wide pH range (6.0–10.0), with an optimum at pH 9.0. | Optimum activity at 45 °C. | [110] | |
| Serine protease | Aspergillus parasiticus | - | Optimum activity at pH 8. Stable activity after 1 h treatment in pH 6.0–10.0. | Optimum activity at 40 °C. The enzyme was stable at 40 °C for 1 h incubation but was inactivated at temperatures over 40 °C | [111] |
| Parengyodontium album (syn. Engyodontium album) | 10 | Enzyme active in a wide pH range (6.0–12.0), with an optimum at pH 11.0. | Optimum activity at 60 °C. More than 90% of the maximal activity was conserved between 45 and 65 °C. | [112] | |
| Clonostachys rosea (syn. Gliocladium roseum) | 7–7.5 | Optimum activity in the pH range of 9.0–10.0. | Optimum activity at 60 °C. | [113] | |
| Trichoderma reesei | 6 | Enzyme active in a wide pH range (6.0–11.0) with an optimum at pH 8.0. | Optimum activity at 50 °C. | [114] | |
| Dipeptidyl peptidase 4 | Chordomyces antarcticus | 10 | Optimum activity at pH 7.7. Stable in the pH range of 3.0–12.0. | Optimum activity at 37 °C. | [115] |
| Sodiomyces alkalinus | 10 | Optimum activity at pH 7.3. Stable in the pH range of 5.0–12.0 and retained 23% of its activity after an hour of incubation at pH 13.0 | Optimum activity at 37 °C. | [115] | |
| Trypsin-like protease | Cordyceps militaris | 6 | Optimum activity in the pH range of 8.5–12.0. | Optimum activity at 25 °C. | [116] |
| Xylanase | Penicillium citrinum | - | Optimum activity at pH 8.5. | Optimum activity at 50 °C. | [117] |
| Cladosporium oxysporum | 7–8 | Optimum activity at pH 8.0. Stable activity after 2 h treatment in pH 7.0–8.5. | Optimum activity at 50 °C. Stable activity after 2 h treatment below 55 °C. | [118] | |
| Aspergillus fischeri | 9 | Optimum activity pH 6.0. Retained over 50% of maximum activity at pH 8.0. pH stability ranged from 5.5 to 9.5 with retention of more than 85% of the activity. | The optimum temperature was 60°C. | [119] | |
| Aspergillus fumigatus | 8 | Optimum pH 8. Substantial residual activity at alkaline pH 8–9 (56–88%). | Optimum temperature 50 °C. Substantial residual activity at 60–70 °C (53–75%). | [120] | |
| Amylases | Clavispora lusitaniae | 7 | Optimum activity at pH 11. Enzymes retained nearly 80% of activity after being exposed to various detergent components for 2 h. | Optimum activity at 40 °C. The enzyme retained 45% and 98% of their maximum activity at 4 °C and 25 °C. | [121] |
| Sporormiella minima (syn. Preussia mínima) | Optimum pH 9. | Optimum activity at 25 °C. | [122] | ||
| Endoglucanase B | Aspergillus niger (The gene encoding the enzyme was cloned and expressed in Pichia pastoris) | - | Stable for 2 h at alkaline pH (7–10). | Stable for 3 h at temperatures below 60 °C. | [123] |
| Endoglucanases | Mycothermus thermophilus (syn. Humicola insolens) (The gene encoding the enzyme was cloned and expressed in Aspergillus oryzae) | - | Optimal activity between pH 7 and 8.5. | [124] | |
| Cellobiohydrolase II | - | Optimal activity pH 9. | |||
| Laccase | Albifimbria verrucaria (syn. Myrothecium verrucaria) | 9 | Optimum activity at pH 9.0 and retained more than 80% of the initial activity after 17 h incubation at 30 °C at pH 8–11.5. | Stable for 1 h at temperatures below 50 °C and retained more than 80% of the initial activity. | [125] |
| Lipases | Trichoderma lentiforme (The gene encoding the enzyme was cloned and expressed in Pichia pastoris) | - | Poor activity under acidic conditions and maximum activity at pH 9.5. The enzyme was stable at pH 6.0–9.0, retaining more than 80% of the initial activity after 1 h pre- incubation at 37 °C. | Temperature optimum at 50 °C and the enzyme was relatively stable at 40 °C, retaining more than 60% of the initial activity after 1 h incubation. | [126] |
| Aspergillus carneus | 8 | The purified enzyme could tolerate pH 6.0–12.0 and it was stable in this range for 24 h. The optimum pH was 9.0. | The optimum temperature was 37 °C and the enzyme was active in the range of 5- 90 °C. | [127] | |
| Lasiodiplodia theobromae | 8 | Optimum activity pH 8.0. | Optimum temperature was 30 °C. | [128] | |
| Gliocladium sp. | - | Optimum activity pH 10. | The assay temperature was 40 °C. | [129] | |
| Verticillium sp. | - | The enzyme was fully stable for 30 min at pH 9 at temperatures up to 50 °C. The enzyme was stable throughout the pH range 6–10 at 25 °C for 24 h. | [129] |
5. Aspergillus sydowii, a Case Study
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berlemont, R.; Gerday, C. Extremophiles. In Comprehensive Biotechnology; Moo-Young, M., Ed.; Elsevier, B.V.: Liége, Belgium, 2011; pp. 229–242. ISBN 978-0-08-088504-9. [Google Scholar]
- Macelroy, R.D. Some comments on the evolution of extremophiles. BioSystems 1974, 6, 74–75. [Google Scholar] [CrossRef]
- Yun, S.H.; Lee, Y.G.; Choi, C.-W.; Lee, S.-Y.; Kim, S.I. Proteomic Exploration of Extremophiles. Curr. Biotechnol. 2014, 3, 87–99. [Google Scholar] [CrossRef]
- Peeples, T.L. Bioremediation Using Extremophiles. In Microbial Biodegradation and Bioremediation; Das, S., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2014; pp. 251–268. ISBN 978-0-12-800021-2. [Google Scholar]
- Grum-Grzhimaylo, A.A.; Georgieva, M.L.; Bondarenko, S.A.; Debets, A.J.M.; Bilanenko, E.N. On the diversity of fungi from soda soils. Fungal Divers. 2016, 76, 27–74. [Google Scholar] [CrossRef] [Green Version]
- Matkawala, F.; Nighojkar, S.; Kumar, A.; Nighojkar, A. A novel thiol-dependent serine protease from Neocosmospora sp. N1. Heliyon 2019, 5, e02246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devi, R.; Revathi, K.; Arunagirinathan, N.; Yogananth, N. Production and Optimization of α-amylase from Aspergillus ochraceus isolated from Marakkanam Saltpans, Tamil Nadu, India. Bull. Environ. Pharmacol. Life Sci. 2022, 1, 997–1002. [Google Scholar]
- Hasnaoui, I.; Dab, A.; Mechri, S.; Abouloifa, H.; Saalaoui, E.; Jaouadi, B.; Noiriel, A.; Asehraou, A.; Abousalham, A. Purification, Biochemical and Kinetic Characterization of a Novel Alkaline sn-1,3-Regioselective Triacylglycerol Lipase from Penicillium crustosum Thom Strain P22 Isolated from Moroccan Olive Mill Wastewater. Int. J. Mol. Sci. 2022, 23, 1920. [Google Scholar] [CrossRef] [PubMed]
- Horikoshi, K. Alkaliphiles: Some applications of their products for biotechnology. Microbiol. Mol. Biol. Rev. 1999, 63, 735–750. [Google Scholar] [CrossRef] [Green Version]
- Kanekar, P.P.; Joshi, A.A.; Kulkarni, S.O.; Borgave, S.B.; Sarnaik, S.S.; Nilegaonkar, S.S.; Kelkar, A.S.; Thombre, R.S. Biotechnological potential of alkaliphilic microorganisms. In Biotechnology and Bioinformatics, Advances and Applications for Bioenergy, Bioremediation, and Biopharmaceutical Research; Hangadurai, D., Sangeetha, J., Eds.; CRC Press Taylor & Francis Group: Toronto, ON, Canada, 2015; pp. 249–280. ISBN 0101001010101. [Google Scholar]
- Jones, B.E.; Grant, W.D.; Duckworth, A.W.; Owenson, G.G. Microbial diversity of soda lakes. Extremophiles 1998, 2, 191–200. [Google Scholar] [CrossRef]
- Peñalva, M.A.; Arst, H.N. Recent Advances in the Characterization of Ambient pH Regulation of Gene Expression in Filamentous Fungi and Yeasts. Annu. Rev. Microbiol. 2004, 58, 425–451. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Muyzer, G. Desulfurispira natronophila gen. nov. sp. nov.: An obligately anaerobic dissimilatory sulfur-reducing bacterium from soda lakes. Extremophiles 2010, 14, 349–355. [Google Scholar] [CrossRef] [Green Version]
- Sorokin, D.Y.; Panteleeva, A.N.; Tourova, T.P.; Kaparullina, E.N.; Muyzer, G. Natronoflexus pectinivorans gen. nov. sp. nov., an obligately anaerobic and alkaliphilic fermentative member of Bacteroidetes from soda lakes. Extremophiles 2011, 15, 691–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antony, C.P.; Kumaresan, D.; Hunger, S.; Drake, H.L.; Murrell, J.C.; Shouche, Y.S. Microbiology of Lonar Lake and other soda lakes. ISME J. 2013, 7, 468–476. [Google Scholar] [CrossRef]
- Lanzén, A.; Simachew, A.; Gessesse, A.; Chmolowska, D.; Jonassen, I.; Øvreas, L. Surprising Prokaryotic and Eukaryotic Diversity, Community Structure and Biogeography of Ethiopian Soda Lakes. PLoS ONE 2013, 8, e0072577. [Google Scholar] [CrossRef] [PubMed]
- Steiman, R.; Ford, L.; Ducros, V.; Lafond, J.-L.; Guiraud, P. First survey of fungi in hypersaline soil and water of Mono Lake area (California). Antonie Van Leeuwenhoek 2004, 85, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Grum-Grzhimaylo, A.A.; Debets, A.J.M.; van Diepeningen, A.D.; Georgieva, M.L.; Bilanenko, E.N. Sodiomyces alkalinus, a new holomorphic alkaliphilic ascomycete within the Plectosphaerellaceae. Persoonia 2013, 31, 147–158. [Google Scholar] [CrossRef] [Green Version]
- Grum-Grzhimaylo, A.A.; Georgieva, M.L.; Debets, A.J.M.; Bilanenko, E.N. Are alkalitolerant fungi of the Emericellopsis lineage (Bionectriaceae) of marine origin? IMA Fungus 2013, 4, 213–228. [Google Scholar] [CrossRef]
- Lisichkina, G.A.; Bab’eva, I.P.; Sorokin, D.Y. Alkalitolerant yeasts from natural biotopes. Microbiology 2003, 72, 618–620. [Google Scholar] [CrossRef]
- Georgieva, M.L.; Lebedeva, M.P.; Bilanenko, E.N. Mycelial fungi in saline soils of the western Transbaikal region. Eurasian Soil Sci. 2012, 45, 1159–1168. [Google Scholar] [CrossRef]
- Nagai, K.; Suzuki, K.; Okada, G. Studies on the distribution of alkalophilic and alkali-tolerant soil fungi II: Fungal flora in two limestone caves in Japan. Mycoscience 1998, 39, 293–298. [Google Scholar] [CrossRef]
- Nagai, K.; Sakai, T.; Rantiatmodjo, R.M.; Suzuki, K.; Gams, W.; Okada, G. Studies on the distribution of alkalophilic and alkali-tolerant soil fungi I. Mycoscience 1995, 36, 247–256. [Google Scholar] [CrossRef]
- Sharma, R.; Prakash, O.; Sonawane, M.S.; Nimonkar, Y.; Golellu, P.B.; Sharma, R. Diversity and distribution of phenol oxidase producing fungi from soda lake and description of Curvularia lonarensis sp. nov. Front. Microbiol. 2016, 7, 01847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bondarenko, S.A.; Georgieva, M.L.; Bilanenko, E.N. Fungi Inhabiting the Coastal Zone of Lake Magadi. Contemp. Probl. Ecol. 2018, 11, 439–448. [Google Scholar] [CrossRef]
- Orwa, P.; Mugambi, G.; Wekesa, V.; Mwirichia, R. Isolation of haloalkaliphilic fungi from Lake Magadi in Kenya. Heiyon 2020, 6, e02823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elíades, L.A.; Cabello, M.N.; Voget, C.E. Contribution to the study of alkalophilic and alkali-tolerant Ascomycota from Argentina. Darwiniana 2006, 44, 63–74. [Google Scholar]
- Szabó, A.; Korponai, K.; Kerepesi, C.; Somogyi, B.; Vörös, L.; Bartha, D.; Márialigeti, K.; Felföldi, T. Soda pans of the Pannonian steppe harbor unique bacterial communities adapted to multiple extreme conditions. Extremophiles 2017, 21, 639–649. [Google Scholar] [CrossRef] [Green Version]
- Pellegrinetti, T.A.; Cotta, S.R.; Sarmento, H.; Costa, J.S.; Delbaje, E.; Montes, C.R.; Camargo, P.B.; Barbiero, L.; Rezende-Filho, A.T.; Fiore, M.F. Bacterial Communities along Environmental Gradients in Tropical Soda Lakes. Microb. Ecol. 2022; under review. [Google Scholar] [CrossRef]
- Chakraborty, J.; Rajput, V.; Sapkale, V.; Kamble, S.; Dharne, M. Spatio-temporal resolution of taxonomic and functional microbiome of Lonar soda lake of India reveals metabolic potential for bioremediation. Chemosphere 2021, 264, 128574. [Google Scholar] [CrossRef]
- Horikoshi, K. Extremophiles. Where It All Began; First; Springer: Tokyo, Japan, 2016; ISBN 9784431554073. [Google Scholar]
- Bignell, E. The Molecular Basis of pH Sensing, Signaling, and Homeostasis in Fungi. In Advances in Applied Microbiology Vol 79; Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2012; pp. 1–18. ISBN 9780123943187. [Google Scholar]
- Scervino, J.M.; Mesa, M.P.; Della Mónica, I.; Recchi, M.; Moreno, N.S.; Godeas, A. Soil fungal isolates produce different organic acid patterns involved in phosphate salts solubilization. Biol. Fertil. Soils 2010, 46, 755–763. [Google Scholar] [CrossRef]
- Wei, Y.; Zhang, S.H. Abiostress resistance and cellulose degradation abilities of haloalkaliphilic fungi: Applications for saline–alkaline remediation. Extremophiles 2017, 22, 155–164. [Google Scholar] [CrossRef]
- Khan, S.; Ivey, D.M.; Krulwich, T.A. Membrane ultrastructure of alkaliphilic Bacillus species studied by rapid-freeze electron microscopy. J. Bacteriol. 1992, 174, 5123–5126. [Google Scholar] [CrossRef] [Green Version]
- Tilburn, J.; Arst, H.N.; Peñalva, M.A. Regulation of Gene Expression by Ambient pH. In Cellular and Molecular Biology of Filamentous Fungi; Borkovich, K.A., Ebbole, D.J., Eds.; ASM Press: Washington, DC, USA, 2010; pp. 480–487. ISBN 9781683671299. [Google Scholar]
- Li, W.; Mitchell, A.P. Proteolytic Activation of Rim1p, a Positive Regulator of Yeast Sporulation and Invasive Growth. Genetics 1997, 145, 63–77. [Google Scholar] [CrossRef]
- Caddick, M.X.; Brownlee, A.G.; Arst, H.N. Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol. Gen. Genet. 1986, 203, 346–353. [Google Scholar] [CrossRef]
- Arst, H.N.; Bignell, E.; Tilburn, J. Two new genes involved in signalling ambient pH in Aspergillus nidulans. Mol. Gen. Genet. 1994, 245, 787–790. [Google Scholar] [CrossRef] [PubMed]
- Mingot, J.M.; Tilburn, J.; Diez, E.; Bignell, E.; Orejas, M.; Widdick, D.A.; Sarkar, S.; Brown, C.V.; Caddick, M.X.; Espeso, E.A.; et al. Specificity determinants of proteolytic processing of Aspergillus PacC transcription factor are remote from the processing site, and processing occurs in yeast if pH signalling is bypassed. Mol. Cell. Biol. 1999, 19, 1390–1400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tilburn, J.; Sarkar, S.; Widdick, D.A.; Espeso, E.A.; Orejas, M.; Mungroo, J.; Peñalva, M.A.; Arst, H.N. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995, 14, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Espeso, E.A.; Arst, H.N. On the Mechanism by which Alkaline pH Prevents Expression of an Acid-Expressed Gene. Mol. Cell. Biol. 2000, 20, 3355–3363. [Google Scholar] [CrossRef]
- Díez, E.; Álvaro, J.; Espeso, E.A.; Rainbow, L.; Suárez, T.; Tilburn, J.; Arst, H.N.; Peñalva, M.Á. Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 2002, 21, 1350–1359. [Google Scholar] [CrossRef] [Green Version]
- Orejas, M.; Espeso, E.A.; Tilburn, J.; Sarkar, S.; Arst, H.N.; Peñalva, M.A. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995, 9, 1622–1632. [Google Scholar] [CrossRef] [Green Version]
- Peñalva, M.A.; Tilburn, J.; Bignell, E.; Arst, H.N. Ambient pH gene regulation in fungi: Making connections. Cell Press 2008, 16, 291–300. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Martínez, J.; Brown, C.V.; Díez, E.; Tilburn, J.; Arst, H.N.; Peñalva, M.Á.; Espeso, E.A. Overlap of nuclear localisation signal and specific DNA-binding residues within the zinc finger domain of PacC. J. Mol. Biol. 2003, 334, 667–684. [Google Scholar] [CrossRef]
- Mingot, J.M.; Espeso, E.A.; Díez, E.; Peñalva, M. Ambient pH Signaling Regulates Nuclear Localization of the Aspergillus nidulans PacC Transcription Factor. Mol. Cell. Biol. 2001, 21, 1688–1699. [Google Scholar] [CrossRef] [Green Version]
- Espeso, E.A.; Tilburn, J.; Sánchez-Pulido, L.; Brown, C.V.; Valencia, A.; Arst, H.N.; Peñalva, M.A. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J. Mol. Biol. 1997, 274, 466–480. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Mitchell, A.P. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 2001, 183, 6917–6923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Futai, E.; Maeda, T.; Sorimachi, H.; Kitamoto, K.; Ishiura, S.; Suzuki, K. The protease activity of a calpain-like cysteine protease in Saccharomyces cerevisiae is required for alkaline adaptation and sporulation. Mol. Gen. Genet. 1999, 260, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Peñalva, M.A.; Arst, H.N. Regulation of Gene Expression by Ambient pH in Filamentous Fungi and Yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 426–446. [Google Scholar] [CrossRef] [Green Version]
- Calcagno-Pizarelli, A.M.; Negrete-Urtasun, S.; Denison, S.H.; Rudnicka, J.D.; Bussink, H.J.; Múnera-Huertas, T.; Stanton, L.; Hervás-Aguilar, A.; Espeso, E.A.; Tilburn, J.; et al. Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot. Cell 2007, 6, 2365–2375. [Google Scholar] [CrossRef] [Green Version]
- Herranz, S.; Rodriguez, J.M.; Bussink, H.-J.; Sanchez-Ferrero, J.C.; Arst, H.N.; Peñalva, M.A.; Vincent, O. Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. USA 2005, 102, 12141–12146. [Google Scholar] [CrossRef] [Green Version]
- Lefkowitz, R.J.; Shenoy, S.K. Transduction of receptor signals by β-arrestins. Science 2005, 308, 512–517. [Google Scholar] [CrossRef]
- Galindo, A.; Hervás-Aguilar, A.; Rodríguez-Galán, O.; Vincent, O.; Arst, H.N.; Tilburn, J.; Peñalva, M.A. PalC, one of two Bro1 domain proteins in the fungal pH signalling pathway, localizes to cortical structures and binds Vps32. Traffic 2007, 8, 1346–1364. [Google Scholar] [CrossRef] [Green Version]
- Lucena-Agell, D.; Galindo, A.; Arst, H.N.; Peñalva, M.A. Aspergillus nidulans Ambient pH Signaling Does Not Require Endocytosis. Eukaryot. Cell 2015, 14, 545–553. [Google Scholar] [CrossRef] [Green Version]
- Vincent, O.; Rainbow, L.; Tilburn, J.; Arst, H.N.; Peñalva, M.A. YPXL/I Is a Protein Interaction Motif Recognized by Aspergillus PalA and Its Human Homologue, AIP1/Alix. Mol. Cell. Biol. 2003, 23, 1647–1655. [Google Scholar] [CrossRef] [Green Version]
- Denison, S.H.; Orejas, M.; Arst, H.N. Signaling of ambient pH in Aspergillus involves a cysteine protease. J. Biol. Chem. 1995, 270, 28519–28522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conner, S.D.; Schmid, S.L. Regulated portals of enrty into the cell. Nature 2003, 422, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Smith, F.J.; Subaran, R.; Mitchell, A.P. Multivesicular Body-ESCRT Components Function in pH Response Regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 2004, 15, 5528–5537. [Google Scholar] [CrossRef] [Green Version]
- MacDonald, C.; Buchkovich, N.J.; Stringer, D.K.; Emr, S.D.; Piper, R.C. Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep. 2012, 13, 331–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT Pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef] [Green Version]
- Nishino, K.; Obara, K.; Kihara, A. The C-terminal cytosolic region of rim21 senses alterations in plasma membrane lipid composition: Insights into sensing mechanisms for plasma membrane lipid asymmetry. J. Biol. Chem. 2015, 290, 30797–30805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grum-Grzhimaylo, A.A.; Falkoski, D.L.; van den Heuvel, J.; Valero-Jiménez, C.A.; Min, B.; Choi, I.G.; Lipzen, A.; Daum, C.G.; Aanen, D.K.; Tsang, A.; et al. The obligate alkalophilic soda-lake fungus Sodiomyces alkalinus has shifted to a protein diet. Mol. Ecol. 2018, 27, 4808–4819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consortium, T.U. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, 523–531. [Google Scholar] [CrossRef]
- Forgac, M. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007, 8, 917–929. [Google Scholar] [CrossRef]
- Lee, L.K.; Stewart, A.G.; Donohoe, M.; Bernal, R.A.; Stock, D. The structure of the peripheral stalk of Thermus thermophilus H +-ATPase/synthase. Nat. Struct. Mol. Biol. 2010, 17, 373–378. [Google Scholar] [CrossRef] [Green Version]
- Harold, F.M. Conservation and transformation of energy by bacterial membranes. Bacteriol. Rev. 1972, 36, 172–230. [Google Scholar] [CrossRef] [PubMed]
- Krulwich, T.A. Bioenergetics of alkalophilic bacteria. J. Membr. Biol. 1986, 89, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Kaieda, N.; Wakagi, T.; Koyama, N. Presence of Na+-stimulated V-type ATPase in the membrane of a facultatively anaerobic and halophilic alkaliphile. FEMS Microbiol. Lett. 1998, 167, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Pomorski, T.; Holthuis, J.C.M.; Herrmann, A.; Meer, G. Van Tracking down lipid flippases and their biological functions. J. Cell Sci. 2004, 117, 805–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, M.; Kihara, A.; Igarashi, Y. Lipid Asymmetry of the Eukaryotic Plasma Membrane: Functions and Related Enzymes. Biol. Pharm. Bull. 2006, 29, 1542–1546. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, M.; Kihara, A.; Denpoh, A.; Igarashi, Y. The Rim101 Pathway Is Involved in Rsb1 Expression Induced by Altered Lipid Asymmetry. Mol. Biol. Cell 2008, 19, 1922–1931. [Google Scholar] [CrossRef] [Green Version]
- Daleke, D.L. Phospholipid Flippases. J. Biol. Chem. 2007, 282, 821–825. [Google Scholar] [CrossRef] [Green Version]
- Sahu, S.K.; Gummadi, S.N.; Manoj, N.; Aradhyam, G.K. Phospholipid scramblases: An overview. Arch. Biochem. Biophys. 2007, 462, 103–114. [Google Scholar] [CrossRef]
- Hankins, H.M.; Baldridge, R.D.; Xu, P.; Graham, T.R. Role of Flippases, Scramblases and Transfer Proteins in Phosphatidylserine Subcellular Distribution. Traffic 2015, 16, 35–47. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, J.; Colombo, A.C.; Zamith-Miranda, D.; Silva, V.K.A.; Allegood, J.C.; Casadevall, A.; Del Poeta, M.; Nosanchuk, J.D.; Kronstad, J.W.; Rodrigues, M.L. The putative flippase Apt1 is required for intracellular membrane architecture and biosynthesis of polysaccharide and lipids in Cryptococcus neoformans. Biochim. Biophys. Acta 2018, 1865, 532–541. [Google Scholar] [CrossRef]
- Mioka, T.; Fujimura-Kamada, K.; Mizugaki, N.; Kishimoto, T.; Sano, T.; Nunome, H.; Williams, D.E.; Andersen, R.J.; Tanaka, K. Phospholipid flippases and Sfk1p, a novel regulator of phospholipid asymmetry, contribute to low permeability of the plasma membrane. Mol. Biol. Cell 2018, 29, 1203–1218. [Google Scholar] [CrossRef] [PubMed]
- Pomorski, T.; Lombardi, R.; Riezman, H.; Devaux, P.F.; van Meer, G.; Holthuis, J.C.M. Drs2p-related P-type ATPases Dnf1p and Dnf2p Are Required for Phospholipid Translocation across the Yeast Plasma Membrane and Serve a Role in Endocytosis. Mol. Biol. Cell 2003, 14, 1240–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, K.; Fujimura-Kamada, K.; Furuta, N.; Kato, U.; Umeda, M.; Tanaka, K. Cdc50p, a Protein Required for Polarized Growth, Associates with the Drs2p P-Type ATPase Implicated in Phospholipid Translocation in Saccharomyces cerevisiae. Mol. Biol. Cell 2004, 15, 3418–3432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuta, N.; Fujimura-Kamada, K.; Saito, K.; Yamamoto, T.; Tanaka, K. Endocytic Recycling in Yeast Is Regulated by Putative Phospholipid Translocases and the Ypt31p/32p–Rcy1p Pathway. Mol. Biol. Cell 2007, 18, 295–312. [Google Scholar] [CrossRef]
- Prasad, R.; Khandelwal, N.K.; Banerjee, A. Yeast ABC Transporters in Lipid Trafficking. Fungal Genet. Biol. 2016, 93, 25–34. [Google Scholar] [CrossRef]
- Decottignies, A.; Grant, A.M.; Nichols, J.W.; De Wet, H.; McIntosh, D.B.; Goffeau, A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 1998, 273, 12612–12622. [Google Scholar] [CrossRef] [Green Version]
- Kihara, A.; Igarashi, Y. Cross Talk between Sphingolipids and Glycerophospholipids in the Establishment of Plasma Membrane Asymmetry. Mol. Biol. Cell 2004, 15, 4949–4959. [Google Scholar] [CrossRef] [Green Version]
- Kihara, A.; Igarashi, Y. Identification and Characterization of a Saccharomyces cerevisiae Gene, RSB1, Involved in Sphingoid Long-chain Base Release. J. Biol. Chem. 2002, 277, 30048–30054. [Google Scholar] [CrossRef] [Green Version]
- Kay, J.G.; Grinstein, S. Phosphatidylserine-Mediated Cellular Signaling. In Lipid-Mediated Protein Signaling; Capelluto, D.G.S., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 991, pp. 177–193. ISBN 978-94-007-6330-2. [Google Scholar]
- Young, B.P.; Shin, J.J.H.; Orij, R.; Chao, J.T.; Li, S.C.; Guan, X.L.; Khong, A.; Jan, E.; Wenk, M.R.; Prinz, W.A.; et al. Phosphatidic Acid Is a pH Biosensor that Links Membrane Biogenesis to Metabolism. Science 2010, 329, 1085–1088. [Google Scholar] [CrossRef]
- Danilova, O.A.; Ianutsevich, E.A.; Bondarenko, S.A.; Georgieva, M.L.; Vikchizhanina, D.A.; Groza, N.V.; Bilanenko, E.N.; Tereshina, V.M. Osmolytes and membrane lipids in the adaptation of micromycete Emericellopsis alkalina to ambient pH and sodium chloride. Fungal Biol. 2020, 124, 884–891. [Google Scholar] [CrossRef]
- Bondarenko, S.A.; Ianutsevich, E.A.; Danilova, O.A.; Grum-Grzhimaylo, A.A.; Kotlova, E.R.; Kamzolkina, O.V.; Bilanenko, E.N.; Tereshina, V.M. Membrane lipids and soluble sugars dynamics of the alkaliphilic fungus Sodiomyces tronii in response to ambient pH. Extremophiles 2017, 21, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Bondarenko, S.A.; Ianutsevich, E.A.; Sinitsyna, N.A.; Georgieva, M.L.; Bilanenko, E.N.; Tereshina, B.M. Dynamics of the Cytosol Soluble Carbohydrates and Membrane Lipids in Response to Ambient pH in Alkaliphilic and Alkalitolerant Fungi. Microbiology 2018, 87, 21–32. [Google Scholar] [CrossRef]
- Jaenicke, R.; Böhm, G. The stability of proteins in extreme environments. Curr. Opin. Struct. Biol. 1998, 8, 738–748. [Google Scholar] [CrossRef]
- Finch, A.; Kim, J. Thermophilic Proteins as Versatile Scaffolds for Protein Engineering. Microorganisms 2018, 6, 97. [Google Scholar] [CrossRef] [Green Version]
- Haki, G.D.; Rakshit, S.K. Developments in industrially important thermostable enzymes: A review. Bioresour. Technol. 2003, 89, 17–34. [Google Scholar] [CrossRef]
- Madern, D.; Ebel, C.; Zaccai, G. Halophilic adaptation of enzymes. Extremophiles 2000, 4, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Plemenitaš, A.; Lenassi, M.; Konte, T.; Kejžar, A.; Zajc, J.; Gostinčar, C.; Gunde-Cimerman, N. Adaptation to high salt concentrations in halotolerant/halophilic fungi: A molecular perspective. Front. Microbiol. 2014, 5, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulkarni, N.; Shendye, A.; Rao, M. Molecular and Biotechnological Aspects of Xilanases. FEMS Microbiol. Rev. 1999, 23, 411–456. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.D.; Sidhu, G.; Pot, I.; Brayer, G.D.; Withers, S.G.; McIntosh, L.P. Hydrogen bonding and catalysis: A novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase. J. Mol. Biol. 2000, 299, 255–279. [Google Scholar] [CrossRef] [Green Version]
- Hakulinen, N.; Turunen, O.; Jänis, J.; Leisola, M.; Rouvinen, J. Three-dimensional structures of thermophilic β-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa: Comparison of twelve xylanases in relation to their thermal stability. Eur. J. Biochem. 2003, 270, 1399–1412. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Liu, X.; Yu, Q.; Zhang, X.; Qu, Y.; Gao, P.; Wang, T. Directed evolution for engineering pH profile of endoglucanase III from Trichoderma reesei. Biomol. Eng. 2005, 22, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Turunen, O.; Vuorio, M.; Fenel, F.; Leisola, M. Engineering of multiple arginines into the Ser/Thr surface of Trichoderma reesei endo-1,4-β-xylanase II increases the thermotolerance and shifts the pH optimum towards alkaline pH. Protein Eng. 2002, 15, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Wang, K.; Huang, H.; Shi, P.; Yang, P.; Yao, B. Gene cloning, expression, and biochemical characterization of an alkali-tolerant β-mannanase from Humicola insolens Y1. J. Ind. Microbiol. Biotechnol. 2012, 39, 547–555. [Google Scholar] [CrossRef]
- Wohlfahrt, G.; Pellikka, T.; Boer, H.; Teeri, T.T.; Koivula, A. Probing pH-dependent functional elements in proteins: Modification of carboxylic acid pairs in Trichoderma reesei cellobiohydrolase ce16A. Biochemistry 2003, 42, 10095–10103. [Google Scholar] [CrossRef] [PubMed]
- Dalmaso, G.Z.L.; Ferreira, D.; Vermelho, A.B. Marine extremophiles a source of hydrolases for biotechnological applications. Mar. Drugs 2015, 13, 1925–1965. [Google Scholar] [CrossRef] [Green Version]
- Dhakar, K.; Pandey, A. Wide pH range tolerance in extremophiles: Towards understanding an important phenomenon for future biotechnology. Appl. Microbiol. Biotechnol. 2016, 100, 2499–2510. [Google Scholar] [CrossRef]
- Kumar, C.G.; Takagi, H. Microbial alkaline proteases: From a bioindustrial viewpoint. Biotechnol. Adv. 1999, 17, 561–594. [Google Scholar] [CrossRef]
- Never, Z.; Cecilia, C.; Suleimain, M.; Madundo, M. Dehairing of animal hides and skins by alkaline proteases of Aspergillus oryzae for efficient processing to leather products in Tanzania. Afr. J. Biotechnol. 2019, 18, 426–434. [Google Scholar] [CrossRef] [Green Version]
- Malathi, S.; Chakraborty, R. Production of alkaline protease by a new Aspergillus flavus isolate under solid-substrate fermentation conditions for use as a depilation agent. Appl. Environ. Microbiol. 1991, 57, 712–716. [Google Scholar] [CrossRef] [Green Version]
- Hajji, M.; Kanoun, S.; Nasri, M.; Gharsallah, N. Purification and characterization of an alkaline serine-protease produced by a new isolated Aspergillus clavatus ES1. Process Biochem. 2007, 42, 791–797. [Google Scholar] [CrossRef]
- Hajji, M.; Rebai, A.; Gharsallah, N.; Nasri, M. Optimization of alkaline protease production by Aspergillus clavatus ES1 in Mirabilis jalapa tuber powder using statistical experimental design. Appl. Microbiol. Biotechnol. 2008, 79, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Damare, S.; Raghukumar, C.; Muraleedharan, U.D.; Raghukumar, S. Deep-sea fungi as a source of alkaline and cold-tolerant proteases. Enzym. Microb. Technol. 2006, 39, 172–181. [Google Scholar] [CrossRef]
- Tunga, R.; Shrivastava, B.; Banerjee, R. Purification and characterization of a protease from solid state cultures of Aspergillus parasiticus. Process Biochem. 2003, 38, 1553–1558. [Google Scholar] [CrossRef]
- Chellappan, S.; Jasmin, C.; Basheer, S.M.; Elyas, K.K.; Bhat, S.G.; Chandrasekaran, M. Production, purification and partial characterization of a novel protease from marine Engyodontium album BTMFS10 under solid state fermentation. Process Biochem. 2006, 41, 956–961. [Google Scholar] [CrossRef]
- Li, J.; Yang, J.; Huang, X.; Zhang, K.Q. Purification and characterization of an extracellular serine protease from Clonostachys rosea and its potential as a pathogenic factor. Process Biochem. 2006, 41, 925–929. [Google Scholar] [CrossRef]
- Dienes, D.; Börjesson, J.; Hägglund, P.; Tjerneld, F.; Lidén, G.; Réczey, K.; Stålbrand, H. Identification of a trypsin-like serine protease from Trichoderma reesei QM9414. Enzym. Microb. Technol. 2007, 40, 1087–1094. [Google Scholar] [CrossRef]
- Alkin, N.; Dunaevsky, Y.; Elpidina, E.; Beljakova, G.; Tereshchenkova, V.; Filippova, I.; Belozersky, M. Proline-specific fungal peptidases: Genomic analysis and identification of secreted dpp4 in alkaliphilic and alkalitolerant fungi. J. Fungi 2021, 7, 744. [Google Scholar] [CrossRef]
- Hattori, M.; Isomura, S.; Yokoyama, E.; Ujita, M.; Hara, A. Extracellular trypsin-like proteases produced by Cordyceps militaris. J. Biosci. Bioeng. 2005, 100, 631–636. [Google Scholar] [CrossRef]
- Dutta, T.; Sengupta, R.; Sahoo, R.; Sinha Ray, S.; Bhattacharjee, A.; Ghosh, S. A novel cellulase free alkaliphilic xylanase from alkali tolerant Penicillium citrinum: Production, purification and characterization. Lett. Appl. Microbiol. 2007, 44, 206–211. [Google Scholar] [CrossRef]
- Guan, G.Q.; Zhao, P.X.; Zhao, J.; Wang, M.J.; Huo, S.H.; Cui, F.J.; Jiang, J.X. Production and Partial Characterization of an Alkaline Xylanase from a Novel Fungus Cladosporium oxysporum. Biomed Res. Int. 2016, 2016, 4575024. [Google Scholar] [CrossRef] [Green Version]
- Chandra Raj, K.; Chandra, T.S. Purification and characterization of xylanase from alkali-tolerant Aspergillus fischeri Fxn1. FEMS Microbiol. Lett. 1996, 145, 457–461. [Google Scholar] [CrossRef]
- Bajaj, B.K.; Abbass, M. Studies on an alkali-thermostable xylanase from Aspergillus fumigatus MA28. 3 Biotech 2011, 1, 161–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranjan, K.; Lone, M.A.; Sahay, S. Detergent compatible cold-active alkaline amylases from Clavispora lusitaniae CB13. J. Microbiol. Biotechnol. Food Sci. 2016, 5, 306–310. [Google Scholar] [CrossRef] [Green Version]
- Zaferanloo, B.; Bhattacharjee, S.; Ghorbani, M.M.; Mahon, P.J.; Palombo, E.A. Amylase production by Preussia minima, a fungus of endophytic origin: Optimization of fermentation conditions and analysis of fungal secretome by LC-MS. BMC Microbiol. 2014, 14, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.H.; Wang, H.R.; Yan, T.R. Cloning, purification, and characterization of a heat- and alkaline-stable endoglucanase B from Aspergillus niger BCRC31494. Molecules 2012, 17, 9774–9789. [Google Scholar] [CrossRef] [Green Version]
- Schülein, M. Enzymatic properties of cellulases from Humicola insolens. J. Biotechnol. 1997, 57, 71–81. [Google Scholar] [CrossRef]
- Sulistyaningdyah, W.T.; Ogawa, J.; Tanaka, H.; Maeda, C.; Shimizu, S. Characterization of alkaliphilic laccase activity in the culture supernatant of Myrothecium verrucaria 24G-4 in comparison with bilirubin oxidase. FEMS Microbiol. Lett. 2004, 230, 209–214. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Ma, R.; Li, S.; Gong, M.; Yao, B.; Bai, Y.; Gu, J. An alkaline and surfactant-tolerant lipase from Trichoderma lentiforme ACCC30425 with high application potential in the detergent industry. AMB Express 2018, 8, 95. [Google Scholar] [CrossRef]
- Saxena, R.K.; Davidson, W.S.; Sheoran, A.; Giri, B. Purification and characterization of an alkaline thermostable lipase from Aspergillus carneus. Process Biochem. 2003, 39, 239–247. [Google Scholar] [CrossRef]
- Venkatesagowda, B.; Ponugupaty, E.; Barbosa-Dekker, A.M.; Dekker, R.F.H. The Purification and Characterization of Lipases from Lasiodiplodia theobromae, and Their Immobilization and Use for Biodiesel Production from Coconut Oil. Appl. Biochem. Biotechnol. 2018, 185, 619–640. [Google Scholar] [CrossRef]
- Hirayama, S.; Taira, R.; Borch, K.; Sandal, T.; Halkier, T.; Oxenboll, K.M.; Nielsen, B.R. Alkaline Lipolytic Enzyme. U.S. Patent 6,350,604, 28 February 2002. [Google Scholar]
- Mamo, G.; Mattiasson, B. Alkaliphilic Microorganisms in Biotechnology. In Biotechnology of Extremophiles Advances and Challenges; Rampelotto, P.H., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 243–272. ISBN 978-3-319-13520-5. [Google Scholar]
- Lübeck, M.; Lübeck, P.S. Fungal Cell Factories for Efficient and Sustainable Production of Proteins and Peptides. Microorganisms 2022, 10, 753. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, M.; Ray, R.R. Current Trends in Research and Application of Microbial Cellulases. Res. J. Microbiol. 2011, 6, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Cavaco-Paulo, A.; Morgado, J.; Almeida, L.; Kilburn, D. Indigo Backstaining During Cellulase Washing. Text. Res. J. 1998, 68, 398–401. [Google Scholar] [CrossRef] [Green Version]
- Ibarra, D.; Monte, M.C.; Blanco, A.; Martínez, A.T.; Martínez, M.J. Enzymatic deinking of secondary fibers: Cellulases/hemicellulases versus laccase-mediator system. J. Ind. Microbiol. Biotechnol. 2012, 39, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Pokhrel, D.; Viraraghavan, T. Treatment of pulp and paper mill wastewater—A review. Sci. Total Environ. 2004, 333, 37–58. [Google Scholar] [CrossRef]
- De Aquino, F.X.; De Sousa, M.V.; Puls, J.; Da Silva, F.G.; Ferreira Filho, E.X. Purification and characterization of a low-molecular-weight xylanase produced by Acrophialophora nainiana. Curr. Microbiol. 1999, 38, 18–21. [Google Scholar] [CrossRef]
- Karlen, D.L.; Andrews, S.S.; Wienhold, B.J.; Zobeck, T.M. Soil Quality Assessment: Past, Present and Future. J. Integr. Biosci. 2014, 6, 3–14. [Google Scholar]
- Rietz, D.N.; Haynes, R.J. Effects of irrigation-induced salinity and sodicity on soil microbial activity. Soil Biol. Biochem. 2003, 35, 845–854. [Google Scholar] [CrossRef]
- Serrano, R.; Gaxiola, R. Microbial Models and Salt Stress Tolerance in Plants. CRC. Crit. Rev. Plant Sci. 1994, 13, 121–138. [Google Scholar] [CrossRef]
- Liu, X.; Liu, J.; Wei, Y.; Tian, Y.; Fan, F.; Pan, H.; Zhang, S.H. Isolation, identification and biologic characteristics of an extreme halotolerant Aspergillus sp. J. Jilin Univ. 2011, 49, 548–552. [Google Scholar]
- Shi, Y.; Zhu, J. Saline–Alkali Soil Improvement Fertilizer and Preparation Method and Use Method Thereof. Chinese Patent CN 105237293 A, 13 January 2016. [Google Scholar]
- Jain, D.; Katyal, P. Optimization of gluco-amylase production from Aspergillus spp. for its use in saccharification of liquefied corn starch. 3 Biotech 2018, 8, 101. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Guilfoyle, E.; Stinson, J.; Skogerson, L. Sugar-Producing and Texture-Improving Bakery Methods and Products Formed Therefrom. United States Patent Application Publication US2018/0242598A1, 30 August 2018. [Google Scholar]
- Mojsov, K.; Andronikov, D.; Janevski, A.; Jordeva, S.; Kertakova, M.; Golomeova, S.; Gaber, S.; Ignjatov, I. Production and application of α-amylase enzyme in textile industry. Tekst. Ind. 2018, 66, 23–28. [Google Scholar]
- Bartelme, M.; Marquardt, J.; Man, V.F.; Leafblad, B. Fast Dissolving Solid Detergent. United States Patent US 10005986, 26 June 2018. [Google Scholar]
- Singh, S.; Singh, S.; Bali, V.; Sharma, L.; Mangla, J. Production of Fungal Amylases Using Cheap, Readily Available Agriresidues, for Potential Application in Textile Industry. Biomed Res. Int. 2014, 2014, 215748. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.; Chisti, Y.; Chand, U. Production, purification, characterization, and applications of lipases. Biotechnol. Adv. 2001, 19, 627–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verger, R. “Interfacial activation” of lipases: Facts and artifacts. Trends Biotechnol. 1997, 15, 32–38. [Google Scholar] [CrossRef]
- Azeem, F.; Afzal, M.; Javed, S.; Riaz, M.; Kouser, A.; Hussain, S.; Siddique, M.H.; Rasul, I.; Nadeem, H. Bacterial lipases: A review on purification and characterization. Prog. Biophys. Mol. Biol. 2018, 132, 23–34. [Google Scholar] [CrossRef]
- Villeneuve, P. Plant lipases and their applications in oils and fats modification. Eur. J. Lipid Sci. Technol. 2003, 105, 308–317. [Google Scholar] [CrossRef]
- Casas-Godoy, L.; Gasteazoro, F.; Duquesne, S.; Bordes, F.; Marty, A.; Sandoval, G. Lipases: An Overview. In Lipases and Phospholipases: Methods and Protocols, Methods in Molecular Biology; Sandoval, G., Ed.; Humana Press: New York, NY, USA, 2018; Volume 1835, pp. 3–38. ISBN 9781493986729. [Google Scholar]
- Su, F.; Peng, C.; Li, G.L.; Xu, L.; Yan, Y.J. Biodiesel production from woody oil catalyzed by Candida rugosa lipase in ionic liquid. Renew. Energy 2016, 90, 329–335. [Google Scholar] [CrossRef]
- Rogozhin, E.A.; Sadykova, V.S.; Baranova, A.A.; Vasilchenko, A.S.; Lushpa, V.A.; Mineev, K.S.; Georgieva, M.L.; Kul’ko, A.B.; Krasheninnikov, M.E.; Lyundup, A.V.; et al. A novel lipopeptaibol Emericellipsin A with antimicrobial and antitumor activity produced by the extremophilic fungus Emericellopsis alkalina. Molecules 2018, 23, 2785. [Google Scholar] [CrossRef] [Green Version]
- Kuvarina, A.E.; Gavryushina, I.A.; Kulko, A.B.; Ivanov, I.A.; Rogozhin, E.A.; Georgieva, M.L.; Sadykova, V.S. The Emericellipsins A-E from an alkalophilic fungus Emericellopsis alkalina show potent activity against multidrug-resistant pathogenic fungi. J. Fungi 2021, 7, 153. [Google Scholar] [CrossRef]
- Kuvarina, A.E.; Georgieva, M.L.; Rogozhin, E.A.; Kulko, A.B.; Gavryushina, I.A.; Sadykova, V.S. Antimicrobial Potential of the Alkalophilic Fungus Sodiomyces alkalinus and Selection of Strains–Producers of New Antimicotic Compound. Appl. Biochem. Microbiol. 2021, 57, 86–93. [Google Scholar] [CrossRef]
- Kuvarina, A.E.; Rogozhin, E.A.; Sykonnikov, M.A.; Timofeeva, A.V.; Serebryakova, M.V.; Fedorova, N.V.; Kokaeva, L.Y.; Efimenko, T.A.; Georgieva, M.L.; Sadykova, V.S. Isolation and Characterization of a Novel Hydrophobin, Sa-HFB1, with Antifungal Activity from an Alkaliphilic Fungus, Sodiomyces alkalinus. J. Fungi 2022, 8, 659. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batista-García, R.A.; Balcázar-López, E.; Miranda-Miranda, E.; Sánchez-Reyes, A.; Cuervo-Soto, L.; Aceves-Zamudio, D.; Atriztán-Hernández, K.; Morales-Herrera, C.; Rodríguez-Hernández, R.; Folch-Mallol, J. Characterization of lignocellulolytic activities from a moderate halophile strain of Aspergillus caesiellus isolated from a sugarcane bagasse fermentation. PLoS ONE 2014, 9, e0105893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Abradelo, D.; Pérez-Llano, Y.; Peidro-Guzmán, H.; Sánchez-Carbente, M.d.R.; Folch-Mallol, J.L.; Aranda, E.; Vaidyanathan, V.K.; Cabana, H.; Gunde-Cimerman, N.; Batista-García, R.A. First demonstration that ascomycetous halophilic fungi (Aspergillus sydowii and Aspergillus destruens) are useful in xenobiotic mycoremediation under high salinity conditions. Bioresour. Technol. 2019, 279, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Holliday, R. The genetics of Ustilago maydis. Genet. Res. 1961, 2, 204–230. [Google Scholar] [CrossRef]
- Cao, X.; An, T.; Fu, W.; Zhang, J.; Zhao, H.; Li, D.; Jin, X.; Liu, B. Genome-Wide Identification of Cellular Pathways and Key Genes that Respond to Sodium Bicarbonate Stress in Saccharomyces cerevisiae. Front. Microbiol. 2022, 13, 831973. [Google Scholar] [CrossRef]
- Jiménez-Gómez, I.; Valdés-Muñoz, G.; Moreno-Ulloa, A.; Pérez-Llano, Y.; Moreno-Perlín, T.; Silva-Jiménez, H.; Barreto-Curiel, F.; Sánchez-Carbente, M.d.R.; Folch-Mallol, J.L.; Gunde-Cimerman, N.; et al. Surviving in the Brine: A Multi-Omics Approach for Understanding the Physiology of the Halophile Fungus Aspergillus sydowii at Saturated NaCl Concentration. Front. Microbiol. 2022, 13, 840408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-López, M.G.; Batista-García, R.A.; Aréchiga-Carvajal, E.T. Alkaliphilic/Alkali-Tolerant Fungi: Molecular, Biochemical, and Biotechnological Aspects. J. Fungi 2023, 9, 652. https://doi.org/10.3390/jof9060652
Fernández-López MG, Batista-García RA, Aréchiga-Carvajal ET. Alkaliphilic/Alkali-Tolerant Fungi: Molecular, Biochemical, and Biotechnological Aspects. Journal of Fungi. 2023; 9(6):652. https://doi.org/10.3390/jof9060652
Chicago/Turabian StyleFernández-López, Maikel Gilberto, Ramón Alberto Batista-García, and Elva Teresa Aréchiga-Carvajal. 2023. "Alkaliphilic/Alkali-Tolerant Fungi: Molecular, Biochemical, and Biotechnological Aspects" Journal of Fungi 9, no. 6: 652. https://doi.org/10.3390/jof9060652






