Reference Genes Selection of Gymnosporangium yamadae during the Interaction with Apple Leaves
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Artificial Inoculation
2.2. RNA Extraction and cDNA Synthesis
2.3. Reference Gene Selection, Primer Design and RT-qPCR Parameters
2.4. Data Analysis
3. Results
3.1. Primer Specificity and Efficiency Evaluation
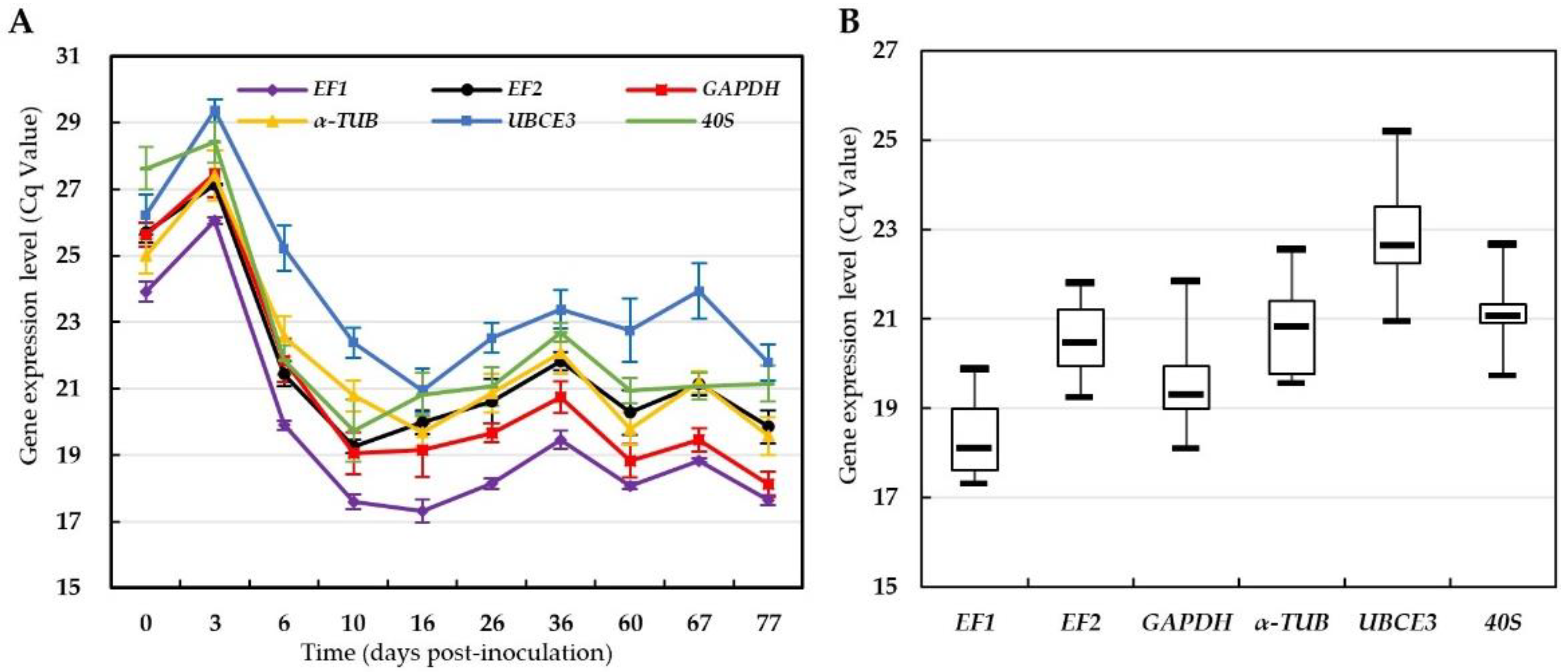
3.2. Expression Levels of Candidate Reference Genes in Different Samples
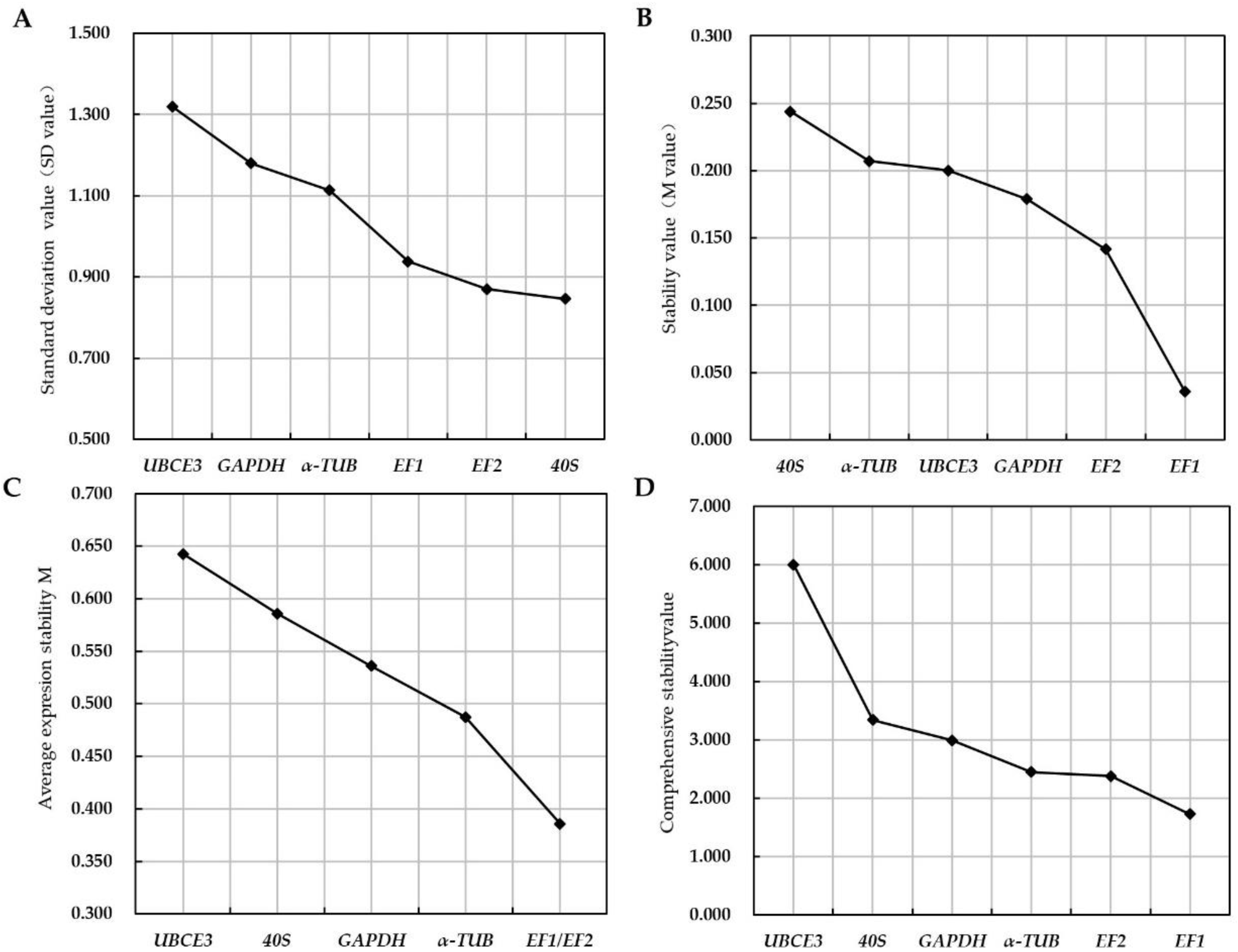
3.3. Stability of Candidate Reference Genes
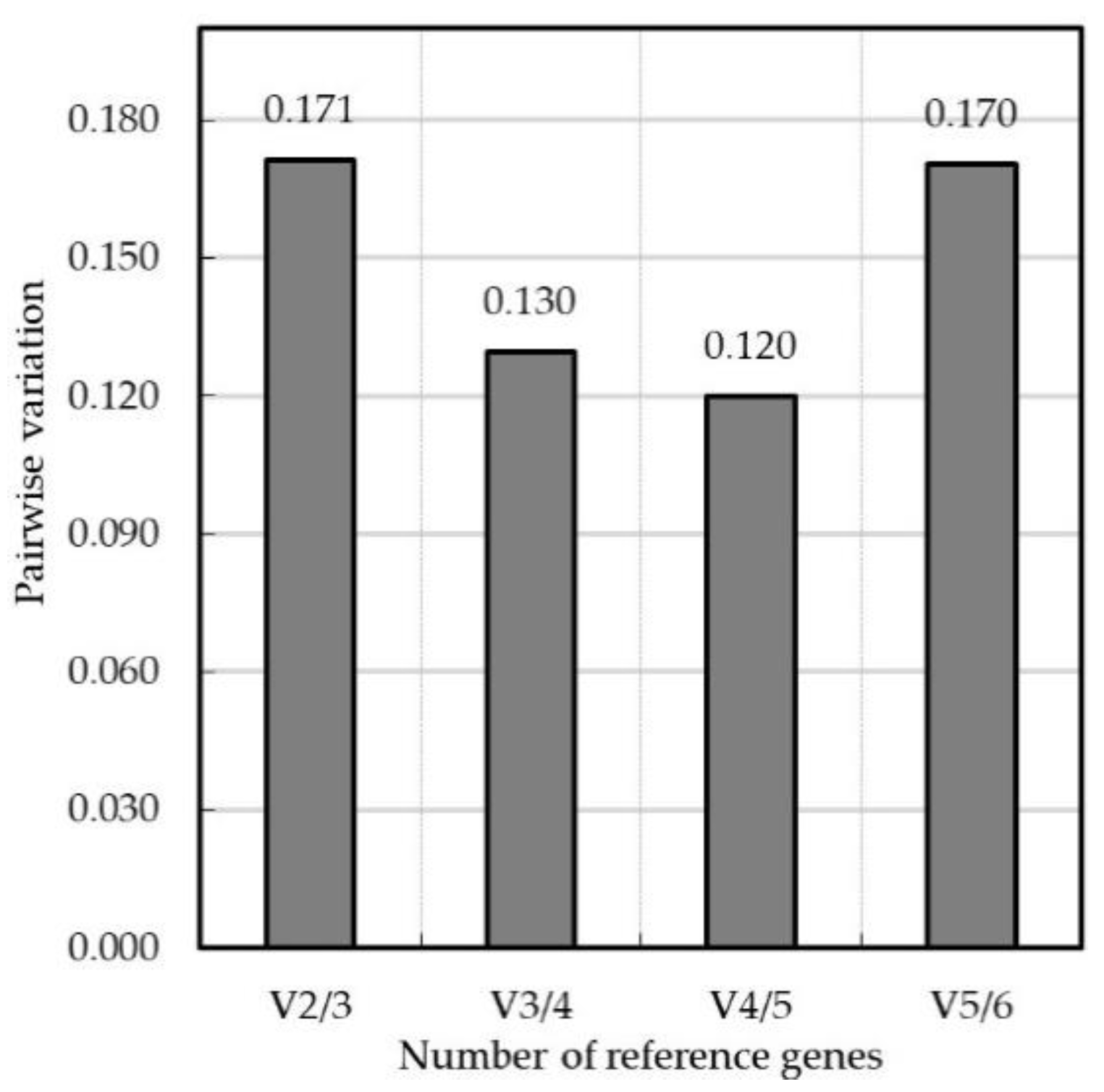
3.4. Validation of Candidate Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, B.H.; Wang, C.X.; Dong, X.L. Research progress in apple diseases and problems in the disease management in China. Plant Prot. 2013, 39, 46–54. (In Chinese) [Google Scholar] [CrossRef]
- Lee, D.K.; Ahn, S.; Cho, H.Y.; Yun, H.Y.; Park, J.H.; Lim, J.; Lee, J.; Kwon, S.W. Metabolic response induced by parasitic plant-fungus interactions hinder amino sugar and nucleotide sugar metabolism in the host. Sci. Rep. 2016, 6, 37434. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.Q.; Cao, B.; Tian, C.M.; Liang, Y.M. Comparative transcriptome analysis and identification of candidate effectors in two related rust species (Gymnosporangium yamadae and Gymnosporangium asiaticum). BMC Genom. 2017, 18, 651. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.Q.; Cao, B.; Kakishima, M.; Liang, Y.M. Species diversity, taxonomy, and phylogeny of Gymnosporangium in China. Mycologia 2020, 112, 941–973. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Kakishima, M. Surface structures of peridial cells of Gymnosporangium and Roestelia (Uredinales). Mycoscience 1999, 40, 121–131. [Google Scholar] [CrossRef]
- Dong, X.L.; Li, H.Y.; Sun, L.J.; Wang, C.X.; Zhang, H.H.; Li, B.H. Control effects and optimal spraying time of fungicides to apple rust caused by Gymnosporangium yamadae. Plant Prot. 2013, 39, 6. (In Chinese) [Google Scholar] [CrossRef]
- Duplessis, S.; Cuomo, C.A.; Lin, Y.C.; Aerts, A.; Tisserant, E.; Veneault-Fourrey, C.; Joly, D.L.; Hacquard, S.; Amselem, J.; Cantarel, B.L.; et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA 2011, 108, 9166–9171. [Google Scholar] [CrossRef]
- Duplessis, S.; Lorrain, C.; Petre, B.; Figueroa, M.; Dodds, P.N.; Aime, M.C. Host adaptation and virulence in heteroecious rust fungi. Annu. Rev. Phytopathol. 2021, 59, 403–422. [Google Scholar] [CrossRef]
- Sanchez-Vallet, A.; Fouche, S.; Fudal, I.; Hartmann, F.E.; Soyer, J.L.; Tellier, A.; Croll, D. The genome biology of effector gene evolution in filamentous plant pathogens. Annu. Rev. Phytopathol. 2018, 56, 21–40. [Google Scholar] [CrossRef]
- Duplessis, S.; Hacquard, S.; Delaruelle, C.; Tisserant, E.; Frey, P.; Martin, F.; Kohler, A. Melampsora larici-populina transcript profiling during germination and timecourse infection of poplar leaves reveals dynamic expression patterns associated with virulence and biotrophy. Mol. Plant-Microbe Interact. 2011, 24, 808–818. [Google Scholar] [CrossRef]
- Hacquard, S.; Joly, D.L.; Lin, Y.C.; Tisserant, E.; Feau, N.; Delaruelle, C.; Legué, V.; Kohler, A.; Tanguay, P.; Petre, B. A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici-populina (poplar leaf rust). Mol. Plant-Microbe Interact. 2012, 25, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Guo, J.; Liu, P.; He, F.; Wan, C.; Islam, M.A.; Tyler, B.M.; Kang, Z.; Guo, J. Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS-promoting transcription factor TaLOL2 to defeat ROS-induced defense in wheat. Mol. Plant 2019, 12, 1624–1638. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tao, S.Q.; Weng, H.; Liang, Y.M. Construction of haustorial isolation systems of Gymnosporangium yamadae and G. asiaticum. Mycosystema 2019, 38, 1430–1439. (In Chinese) [Google Scholar] [CrossRef]
- Tao, S.Q.; Auer, L.; Morin, E.; Liang, Y.M.; Duplessis, S. Transcriptome analysis of apple leaves infected by the rust fungus Gymnosporangium yamadae at two sporulation stages. Mol. Plant-Microbe Interact. 2020, 33, 444–461. [Google Scholar] [CrossRef]
- Vieira, A.; Talhinhas, P.; Loureiro, A.; Duplessis, S.; Fernandez, D.; Silva Mdo, C.; Paulo, O.S.; Azinheira, H.G. Validation of RT-qPCR reference genes for in planta expression studies in Hemileia vastatrix, the causal agent of coffee leaf rust. Fungal Biol. 2011, 115, 891–901. [Google Scholar] [CrossRef]
- Li, H.; Niu, H.W.; Liu, N.; Chen, Y. Reference Gene Selection for RT-qPCR and Expression Analysis of TaSGR Gene in Defense Response of Wheat to Puccinia triticina. Plant Physiol. J. 2015, 51, 642–648. [Google Scholar] [CrossRef]
- Song, Y.; Wang, Y.; Guo, D.; Jing, L. Selection of reference genes for quantitative real-time PCR normalization in the plant pathogen Puccinia helianthi Schw. BMC Plant Biol. 2019, 19, 20. [Google Scholar] [CrossRef]
- Link, T.I. Testing reference genes for transcript profiling in Uromyces appendiculatus during urediospore infection of common bean. PLoS ONE 2020, 15, e0237273. [Google Scholar] [CrossRef]
- Kim, H.K.; Yun, S.H. Evaluation of potential reference genes for quantitative RT-PCR analysis in Fusarium graminearum under different culture conditions. Plant Pathol. J. 2011, 27, 301–309. [Google Scholar] [CrossRef]
- Omar, S.C.; Bentley, M.A.; Morieri, G.; Preston, G.M.; Gurr, S.J. Validation of reference genes for robust qRT-PCR gene expression analysis in the rice blast fungus Magnaporthe oryzae. PLoS ONE 2016, 11, e0160637. [Google Scholar] [CrossRef]
- Hacquard, S.; Veneault-Fourrey, C.; Delaruelle, C.; Frey, P.; Martin, F.; Duplessis, S. Validation of Melampsora larici-populina reference genes for in planta RT-quantitative PCR expression profiling during time-course infection of poplar leaves. Physiol. Mol. Plant Pathol. 2011, 75, 106–112. [Google Scholar] [CrossRef]
- Huang, X.L.; Feng, H.; Kang, Z.S. Selection of reference genes for quantitative real-time PCR normalization in Puccinia Striiformis f.sp.tritici. J. Agricutural Biotechnol. 2012, 20, 181–187. (In Chinese) [Google Scholar] [CrossRef]
- Voegele, R.T.; Schmid, A. RT real-time PCR-based quantification of Uromyces fabae in planta. FEMS Microbiol. Lett. 2011, 322, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Scholtz, J.J.; Visser, B. Reference gene selection for qPCR gene expression analysis of rust-infected wheat. Physiol. Mol. Plant Pathol. 2013, 81, 22–25. [Google Scholar] [CrossRef]
- Hirschburger, D.; Muller, M.; Voegele, R.T.; Link, T. Reference genes in the pathosystem Phakopsora pachyrhizi/ soybean suitable for normalization in transcript profiling. Int. J. Mol. Sci. 2015, 16, 23057–23075. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Boil. 2002, 3, research0034. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-Based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Ward, D.S.; Jutta, D.W.; Roswitha, W.; Valérie, S. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, M.; Meng, Y. Identification and validation of reference genes for RT-qPCR analysis in Switchgrass under heavy metal stresses. Genes 2020, 11, 502. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Lee, Y.C.; Torres-Jerez, I.; Wang, M.; Yin, Y.; Chou, W.C.; He, J.; Shen, H.; Srivastava, A.C.; Pennacchio, C.; et al. Development of an integrated transcript sequence database and a gene expression atlas for gene discovery and analysis in switchgrass (Panicum virgatum L.). Plant J. 2013, 74, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Fu, H.; Qiao, H.; Sun, S.; Zhang, W.; Jin, S.; Jiang, S.; Gong, Y.; Xiong, Y.; Wu, Y. Validation and evaluation of reference genes for quantitative real-time PCR in Macrobrachium Nipponense. Int. J. Mol. Sci. 2018, 19, 2258. [Google Scholar] [CrossRef] [PubMed]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnson, M.A.; Rook, G.; Zumla, A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004, 37, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Banda, M.; Bommineni, A.; Thomas, R.A.; Luckinbill, L.S.; Tucker, J.D. Evaluation and validation of housekeeping genes in response to ionizing radiation and chemical exposure for normalizing RNA expression in real-time PCR. Mutat. Res. 2008, 649, 126–134. [Google Scholar] [CrossRef]
- Perez, S.; Royo, L.J.; Astudillo, A.; Escudero, D.; Alvarez, F.; Rodriguez, A.; Gomez, E.; Otero, J. Identifying the most suitable endogenous control for determining gene expression in hearts from organ donors. BMC Mol. Biol. 2007, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Zhao, Q.; Zhou, A.; Wen, X.; Li, M.; Li, R.; Liao, X.; Xu, T. The antifungal effects of citral on Magnaporthe oryzae occur via modulation of chitin content as revealed by RNA-Seq analysis. J. Fungi. 2021, 7, 1023. [Google Scholar] [CrossRef]
- Yan, H.Z.; Liou, R.F. Selection of internal control genes for real-time quantitative RT-PCR assays in the oomycete plant pathogen Phytophthora parasitica. Fungal Genet. Biol. 2006, 43, 430–438. [Google Scholar] [CrossRef]
- Tao, S.Q.; Cao, B.; Morin, E.; Liang, Y.M.; Duplessis, S. Comparative transcriptomics of Gymnosporangium spp. teliospores reveals a conserved genetic program at this specific stage of the rust fungal life cycle. BMC Genom. 2019, 20, 723. [Google Scholar] [CrossRef]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.R. Eleven golden rules of quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.K.; Yan, H.D.; Jiang, X.M.; Zhang, X.Q.; Zhang, Y.W.; Huang, X.; Zhang, Y.; Miao, J.M.; Xu, B.; Frazier, T.; et al. Evaluation of candidate reference genes for normalization of quantitative RT-PCR in Switchgrass under various abiotic stress conditions. Bioenergy Res. 2014, 7, 1201–1211. [Google Scholar] [CrossRef]
- Exposito-Rodriguez, M.; Borges, A.A.; Borges-Perez, A.; Perez, J.A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008, 8, 131. [Google Scholar] [CrossRef] [PubMed]

| Gene | Description | KOG ID | Sequence (5′-3′) | Amplicon Size (bp) | Efficiency (%) | R2 |
|---|---|---|---|---|---|---|
| ACT a | Actin and related proteins | KOG0676 | GACCATGTACTCGGGCATCTCAGCGAAGCCAGAATAGACC | 139 | - | - |
| EF1 | Translation elongation factor EF-1 alpha | KOG0052 | AGGAGGCTCAATAGCGTCAA | 97 | 89.30 | 0.9970 |
| CAACATGCAATGGTTCAAGG | ||||||
| EF2 | Elongation factor 2 | KOG0469 | AGAAGCGAGGTCACGTGTTT | 155 | 98.40 | 0.9960 |
| GTGGTCGAACACCATCTGTG | ||||||
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | KOG0657 | TCCTGCCTTTGAAATTTTGG | 101 | 114.50 | 0.9910 |
| TTGCTTTACGCTTGATGTGC | ||||||
| 40S | 40S ribosomal protein S7 | KOG3320 | GGATCCATCGGTGTGAAATC | 104 | 91.16 | 0.9976 |
| CTCGGTCACGAACACTCAAA | ||||||
| 60S a | 60S ribosomal protein L14/L17/L23 | KOG0901 | ATAGACGCGTTTTCGCTTGT | 195 | - | - |
| CGGTAAAAGGCTCGTCTCAG | ||||||
| β-TUB a | Beta tubulin | KOG1668 | CCGATCAATTCACAGCAATG | 195 | - | - |
| CAGGGGGTACCTCATCTTCA | ||||||
| α-TUB | Alpha tubulin | KOG1376 | ATATTTCCCGGAGCCAGTCT | 106 | 95.50 | 0.9950 |
| TACCATCGAGCATGGTTTGA | ||||||
| UBCE3 | E3 ubiquitin-protein ligase | KOG0939 | CCTTTGCTGGACTTTGAAGC | 183 | 103.10 | 0.9910 |
| GCCTACACCAGAGGAGCTTG | ||||||
| Cluster-3395.48660 b | - | - | GGAAGTGGCACCAACAAAGT | 84 | - | - |
| ATCCACGACATTCGCATACA |
| Parameters | Gene | |||||
|---|---|---|---|---|---|---|
| EF1 | EF2 | GAPDH | α-TUB | UBCE3 | 40S | |
| Geo Mean | 18.35 | 20.53 | 19.58 | 20.79 | 22.83 | 21.15 |
| AR Mean | 18.37 | 20.55 | 19.61 | 20.81 | 22.87 | 21.17 |
| Min | 17.31 | 19.25 | 18.11 | 19.57 | 20.94 | 19.74 |
| Max | 19.90 | 21.81 | 21.85 | 22.56 | 25.22 | 22.69 |
| SD | 0.77 | 0.71 | 0.86 | 0.86 | 0.98 | 0.55 |
| CV (%) | 4.21 | 3.43 | 4.38 | 4.12 | 4.30 | 2.61 |
| x-fold Min [x-fold] | −2.04 | −2.43 | −2.76 | −2.33 | −3.71 | −2.66 |
| x-fold Max [x-fold] | 2.94 | 2.43 | 4.85 | 3.43 | 5.22 | 2.89 |
| x-fold SD [± x-fold] | 1.71 | 1.63 | 1.81 | 1.81 | 1.98 | 1.47 |
| CC [r] | 0.991 | 0.915 | 0.930 | 0.938 | 0.892 | 0.768 |
| p-value | 0.001 | 0.001 | 0.001 | 0.001 | 0.003 | 0.026 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, C.; Lao, W.; Liang, Y. Reference Genes Selection of Gymnosporangium yamadae during the Interaction with Apple Leaves. J. Fungi 2022, 8, 830. https://doi.org/10.3390/jof8080830
Shao C, Lao W, Liang Y. Reference Genes Selection of Gymnosporangium yamadae during the Interaction with Apple Leaves. Journal of Fungi. 2022; 8(8):830. https://doi.org/10.3390/jof8080830
Chicago/Turabian StyleShao, Chenxi, Wenhao Lao, and Yingmei Liang. 2022. "Reference Genes Selection of Gymnosporangium yamadae during the Interaction with Apple Leaves" Journal of Fungi 8, no. 8: 830. https://doi.org/10.3390/jof8080830





