Metabolomic Profiling Revealed Diversion of Cytidinediphosphate-Diacylglycerol and Glycerol Pathway towards Denovo Triacylglycerol Synthesis in Rhodosporidium toruloides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast and Culture Conditions
2.2. Biochemical Analysis
2.3. Lipid Quantification and Profiling
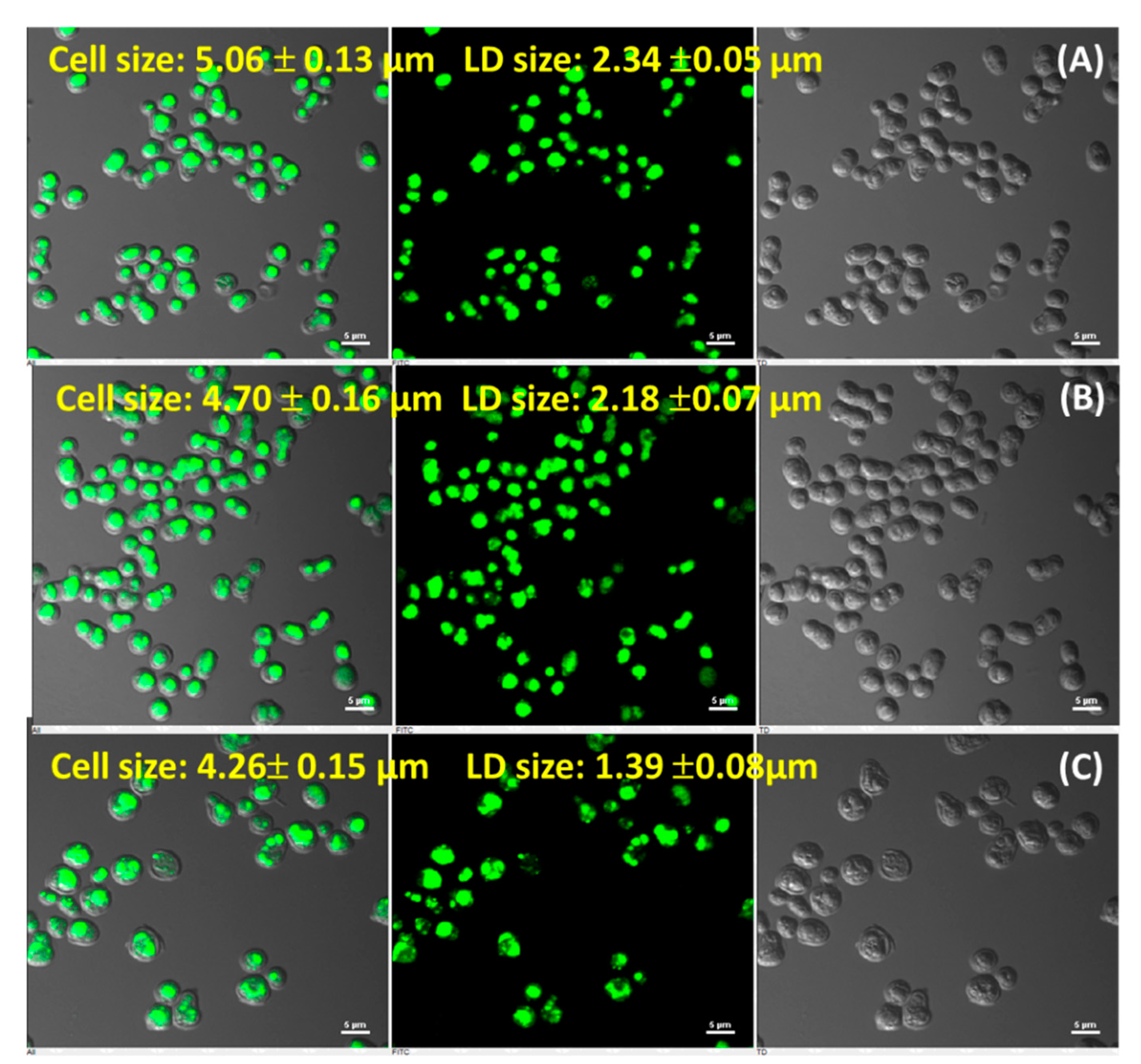
2.4. Lipid Droplet Size Estimation
2.5. Qualitative Metabolomics Using GC-MS
2.6. LC–MS Based Untargeted Metabolite Analysis
2.7. Statistical Analysis
3. Results
3.1. Cultivation of R. toruloides at Variable Carbon Concentrations
3.2. Time Course of Lipid Accumulation under Nitrogen-Limited Condition
3.3. R. toruloideslipid Droplet Formation at Nitrogen Stress Condition
3.4. Fatty Acid Profiling of R. toruloides under Nutrient Stress Condition
3.5. GC–MS and LC-MS/MS Based Untargeted Metabolome Analysis
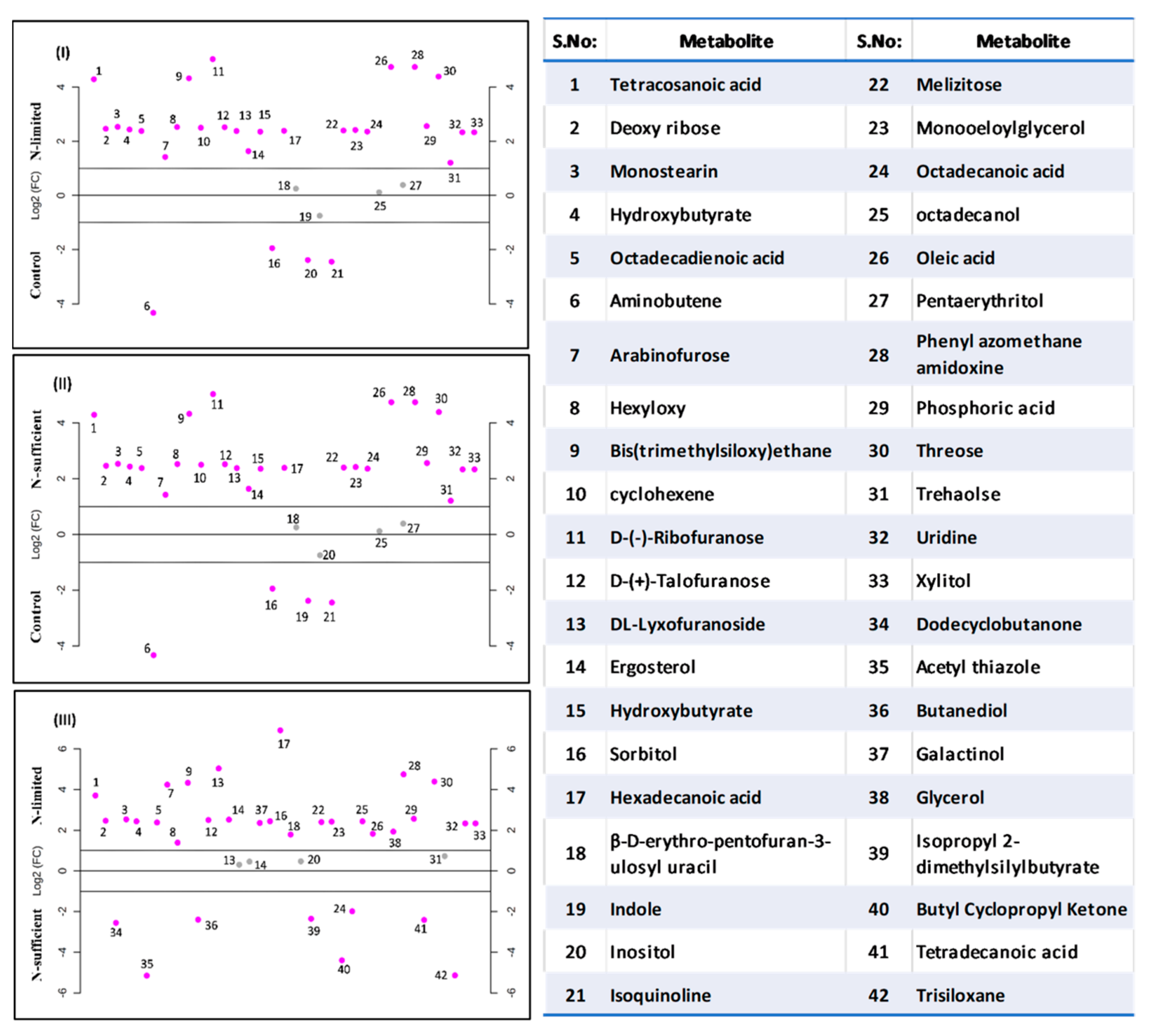
3.5.1. GC-MS Analysis
3.5.2. LC-MS/MS Analysis
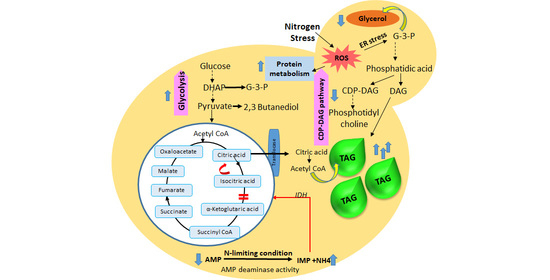
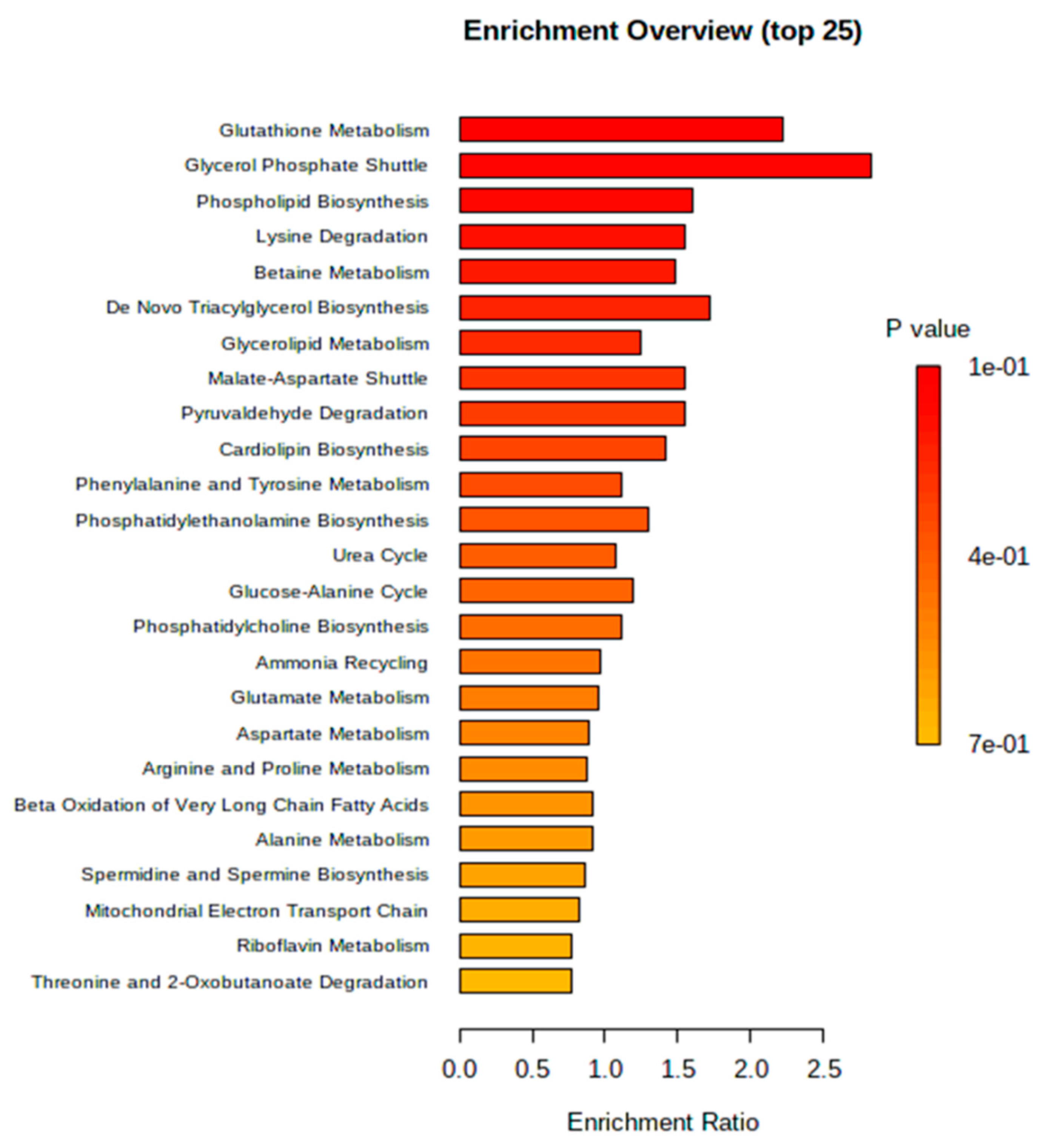
3.6. Metabolic Pathway Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
References
- Oliveira, J.; Antunes, M.; Godinho, C.P.; Teixeira, M.C.; Sá-Correia, I.; Monteiro, P.T. From a genome assembly to full regulatory network prediction: The case study of Rhodotorula toruloides putative Haa1-regulon. BMC Bioinform. 2021, 22, 399. [Google Scholar] [CrossRef]
- Lee, J.J.L.; Chen, L.; Shi, J.; Trzcinski, A.; Chen, W. Metabolomic Pro fi ling of Rhodosporidium toruloides Grown on Glycerol for Carotenoid Production during Different Growth Phases. J. Agric. Food Chem. 2014, 62, 10203–10209. [Google Scholar] [CrossRef]
- Wen, Z.; Zhang, S.; Odoh, C.K.; Jin, M.; Zhao, Z.K. Rhodosporidium toruloides—A potential red yeast chassis for lipids and beyond. FEMS Yeast Res. 2020, 20, foaa038. [Google Scholar] [CrossRef]
- Martins, L.C.; Palma, M.; Angelov, A.; Nevoigt, E.; Liebl, W.; Sá-Correia, I. Complete utilization of the major carbon sources present in sugar beet pulp hydrolysates by the oleaginous red yeasts Rhodotorula toruloides and R. Mucilaginosa. J. Fungi 2021, 7, 215. [Google Scholar] [CrossRef]
- Saran, S.; Mathur, A.; Dalal, J.; Saxena, R.K. Process optimization for cultivation and oil accumulation in an oleaginous yeast Rhodosporidium toruloides A29. Fuel 2017, 188, 324–331. [Google Scholar] [CrossRef]
- Ljungdahl, P.O.; Bertrand, D.-F. Regulation of Amino Acid, Nucleotide, and Phosphate Metabolism in Saccharomyces cerevisiae. Genetics 2012, 190, 885–929. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.; Nicaud, J.; Ledesma-amaro, R. The Engineering Potential of Rhodosporidium toruloides as a Workhorse for Biotechnological Applications. Trends Biotechnol. 2018, 36, 304–317. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, S.; Liu, H.; Shen, H.; Lin, X.; Yang, F.; Zhou, Y.J.; Jin, G.; Ye, M.; Zou, H.; et al. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat. Commun. 2012, 3, 1111–1112. [Google Scholar] [CrossRef] [Green Version]
- Jagtap, S.S.; Deewan, A.; Jing, J.; Hanna, L.; Yun, E.J.; Jin, Y.S.; Rao, C. V Integrating transcriptomic and metabolomic analysis of the oleaginous yeast Rhodosporidium toruloides IFO0880 during growth under different carbon sources. Appl. Microbiol. Biotechnol. 2021, 105, 7411–7425. [Google Scholar] [CrossRef]
- Lopes, H.J.S.; Bonturi, N.; Kerkhoven, E.J.; Miranda, E.A.; Lahtvee, P. C/N ratio and carbon source-dependent lipid production profiling in Rhodotorula toruloides. Appl. Microbiol. Biotechnol. 2020, 104, 2639–2649. [Google Scholar] [CrossRef] [Green Version]
- Kumar, K.K.; Deeba, F.; Negi, Y.S.; Gaur, N.A. Harnessing pongamia shell hydrolysate for triacylglycerol agglomeration by novel oleaginous yeast Rhodotorula pacifica INDKK. Biotechnol. Biofuels 2020, 13, 175. [Google Scholar] [CrossRef]
- Pandey, A.K.; Kumar, M.; Kumari, S.; Kumari, P.; Yusuf, F.; Jakeer, S.; Naz, S.; Chandna, P.; Bhatnagar, I.; Gaur, N.A. Evaluation of divergent yeast genera for fermentation-associated stresses and identification of a robust sugarcane distillery waste isolate Saccharomyces cerevisiae NGY10 for lignocellulosic ethanol production in SHF and SSF. Biotechnol. Biofuels 2019, 12, 40. [Google Scholar] [CrossRef] [Green Version]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Deeba, F.; Pruthi, V.; Negi, Y.S. Fostering triacylglycerol accumulation in novel oleaginous yeast Cryptococcus psychrotolerans IITRFD utilizing groundnut shell for improved biodiesel production. Bioresour. Technol. 2017, 242, 113–120. [Google Scholar] [CrossRef]
- Liu, B.; Zhao, Z. (Kent) Biodiesel production by direct methanolysis of oleaginous microbial biomass. J. Chem. Technol. Biotechnol. 2007, 82, 775–780. [Google Scholar] [CrossRef]
- Patel, A.; Sindhu, D.K.; Arora, N.; Singh, R.P.; Pruthi, V.; Pruthi, P.A. Biodiesel production from non-edible lignocellulosic biomass of Cassia fistula L. fruit pulp using oleaginous yeast Rhodosporidium kratochvilovae HIMPA1. Bioresour. Technol. 2015, 197, 91–98. [Google Scholar] [CrossRef]
- Mariam, I.; Kareya, M.S.; Nesamma, A.A.; Jutur, P.P. Delineating metabolomic changes in native isolate Aurantiochytrium for production of docosahexaenoic acid in presence of varying carbon substrates. Algal Res. 2021, 55, 102285. [Google Scholar] [CrossRef]
- Bhatnagar, V.S.; Bandyopadhyay, P.; Rajacharya, G.H.; Sarkar, S.; Poluri, K.M.; Kumar, S. Amelioration of biomass and lipid in marine alga by an endophytic fungus Piriformospora indica. Biotechnol. Biofuels 2019, 12, 176. [Google Scholar] [CrossRef] [Green Version]
- González-García, Y.; Rábago-Panduro, L.M.; French, T.; Camacho-Córdova, D.I.; Gutiérrez-González, P.; Córdova, J. High lipids accumulation in Rhodosporidium toruloides by applying single and multiple nutrients limitation in a simple chemically defined medium. Ann. Microbiol. 2017, 67, 519–527. [Google Scholar] [CrossRef]
- Tchakouteu, S.S.; Kopsahelis, N.; Chatzifragkou, A.; Kalantzi, O.; Stoforos, N.G.; Koutinas, A.A.; Aggelis, G.; Papanikolaou, S. Rhodosporidium toruloides cultivated in NaCl-enriched glucose-based media: Adaptation dynamics and lipid production. Eng. Life Sci. 2017, 17, 237–248. [Google Scholar] [CrossRef] [Green Version]
- Petelenz-Kurdziel, E.; Kuehn, C.; Nordlander, B.; Klein, D.; Hong, K.K.; Jacobson, T.; Dahl, P.; Schaber, J.; Nielsen, J.; Hohmann, S.; et al. Quantitative Analysis of Glycerol Accumulation, Glycolysis and Growth under Hyper Osmotic Stress. PLoS Comput. Biol. 2013, 9, e1003084. [Google Scholar] [CrossRef]
- Lu, H.; Chen, H.; Tang, X.; Yang, Q.; Zhang, H.; Chen, Y.Q.; Chen, W. Metabolomics analysis reveals the role of oxygen control in the nitrogen limitation induced lipid accumulation in Mortierella alpina. J. Biotechnol. 2021, 325, 325–333. [Google Scholar] [CrossRef]
- Capusoni, C.; Rodighiero, V.; Cucchetti, D.; Galafassi, S.; Bianchi, D.; Franzosi, G.; Compagno, C. Characterization of lipid accumulation and lipidome analysis in the oleaginous yeasts Rhodosporidium azoricum and Trichosporon oleaginosus. Bioresour. Technol. 2017, 238, 281–289. [Google Scholar] [CrossRef]
- Tang, X.; Lee, J.; Chen, W.N. Engineering the fatty acid metabolic pathway in Saccharomyces cerevisiae for advanced biofuel production. Metab. Eng. Commun. 2015, 2, 58–66. [Google Scholar] [CrossRef]
- Patel, A.; Arora, N.; Mehtani, J.; Pruthi, V.; Pruthi, P.A. Assessment of fuel properties on the basis of fatty acid pro fi les of oleaginous yeast for potential biodiesel production. Renew. Sustain. Energy Rev. 2017, 77, 604–616. [Google Scholar] [CrossRef] [Green Version]
- Hakizimana, O.; Matabaro, E.; Lee, B.H. The current strategies and parameters for the enhanced microbial production of 2,3-butanediol. Biotechnol. Rep. 2020, 25, e00397. [Google Scholar] [CrossRef]
- Yodsuwan, N.; Sawayama, S.; Sirisansaneeyakul, S. Effect of nitrogen concentration on growth, lipid production and fatty acid profiles of the marine diatom Phaeodactylum tricornutum. Agric. Nat. Resour. 2017, 51, 190–197. [Google Scholar] [CrossRef]
- Martin, G.J.O.; Hill, D.R.A.; Olmstead, I.L.D.; Bergamin, A.; Shears, M.J.; Dias, D.A.; Kentish, S.E.; Scales, P.J.; Botté, C.Y.; Callahan, D.L. Lipid profile remodeling in response to nitrogen deprivation in the microalgae Chlorella sp. (Trebouxiophyceae) and Nannochloropsis sp. (Eustigmatophyceae). PLoS ONE 2014, 9, e103389. [Google Scholar] [CrossRef] [Green Version]
- Sitepu, I.R.; Sestric, R.; Ignatia, L.; Levin, D.; German, J.B.; Gillies, L.A.; Almada, L.A.G.; Boundy-mills, K.L. Manipulation of culture conditions alters lipid content and fatty acid profiles of a wide variety of known and new oleaginous yeast species. Bioresour. Technol. 2013, 144, 360–369. [Google Scholar] [CrossRef] [Green Version]
- Knothe, G. A comprehensive evaluation of the cetane numbers of fatty acid methyl esters. Fuel 2014, 119, 6–13. [Google Scholar] [CrossRef]
- Patel, A.; Pruthi, V.; Pruthi, P.A. Synchronized nutrient stress conditions trigger the diversion of CDP-DG pathway of phospholipids synthesis towards de novo TAG synthesis in oleaginous yeast escalating biodiesel production. Energy 2017, 139, 962–974. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Liu, D. Exploitation of genus Rhodosporidium for microbial lipid production. World J. Microbiol. Biotechnol. 2017, 33, 54. [Google Scholar] [CrossRef]
- Hoekman, S.K.; Broch, A.; Robbins, C.; Ceniceros, E.; Natarajan, M. Review of biodiesel composition, properties, and specifications. Renew. Sustain. Energy Rev. 2012, 16, 143–169. [Google Scholar] [CrossRef]
- Knothe, G. Improving biodiesel fuel properties by modifying fatty ester composition. Energy Environ. Sci. 2009, 2, 759–766. [Google Scholar] [CrossRef]
- Shaikh, K.M.; Nesamma, A.A.; Abdin, M.Z.; Jutur, P.P. Biotechnology for Biofuels Molecular profiling of an oleaginous trebouxiophycean alga Parachlorella kessleri subjected to nutrient deprivation for enhanced biofuel production. Biotechnol. Biofuels 2019, 12, 182. [Google Scholar] [CrossRef] [Green Version]
- Arora, N.; Dubey, D.; Sharma, M.; Patel, A.; Guleria, A.; Pruthi, P.A.; Kumar, D.; Pruthi, V.; Poluri, K.M. NMR-Based Metabolomic Approach to Elucidate the Differential Cellular Responses during Mitigation of Arsenic(III, V) in a Green Microalga. ACS Omega 2018, 3, 11847–11856. [Google Scholar] [CrossRef]
- Carsanba, E.; Papanikolaou, S.; Erten, H. Production of oils and fats by oleaginous microorganisms with an emphasis given to the potential of the nonconventional yeast Yarrowia lipolytica. Crit. Rev. Biotechnol. 2018, 38, 1230–1243. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, Y.; Fu, R.; Zhang, T.; Li, J.; Zhang, J. Transcriptomic and metabolomic profiling of a Rhodotorula color mutant to improve its lipid productivity in fed-batch fermentation. World J. Microbiol. Biotechnol. 2021, 37, 77. [Google Scholar] [CrossRef]
- Dissook, S.; Putri, S.P.; Fukusaki, E. Metabolomic analysis of response to nitrogen-limiting conditions in Yarrowia spp. Metabolites 2021, 11, 16. [Google Scholar] [CrossRef]
- Shen, H.; Zhang, X.; Gong, Z.; Wang, Y.; Yu, X. Compositional profiles of Rhodosporidium toruloides cells under nutrient limitation. Appl. Microbiol. Biotechnol. 2017, 101, 3801–3809. [Google Scholar] [CrossRef]






| YNB Medium | DCW (g/L) | Lipid Titer (g/L) | Glycerol (g/L) | Citric Acid (g/L) | Residual Glucose (g/L) | Butanediol (g/L) | FAME Productivity (g/L/h) |
|---|---|---|---|---|---|---|---|
| 3% Glucose, 0.5% (NH4)2SO4 | 6.8 ± 0.18 | 3.4 ± 0.08 | - | - | 1.67 ± 0.005 | - | 0.53 ± 0.02 |
| 4% Glucose, 0.5% (NH4)2SO4 | 8.2 ± 0.15 | 4 ± 0.007 | 3.45 ± 0.038 | 0.52 ± 0.005 | 2.58 ± 0.006 | 0.11 ±0.005 | 0.48 ± 0.04 |
| 5% Glucose, 0.5% (NH4)2SO4 | 9.6 ± 0.12 | 5.8 ± 0.08 | 3.83 ± 0.027 | 0.77 ± 0.008 | 4.26 ± 0.007 | 0.12 ± 0.006 | 0.69 ± 0.02 |
| 6% Glucose, 0.5% (NH4)2SO4 | 10.4 ± 0.28 | 5.3 ± 0.12 | 6.27 ± 0.045 | 0.43 ± 0.007 | 10.29 ± 0.004 | 0.07 ± 0.008 | 0.63 ± 0.05 |
| 7% Glucose, 0.5% (NH4)2SO4 | 9.2 ± 0.13 | 3.8 ± 0.15 | 5.60 ± 0.036 | 0.49 ± 0.006 | 21.28 ± 0.016 | - | 0.45 ± 0.03 |
| 8% Glucose, 0.5% (NH4)2SO4 | 9.2 ± 0.27 | 3.6 ± 0.08 | 6.63 ± 0.042 | 0.48 ± 0.008 | 35.03 ± 0.015 | 0.07 ± 0.008 | 0.43 ± 0.03 |
| 9% Glucose, 0.5% (NH4)2SO4 | 7.41 ± 0.19 | 3.1 ± 0.06 | 3.84 ± 0.054 | 0.67 ± 0.009 | 36.89 ± 0.008 | 0.05 ± 0.004 | 0.37 ± 0.05 |
| 10% Glucose, 0.5% (NH4)2SO4 | 7.25 ± 0.17 | 2.5 ± 0.04 | 3.67 ± 0.044 | 0.70 ± 0.045 | 44.98 ± 0.014 | 0.08 ± 0.006 | 0.3 ± 0.04 |
| 5% Glucose, 0.5% Yeast extract | 10.2 ± 0.28 | 3.1 ± 0.16 | 6.34 ± 0.45 | 1.10 ± 0.52 | 8.75 ± 0.48 | - | 0.37 ± 0.03 |
| 5% Glucose, 0.5% Peptone | 8.1 ± 0.27 | 3.6 ± 0.18 | 2.51 ± 0.34 | 1.02 ± 0.37 | 2.60 ± 0.35 | 0.12 ± 0.06 | 0.43 ± 0.06 |
| 5% Glucose, 0.5% Urea | 5.5 ± 0.14 | 2.8 ± 0.16 | - | - | 10.68 ± 0.57 | - | 0.33 ± 0.04 |
| 5% Glucose, 0.12% (NH4)2SO4, | 13 ± 0.52 | 8.23 ± 0.48 | 1.72 ± 0.08 | 0.08 ± 0.004 | 5.2 ± 0.26 | 0.13 ± 0.05 | 0.98 ± 0.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deeba, F.; Kumar, K.K.; Rajacharya, G.H.; Gaur, N.A. Metabolomic Profiling Revealed Diversion of Cytidinediphosphate-Diacylglycerol and Glycerol Pathway towards Denovo Triacylglycerol Synthesis in Rhodosporidium toruloides. J. Fungi 2021, 7, 967. https://doi.org/10.3390/jof7110967
Deeba F, Kumar KK, Rajacharya GH, Gaur NA. Metabolomic Profiling Revealed Diversion of Cytidinediphosphate-Diacylglycerol and Glycerol Pathway towards Denovo Triacylglycerol Synthesis in Rhodosporidium toruloides. Journal of Fungi. 2021; 7(11):967. https://doi.org/10.3390/jof7110967
Chicago/Turabian StyleDeeba, Farha, Kukkala Kiran Kumar, Girish H. Rajacharya, and Naseem A. Gaur. 2021. "Metabolomic Profiling Revealed Diversion of Cytidinediphosphate-Diacylglycerol and Glycerol Pathway towards Denovo Triacylglycerol Synthesis in Rhodosporidium toruloides" Journal of Fungi 7, no. 11: 967. https://doi.org/10.3390/jof7110967