A Novel Deoxynivalenol-Activated Wheat Arl6ip4 Gene Encodes an Antifungal Peptide with Deoxynivalenol Affinity and Protects Plants against Fusarium Pathogens and Mycotoxins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant and Microbial Materials
2.2. RNA Extraction and Suppression Subtractive Hybridization
2.3. DON Treatment and Fungal Inoculation of Wheat Spikes
2.4. Molecular Cloning and Sequence Analysis
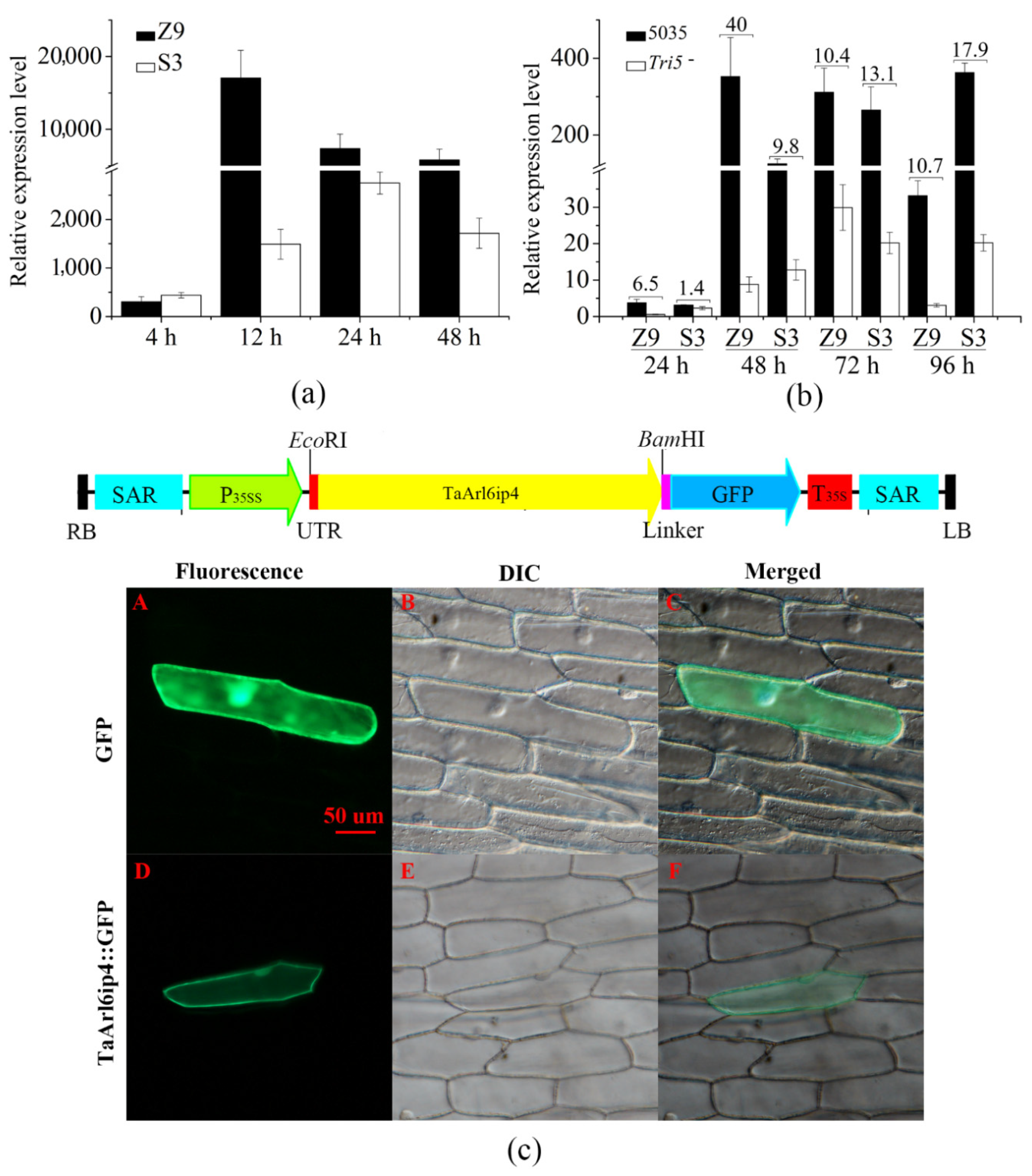
2.5. Subcellular Localization
2.6. Plant Transformation
2.7. Antifungal Activity
2.8. Microscale Thermophoresis Analysis
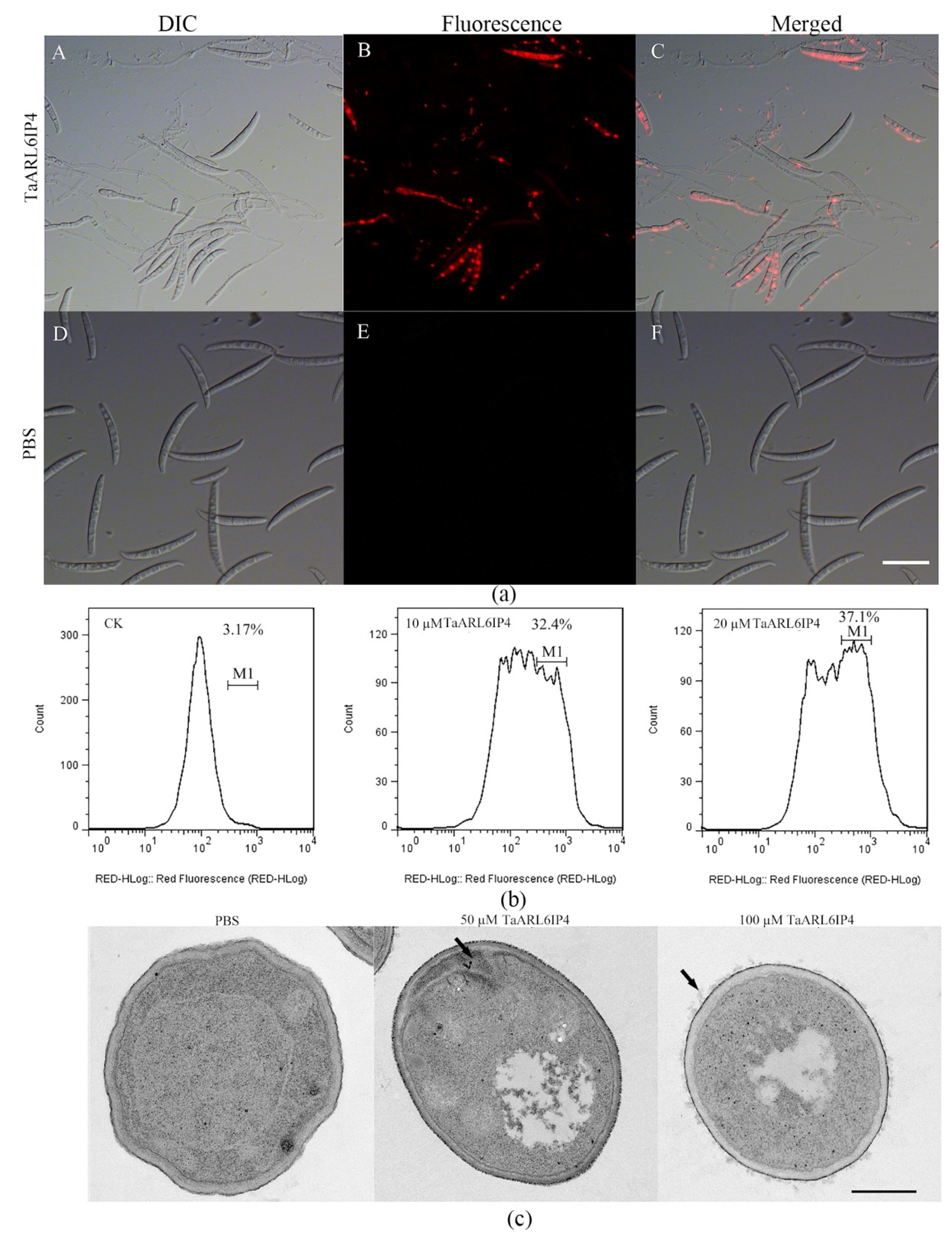
2.9. Fluorescence Microscopy Analysis
2.10. Transmission Electron Microscopy Assay
2.11. DON Tolerance and Fungal Resistance Assays
2.12. Southern Blotting and Northern Blotting
2.13. Statistical Analysis
3. Results
3.1. TaArl6ip4 Encodes an ADP-Ribosylation Factor-Like Protein 6-Interacting Protein 4 in Response to DON
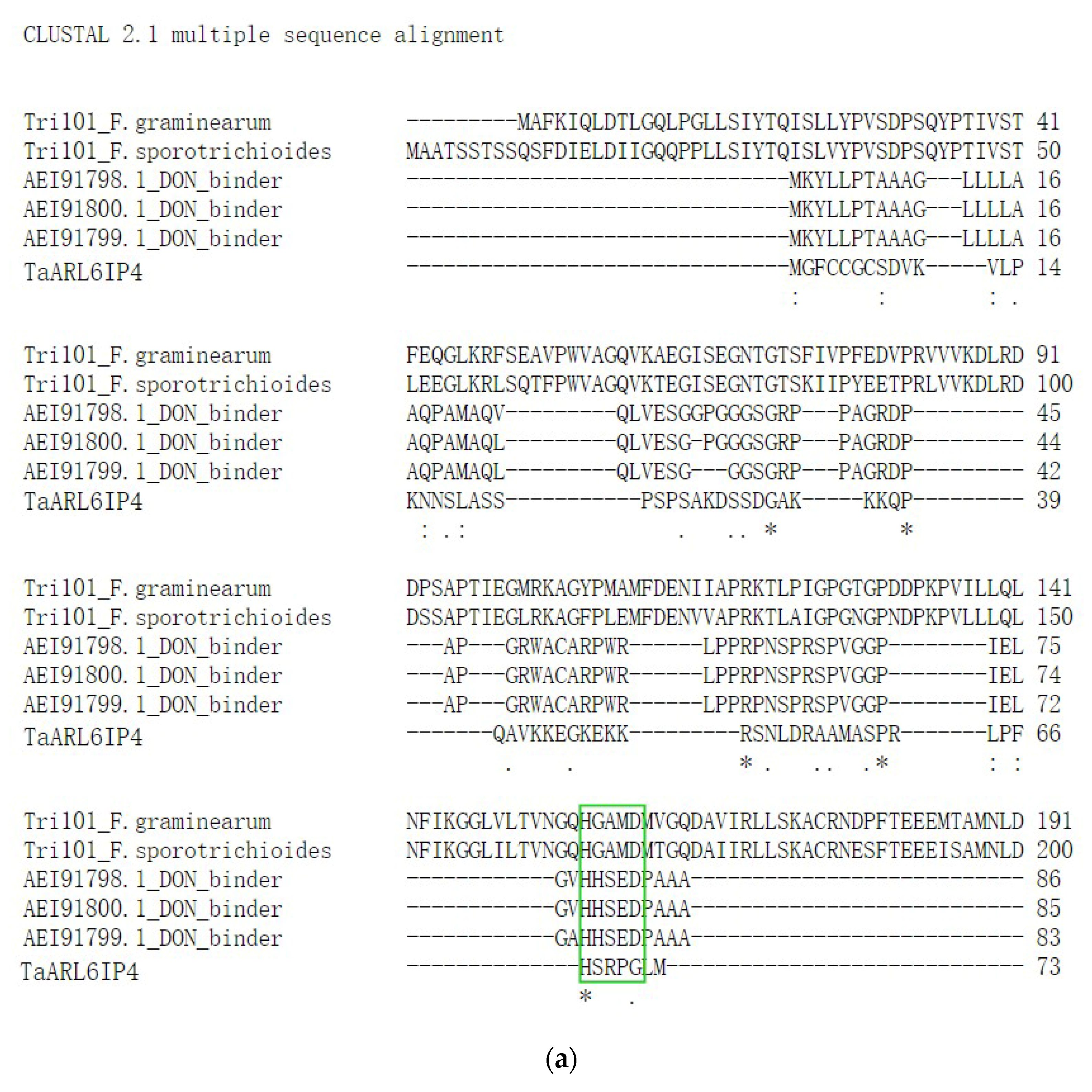
3.2. Sequence Analysis and Functional Prediction of TaARL6IP4
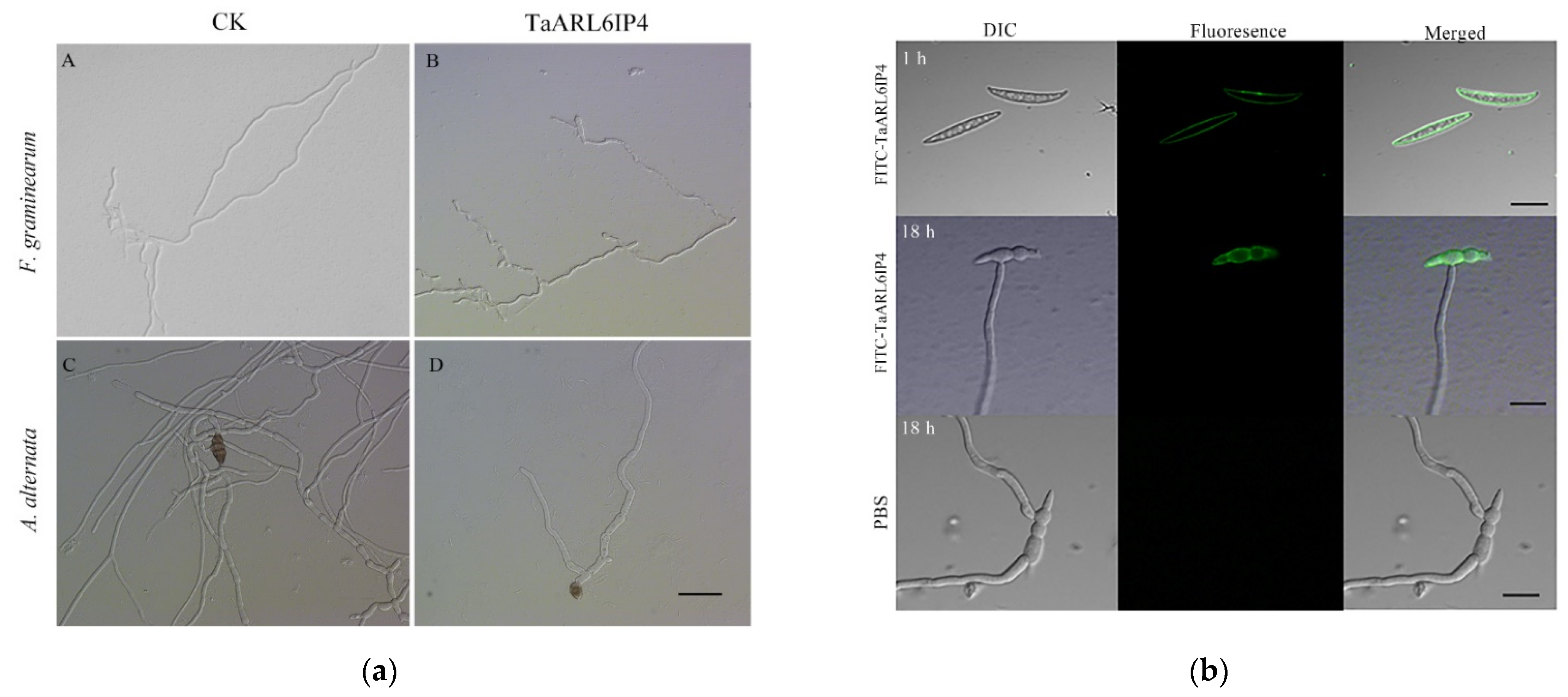
3.3. TaArl6ip4 Encodes an Antifungal Peptide
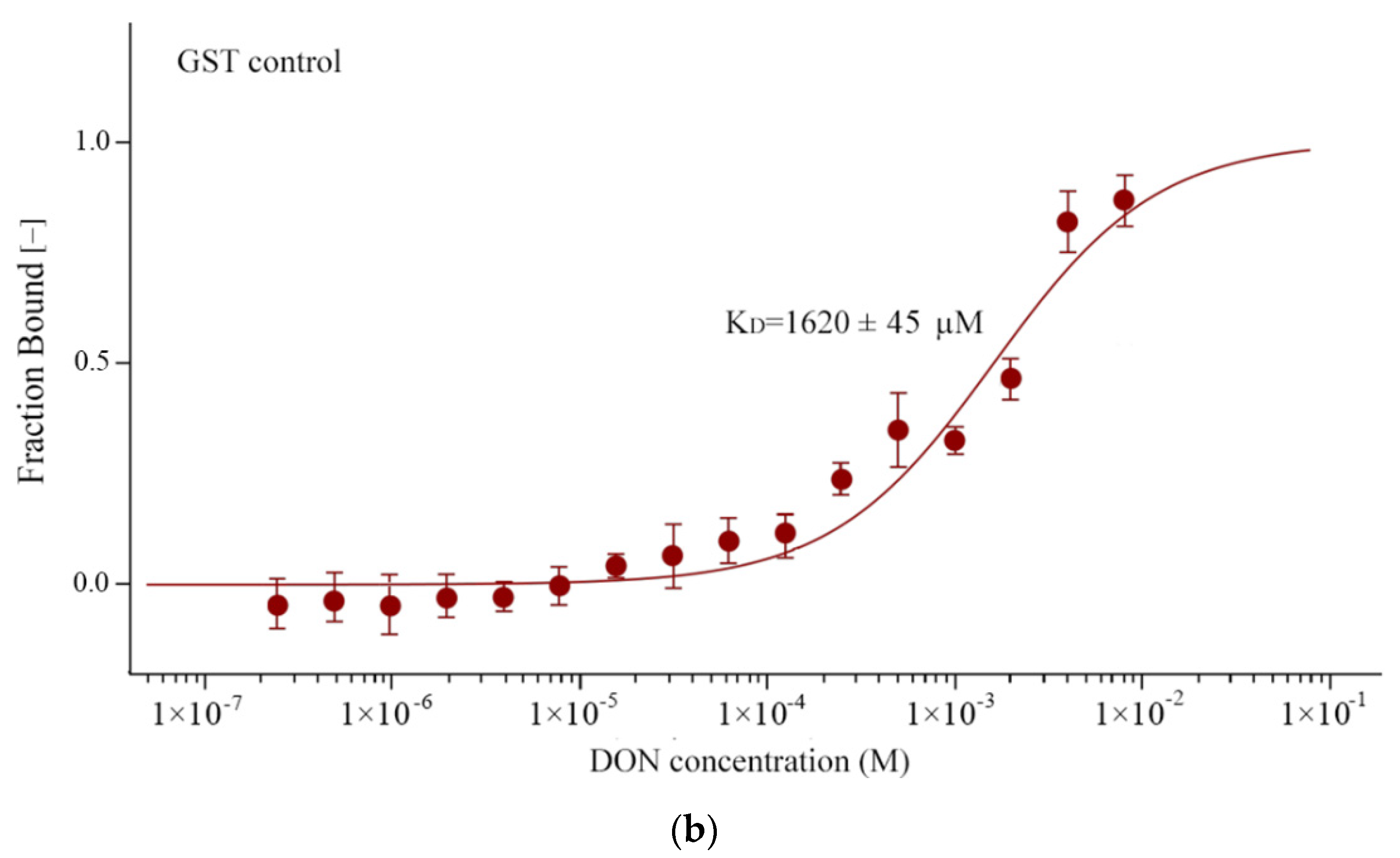
3.4. TaARL6IP4 Exhibits DON Affinity In Vitro
3.5. TaARL6IP4 Disrupts the Membrane Integrity of F. graminearum Spores
3.6. TaArl6ip4 Enhances DON Tolerance and FHB Resistance in Arabidopsis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parry, D.W.; Jenkinson, P.; McLeod, L. Fusarium ear blight (scab) in small grain cereals—A review. Plant Pathol. J. 1995, 44, 207–238. [Google Scholar] [CrossRef]
- Bai, G.; Shaner, G. Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 2004, 42, 135–161. [Google Scholar] [CrossRef]
- Wegulo, S.N.; Baenziger, P.S.; Hernandez Nopsa, J.; Bockus, W.W.; Hallen-Adams, H. Management of Fusarium head blight of wheat and barley. Crop Prot. 2015, 73, 100–107. [Google Scholar] [CrossRef]
- Kazan, K.; Gardiner, D.M.; Manners, J.M. On the trail of a cereal killer: Recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 2012, 13, 399–413. [Google Scholar] [CrossRef]
- McMullen, M.; Bergstrom, G.; De Wolf, E.; Dill-Macky, R.; Hershman, D.; Shaner, G.; Van Sanford, D. A unified effort to fight an enemy of wheat and barley: Fusarium head blight. Plant Dis. 2012, 96, 1712–1728. [Google Scholar] [CrossRef] [Green Version]
- Windels, C.E. Economic and social impacts of Fusarium head blight: Changing farms and rural communities in the northern great plains. Phytopathology 2000, 90, 17–21. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.M.; Parry, D.W.; Nicholson, P.; Thomsett, M.A.; Simpson, D.; Edwards, S.G.; Cooke, B.M.; Doohan, F.M.; Brennan, J.M.; Moretti, A.; et al. Predominance and association of pathogenic fungi causing Fusarium ear blight in wheat in four European countries. Eur. J. Plant Pathol. 2005, 112, 143–154. [Google Scholar] [CrossRef]
- Zhu, Z.W.; Hao, Y.F.; Mergoum, M.; Bai, G.H.; Humphreys, G.; Cloutier, S.; Xia, X.C.; He, Z.H. Breeding wheat for resistance to Fusarium head blight in the Global North: China, USA, and Canada. Crop J. 2019, 7, 730–738. [Google Scholar] [CrossRef]
- Huang, Y.; Haas, M.; Heinen, S.; Steffenson, B.J.; Smith, K.P.; Muehlbauer, G.J. QTL mapping of Fusarium head blight and correlated agromorphological traits in an elite barley cultivar Rasmusson. Front. Plant Sci. 2018, 9, 1260. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Z.; Ma, H.; Huang, L.; Ding, F.; Du, Y.; Jia, H.; Li, G.; Kong, Z.; Ran, C.; et al. Pyramiding of Fusarium Head Blight resistance quantitative trait loci, Fhb1, Fhb4, and Fhb5, in modern Chinese wheat cultivars. Front. Plant Sci. 2021, 12, 694023. [Google Scholar] [CrossRef]
- Audenaert, K.; Vanheule, A.; Höfte, M.; Haesaert, G. Deoxynivalenol: A major player in the multifaceted response of Fusarium to its environment. Toxins 2014, 6, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Karlovsky, P. Biological detoxification of the mycotoxin deoxynivalenol and its use in genetically engineered crops and feed additives. Appl. Microbiol. Biotechnol. 2011, 91, 491–504. [Google Scholar] [CrossRef] [Green Version]
- de Loubresse, N.G.; Prokhorova, I.; Holtkamp, W.; Rodnina, M.V.; Yusupova, G.; Yusupov, M. Structural basis for the inhibition of the eukaryotic ribosome. Nature 2014, 513, 517–522. [Google Scholar] [CrossRef] [Green Version]
- Dellafiora, L.; Dall’Asta, C.; Galaverna, G. Toxicodynamics of mycotoxins in the framework of food risk assessment—An in silico perspective. Toxins 2018, 10, 52. [Google Scholar] [CrossRef] [Green Version]
- Rotter, B.A.; Prelusky, D.B.; Pestka, J.J. Invited review: Toxicology of deoxynivalenol (vomitoxin). J. Toxicol. Environ. Health 1996, 48, 1–34. [Google Scholar] [CrossRef]
- Pestka, J.J. Deoxynivalenol: Toxicity, mechanisms and animal health risks. Anim. Feed Sci. Technol. 2007, 137, 283–298. [Google Scholar] [CrossRef]
- Masuda, D.; Ishida, M.; Yamaguchi, K.; Yamaguchi, I.; Kimura, M.; Nishiuchi, T. Phytotoxic effects of trichothecenes on the growth and morphology of Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 1617–1626. [Google Scholar] [CrossRef] [Green Version]
- Harris, L.J.; Desjardins, A.E.; Plattner, R.D.; Nicholson, P.; Butler, G.; Young, J.C.; Weston, G.; Proctor, R.H.; Hohn, T.M. Possible role of trichothecene mycotoxins in virulence of Fusarium graminearum on maize. Plant Dis. 1999, 83, 954–960. [Google Scholar] [CrossRef] [Green Version]
- Bai, G.H.; Desjardins, A.E.; Plattner, R.D. Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 2002, 153, 91–98. [Google Scholar] [CrossRef]
- Maier, F.J.; Miedaner, T.; Hadeler, B.; Felk, A.; Salomon, S.; Lemmens, M.; Kassner, H.; Schäfer, W. Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 2006, 7, 449–461. [Google Scholar] [CrossRef]
- Hassan, Y.I.; Watts, C.; Li, X.Z.; Zhou, T. A novel Peptide-binding motifs inference approach to understand deoxynivalenol molecular toxicity. Toxins 2015, 7, 1989–2005. [Google Scholar] [CrossRef] [Green Version]
- Garvey, G.S.; McCormick, S.P.; Rayment, I. Structural and functional characterization of the TRI101 trichothecene 3-O-acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: Kinetic insights to combating Fusarium head blight. Int. J. Biol. Chem. 2008, 283, 1660–1669. [Google Scholar] [CrossRef] [Green Version]
- Tu, Z.; Xu, Y.; He, Q.; Fu, J.; Liu, X.; Tao, Y. Isolation and characterisation of deoxynivalenol affinity binders from a phage display library based on single-domain camelid heavy chain antibodies (VHHs). Food Agric. Immunol. 2012, 23, 123–131. [Google Scholar] [CrossRef]
- Stintzi, A.; Heitz, T.; Prasad, V.; Wiedemann-Merdinoglu, S.; Kauffmann, S.; Geoffroy, P.; Legrand, M.; Fritig, B. Plant ’pathogenesis-related’ proteins and their role in defense against pathogens. Biochimie 1993, 75, 687–706. [Google Scholar] [CrossRef]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial peptides from plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef]
- Campos, M.L.; Souza, C.M.; Oliveira, K.B.; Dias, S.C.; Franco, O.L. The role of antimicrobial peptides in plant immunity. J. Exp. Bot. 2018, 69, 4997–5011. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Hu, S.; Jian, W.; Xie, C.; Yang, X. Plant antimicrobial peptides: Structures, functions, and applications. Bot. Stud. 2021, 62, 5. [Google Scholar] [CrossRef]
- Colilla, F.J.; Rocher, A.; Mendez, E. γ-Purothionins: Amino acid sequence of two polypeptides of a new family of thionins from wheat endosperm. FEBS Lett. 1990, 270, 191–194. [Google Scholar] [CrossRef] [Green Version]
- Utkina, L.L.; Andreev, Y.A.; Rogozhin, E.A.; Korostyleva, T.V.; Slavokhotova, A.A.; Oparin, P.B.; Vassilevski, A.A.; Grishin, E.V.; Egorov, T.A.; Odintsova, T.I. Genes encoding 4-Cys antimicrobial peptides in wheat Triticum kiharae Dorof. et Migush.: Multimodular structural organization, instraspecific variability, distribution and role in defence. FEBS J. 2013, 280, 3594–3608. [Google Scholar] [CrossRef]
- Islam, K.T.; Velivelli, S.L.S.; Berg, R.H.; Oakley, B.; Shah, D.M. A novel bi-domain plant defensin MtDef5 with potent broad-spectrum antifungal activity binds to multiple phospholipids and forms oligomers. Sci. Rep. 2017, 7, 16157. [Google Scholar] [CrossRef]
- Safi, H.; Saibi, W.; Alaoui, M.M.; Hmyene, A.; Masmoudi, K.; Hanin, M.; Brini, F. A wheat lipid transfer protein (TdLTP4) promotes tolerance to abiotic and biotic stress in Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 89, 64–75. [Google Scholar] [CrossRef]
- D’Souza-Schorey, C.; Chavrier, P. ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006, 7, 347–358. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Mangu, V.; Zandkarimi, H.; Prasad, M.; Baisakh, N. Structure, organization and evolution of ADP-ribosylation factors in rice and foxtail millet and their expression in rice. Sci. Rep. 2016, 6, 24008. [Google Scholar] [CrossRef] [Green Version]
- Regad, F.; Bardet, C.; Tremousaygue, D.; Moisan, A.; Lescure, B.; Axelos, M. cDNA cloning and expression of an Arabidopsis GTP-binding protein of the ARF family. FEBS Lett. 1993, 316, 133–136. [Google Scholar] [CrossRef] [Green Version]
- Gebbie, L.K.; Burn, J.E.; Hocart, C.H.; Williamson, R.E. Genes encoding ADP-ribosylation factors in Arabidopsis thaliana L. Heyn.; genome analysis and antisense suppression. J. Exp. Bot. 2005, 56, 1079–1091. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.L.; Zhang, N.; Wen, Y.K.; Si, H.J.; Wang, D. Identification of differentially expressed genes in potato associated with tuber dormancy release. Mol. Biol. Rep. 2012, 39, 11277–11287. [Google Scholar] [CrossRef]
- Higo, H.; Kishimoto, N.; Saito, A.; Higo, K.I. Molecular cloning and characterization of a cDNA encoding a small GTP-binding protein related to mammalian ADP-ribosylation factor from rice. Plant Sci. 1994, 100, 41–49. [Google Scholar] [CrossRef]
- Zhou, X.J.; Li, J.; Cheng, W.; Liu, H.; Li, M.M.; Zhang, Y.; Li, W.B.; Han, S.C.; Wang, Y.D. Gene structure analysis of rice ADP-ribosylation factors (OsARFs) and their mRNA expression in developing rice plants. Plant Mol. Biol. Rep. 2010, 28, 692–703. [Google Scholar] [CrossRef]
- Kobayashi-Uehara, A.; Shimosaka, E.; Handa, H. Cloning and expression analyses of cDNA encoding an ADP-ribosylation factor from wheat: Tissue-specific expression of wheat ARF. Plant Sci. 2001, 160, 535–542. [Google Scholar] [CrossRef]
- Takeuchi, M.; Ueda, T.; Yahara, N.; Nakano, A. Arf1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 2002, 31, 499–515. [Google Scholar] [CrossRef] [PubMed]
- Welsh, C.F.; Moss, J.; Vaughan, M. ADP-ribosylation factors: A family of approximately 20-kDa guanine nucleotide-binding proteins that activate cholera toxin. Mol. Cell Biochem. 1994, 138, 157–166. [Google Scholar] [CrossRef] [Green Version]
- Radhakrishna, H.; Donaldson, J.G. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol. 1997, 139, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavenagh, M.M.; Whitney, J.A.; Carroll, K.; Zhang, C.J.; Boman, A.L.; Rosenwald, A.G.; Mellman, I.; Kahn, R.A. Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J. Biol. Chem. 1996, 271, 21767–21774. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.Z.; Heimberg, H.; D’Souza-Schorey, C.; Mueckler, M.M.; Stahl, P.D. Subcellular distribution and differential expression of endogenous ADP-ribosylation factor 6 in mammalian cells. J. Biol. Chem. 1998, 273, 4006–4011. [Google Scholar] [CrossRef] [Green Version]
- Niedergang, F.; Colucci-Guyon, E.; Dubois, T.; Raposo, G.; Chavrier, P. ADP ribosylation factor 6 is activated and controls membrane delivery during phagocytosis in macrophages. J. Cell Biol. 2003, 161, 1143–1150. [Google Scholar] [CrossRef] [Green Version]
- D’Souza-Schorey, C.; Boshans, R.L.; McDonough, M.; Stahl, P.D.; Van Aelst, L. A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO J. 1997, 16, 5445–5454. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, P. SRrp37, a novel splicing regulator located in the nuclear speckles and nucleoli, interacts with SC35 and modulates alternative pre-mRNA splicing in vivo. J. Cell Biochem. 2009, 108, 304–314. [Google Scholar] [CrossRef]
- Sasahara, K.; Yamaoka, T.; Moritani, M.; Tanaka, M.; Iwahana, H.; Yoshimoto, K.; Miyagawa, J.; Kuroda, Y.; Itakura, M. Molecular cloning and expression analysis of a putative nuclear protein, SR-25. Biochem. Biophys. Res. Commun. 2000, 269, 444–450. [Google Scholar] [CrossRef]
- Li, Q.H.; Zhao, H.L.; Jiang, L.; Che, Y.C.; Dong, C.H.; Wang, L.C.; Wang, J.; Liu, L.D. An SR-protein induced by HSVI binding to cells functioning as a splicing inhibitor of viral pre-mRNA. J. Mol. Biol. 2002, 316, 887–894. [Google Scholar] [CrossRef]
- Zuo, D.Y.; Yi, S.Y.; Liu, R.J.; Qu, B.; Huang, T.; He, W.J.; Li, C.; Li, H.P.; Liao, Y.C. A Deoxynivalenol-activated methionyl-tRNA synthetase gene from wheat encodes a nuclear localized protein and protects plants against Fusarium pathogens and mycotoxins. Phytopathology 2016, 106, 614–623. [Google Scholar] [CrossRef] [Green Version]
- Qu, B.; Li, H.P.; Zhang, J.B.; Xu, Y.B.; Huang, T.; Wu, A.B.; Zhao, C.S.; Carter, P.; Nicholson, P.; Liao, Y.C. Geographic distribution and genetic diversity of Fusarium graminearum and F. asiaticum on wheat spikes throughout China. Plant Pathol. 2008, 57, 15–24. [Google Scholar] [CrossRef]
- Li, X.; Zhang, J.B.; Song, B.; Li, H.P.; Xu, H.Q.; Qu, B.; Dang, F.J.; Liao, Y.C. Resistance to Fusarium head blight and seedling blight in wheat is associated with activation of a cytochrome P450 gene. Phytopathology 2010, 100, 183–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fellers, J.P.; Guenzi, A.C.; Taliaferro, C.M. Factors affecting the establishment and maintenance of embryogenic callus and suspension cultures of wheat (Triticum aestivum L.). Plant Cell. Rep. 1995, 15, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Peschen, D.; Li, H.P.; Fischer, R.; Kreuzaler, F.; Liao, Y.C. Fusion proteins comprising a Fusarium-specific antibody linked to antifungal peptides protect plants against a fungal pathogen. Nat. Biotechnol. 2004, 22, 732–738. [Google Scholar] [CrossRef]
- Bernardo, A.; Bai, G.H.; Guo, P.; Guo, P.G.; Xiao, K.; Xiao, K.; Guenzi, A.C.; Ayoubi, P. Fusarium graminearum-induced changes in gene expression between Fusarium head blight-resistant and susceptible wheat cultivars. Funct. Integr. Genomics 2007, 7, 69–77. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2015, 44, D1087–D1093. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.R.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Lacadena, J.; Martínez del Pozo, A.; Gasset, M.; Patiño, B.; Campos-Olivas, R.; Vázquez, C.; Martínez-Ruiz, A.; Mancheño, J.M.; Oñaderra, M.; Gavilanes, J.G. Characterization of the antifungal protein secreted by the mould Aspergillus giganteus. Arch. Biochem. Biophys. 1995, 324, 273–281. [Google Scholar] [CrossRef]
- Li, X.; Saha, P.; Li, J.; Blobel, G.; Pfeffer, S.R. Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. Proc. Natl. Acad. Sci. USA 2016, 113, 10079–10084. [Google Scholar] [CrossRef] [Green Version]
- Kong, X.; Yang, M.; Abbas, H.M.K.; Wu, J.; Li, M.G.; Dong, W.B. Antimicrobial genes from Allium sativum and Pinellia ternata revealed by a Bacillus subtilis expression system. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Islam, S.A.-O.; Guo, P.; Hu, X.; Dong, W.A.-O. Isolation of Antimicrobial genes from oryza rufipogon griff by using a Bacillus subtilis expression system with potential antimicrobial activities. Int. J. Mol. Sci. 2020, 21, 8722. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.; Daniels, S.; Mott, E.; Hammond-Kosack, K. Arabidopsis is susceptible to the cereal ear blight fungal pathogens Fusarium graminearum and Fusarium culmorum. Plant J. 2002, 32, 961–973. [Google Scholar] [CrossRef] [Green Version]
- Van Hemelrijck, W.; Wouters, P.F.W.; Brouwer, M.; Windelinckx, A.; Goderis, I.J.W.M.; De Bolle, M.F.C.; Thomma, B.P.H.J.; Cammue, B.P.A.; Delauré, S.L. The Arabidopsis defense response mutant esa1 as a model to discover novel resistance traits against Fusarium diseases. Plant Sci. 2006, 171, 585–595. [Google Scholar] [CrossRef]
- Cheng, W.; Li, H.P.; Zhang, J.B.; Du, H.J.; Wei, Q.Y.; Huang, T.; Yang, P.; Kong, X.W.; Liao, Y.C. Tissue-specific and pathogen-inducible expression of a fusion protein containing a Fusarium-specific antibody and a fungal chitinase protects wheat against Fusarium pathogens and mycotoxins. Plant Biotechnol. J. 2015, 13, 664–674. [Google Scholar] [CrossRef]
- Kraft, M.L. Plasma membrane organization and function: Moving past lipid rafts. Mol. Biol. Cell. 2013, 24, 2765–2768. [Google Scholar] [CrossRef]
- Zhang, J.-B.; Li, H.-P.; Dang, F.-J.; Qu, B.; Xu, Y.-B.; Zhao, C.-S.; Liao, Y.-C. Determination of the trichothecene mycotoxin chemotypes and associated geographical distribution and phylogenetic species of the Fusarium graminearum clade from China. Mycol. Res. 2007, 111, 967–975. [Google Scholar] [CrossRef]
- Zelazny, E.; Borst, J.W.; Muylaert, M.; Batoko, H.; Hemminga, M.A.; Chaumont, F. FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc. Natl. Acad. Sci. USA 2007, 104, 12359–12364. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Bowra, S.; Vincze, E. The development and evaluation of single cell suspension from wheat and barley as a model system; a first step towards functional genomics application. BMC Plant Biol. 2010, 10, 239. [Google Scholar] [CrossRef] [Green Version]
- Myung, K.; Williams, D.A.; Xiong, Q.; Thornburgh, S. Metabolism of strobilurins by wheat cell suspension cultures. J. Agric. Food Chem. 2013, 61, 47–52. [Google Scholar] [CrossRef]
- Rodríguez-Serrano, M.; Bárány, I.; Prem, D.; Coronado, M.J.; Risueño, M.C.; Testillano, P.S. NO, ROS, and cell death associated with caspase-like activity increase in stress-induced microspore embryogenesis of barley. J. Exp. Bot. 2012, 63, 2007–2024. [Google Scholar] [CrossRef] [Green Version]
- Poppenberger, B.; Berthiller, F.; Lucyshyn, D.; Sieberer, T.; Schuhmacher, R.; Krska, R.; Kuchler, K.; Glössl, J.; Luschnig, C.; Adam, G. Detoxification of the Fusarium Mycotoxin Deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 47905–47914. [Google Scholar] [CrossRef] [Green Version]
- Nishiuchi, T.; Masuda, D.; Nakashita, H.; Ichimura, K.; Shinozaki, K.; Yoshida, S.; Kimura, M.; Yamaguchi, I.; Yamaguchi, K. Fusarium phytotoxin trichothecenes have an elicitor-like activity in Arabidopsis thaliana, but the activity differed significantly among their molecular species. Mol. Plant Microbe Interact. 2006, 19, 512–520. [Google Scholar] [CrossRef] [Green Version]
- Jansen, C.; von Wettstein, D.; Schäfer, W.; Kogel, K.H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woelflingseder, L.; Warth, B.; Vierheilig, I.; Schwartz-Zimmermann, H.; Hametner, C.; Nagl, V.; Novak, B.; Šarkanj, B.; Berthiller, F.; Adam, G.; et al. The Fusarium metabolite culmorin suppresses the in vitro glucuronidation of deoxynivalenol. Arch. Toxicol. 2019, 93, 1729–1743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wipfler, R.; McCormick, S.P.; Proctor, R.; Teresi, J.; Hao, G.; Ward, T.; Alexander, N.; Vaughan, M.M. Synergistic phytotoxic effects of culmorin and trichothecene mycotoxins. Toxins 2019, 11, 555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desmond, O.J.; Manners, J.M.; Stephens, A.E.; Maclean, D.J.; Schenk, P.M.; Gardiner, D.M.; Munn, A.L.; Kazan, K. The Fusarium mycotoxin deoxynivalenol elicits hydrogen peroxide production, programmed cell death and defence responses in wheat. Mol. Plant Pathol. 2008, 9, 435–445. [Google Scholar] [CrossRef] [Green Version]
- Michlmayr, H.; Malachová, A.; Varga, E.; Kleinová, J.; Lemmens, M.; Newmister, S.; Rayment, I.; Berthiller, F.; Adam, G. Biochemical characterization of a recombinant UDP-glucosyltransferase from rice and enzymatic production of deoxynivalenol-3-O-β-D-glucoside. Toxins 2015, 7, 2685–2700. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Michlmayr, H.; Schweiger, W.; Malachova, A.; Shin, S.; Huang, Y.; Dong, Y.; Wiesenberger, G.; McCormick, S.; Lemmens, M.; et al. A barley UDP-glucosyltransferase inactivates nivalenol and provides Fusarium Head Blight resistance in transgenic wheat. J. Exp. Bot. 2017, 68, 2187–2197. [Google Scholar] [CrossRef]
- Planchon, S.M.; Waite, K.A.; Eng, C. The nuclear affairs of PTEN. J. Cell Sci. 2008, 121, 249–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minaguchi, T.; Waite, K.A.; Eng, C. Nuclear localization of PTEN is regulated by Ca(2+) through a tyrosil phosphorylation-independent conformational modification in major vault protein. Cancer Res. 2006, 66, 11677–11682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bong, S.M.; Bae, S.H.; Song, B.; Gwak, H.R.; Yang, S.W.; Kim, S.; Nam, S.; Rajalingam, K.; Oh, S.J.; Kim, T.W.; et al. Regulation of mRNA export through API5 and nuclear FGF2 interaction. Nucleic Acids Res. 2021, 48, 6340–6352. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.Y.; Hong, J.K.; Kim, C.Y.; Chun, H.J.; Park, H.C.; Kim, J.C.; Yun, D.-J.; Chung, W.S.; Lee, S.-H.; Lee, S.Y.; et al. Over-expressed rice ADP-ribosylation factor 1 (RARF1) induces pathogenesis-related genes and pathogen resistance in tobacco plants. Physiol. Plant. 2003, 119, 573–581. [Google Scholar] [CrossRef]
- Gangadharan, A.; Sreerekha, M.V.; Whitehill, J.; Ham, J.H.; Mackey, D. The Pseudomonas syringae pv. tomato type III effector HopM1 suppresses Arabidopsis defenses independent of suppressing salicylic acid signaling and of targeting AtMIN7. PLoS ONE 2013, 8, e82032. [Google Scholar] [CrossRef]
- Nomura, K.; Mecey, C.; Lee, Y.N.; Imboden, L.A.; Chang, J.H.; He, S.Y. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 10774–10779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coemans, B.; Takahashi, Y.; Berberich, T.; Ito, A.; Kanzaki, H.; Matsumura, H.; Saitoh, H.; Tsuda, S.; Kamoun, S.; Sági, L.; et al. High-throughput in planta expression screening identifies an ADP-ribosylation factor (ARF1) involved in non-host resistance and R gene-mediated resistance. Mol. Plant Pathol. 2008, 9, 25–36. [Google Scholar] [CrossRef]
- Ingley, E.; Williams, J.H.; Walker, C.E.; Tsai, S.; Colley, S.; Sayer, M.S.; Tilbrook, P.A.; Sarna, M.; Beaumont, J.G.; Klinken, S.P. A novel ADP-ribosylation like factor (ARL-6), interacts with the protein-conducting channel SEC61beta subunit. FEBS Lett. 1999, 459, 69–74. [Google Scholar] [CrossRef] [Green Version]
- Nawrot, R.; Barylski, J.; Nowicki, G.; Broniarczyk, J.; Buchwald, W.; Goździcka-Józefiak, A. Plant antimicrobial peptides. Folia Microbiol. 2014, 59, 181–196. [Google Scholar] [CrossRef] [Green Version]
- Gao, A.G.; Hakimi, S.M.; Mittanck, C.A.; Wu, Y.; Woerner, B.M.; Stark, D.M.; Shah, D.M.; Liang, J.; Rommens, C.M. Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat. Biotechnol. 2000, 18, 1307–1310. [Google Scholar] [CrossRef]
- Lay, F.T.; Schirra, H.J.; Scanlon, M.J.; Anderson, M.A.; Craik, D.J. The three-dimensional solution structure of NaD1, a new floral defensin from Nicotiana alata and its application to a homology model of the crop defense protein alfAFP. J. Mol. Biol. 2003, 325, 175–188. [Google Scholar] [CrossRef]
- Krishnamurthy, K.; Balconi, C.; Sherwood, J.E.; Giroux, M.J. Wheat puroindolines enhance fungal disease resistance in transgenic rice. Mol. Plant Microbe Interact. 2001, 14, 1255–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rong, W.; Qi, L.; Wang, J.; Du, L.; Xu, H.; Wang, A.; Zhang, Z. Expression of a potato antimicrobial peptide SN1 increases resistance to take-all pathogen Gaeumannomyces graminis var. tritici in transgenic wheat. Funct. Integr. Genomics 2013, 13, 403–409. [Google Scholar] [CrossRef] [PubMed]



| Fungi | IC50 |
|---|---|
| F. graminearum 5035 | 22 ± 1.5 µM |
| A. alternata | 25 ± 2.6 µM |
| F. oxysporum | >200 µM |
| C. higginsianum | >200 µM |
| S. sclerotiorum | >200 µM |
| Genotype | Root Length (mm) | Fresh Weight (mg) | Disease (FAD) | |||||
|---|---|---|---|---|---|---|---|---|
| T2 | T3 | T2 | T3 | T2 | T3 | |||
| 14 dpi | 14 dpi | 14 dpi | 14 dpi | 7 dpi | 10 dpi | 7 dpi | 10 dpi | |
| TaArl6ip4-1 | 17.00 ± 1.63 a | 19.12 ± 2.95 a | 6.71 ± 1.27 a | 7.23 ± 1.39 a | 6.48 ± 2.17 a | 9.15 ± 1.93 a | 4.76 ± 1.73 a | 5.10 ± 2.20 a |
| TaArl6ip4-2 | 14.36 ± 2.38 a | 17.89 ± 1.69 a | 6.15 ± 1.57 a | 6.77 ± 1.23 a | 7.32 ± 1.87 a | 10.10 ± 2.31a | 5.80 ± 2.07 a | 6.20 ± 2.07 a |
| WT | 3.72 ± 0.55 | 4.40 ± 0.71 | 3.17 ± 0.74 | 4.45 ± 0.89 | 11.86 ± 2.32 | 14.74 ± 2.16 | 7.93 ± 1.02 | 9.85 ± 1.87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Zuo, D.-Y.; Yang, P.; He, W.-J.; Yang, Z.; Zhang, J.-B.; Wu, A.-B.; Yi, S.-Y.; Li, H.-P.; Huang, T.; et al. A Novel Deoxynivalenol-Activated Wheat Arl6ip4 Gene Encodes an Antifungal Peptide with Deoxynivalenol Affinity and Protects Plants against Fusarium Pathogens and Mycotoxins. J. Fungi 2021, 7, 941. https://doi.org/10.3390/jof7110941
Liu G, Zuo D-Y, Yang P, He W-J, Yang Z, Zhang J-B, Wu A-B, Yi S-Y, Li H-P, Huang T, et al. A Novel Deoxynivalenol-Activated Wheat Arl6ip4 Gene Encodes an Antifungal Peptide with Deoxynivalenol Affinity and Protects Plants against Fusarium Pathogens and Mycotoxins. Journal of Fungi. 2021; 7(11):941. https://doi.org/10.3390/jof7110941
Chicago/Turabian StyleLiu, Gang, Dong-Yun Zuo, Peng Yang, Wei-Jie He, Zheng Yang, Jing-Bo Zhang, Ai-Bo Wu, Shu-Yuan Yi, He-Ping Li, Tao Huang, and et al. 2021. "A Novel Deoxynivalenol-Activated Wheat Arl6ip4 Gene Encodes an Antifungal Peptide with Deoxynivalenol Affinity and Protects Plants against Fusarium Pathogens and Mycotoxins" Journal of Fungi 7, no. 11: 941. https://doi.org/10.3390/jof7110941






