Abstract
Coccidioidomycosis (Valley Fever) is a disease caused by species of Coccidioides. The disease is endemic to arid regions of the Southwestern US and while most common in CA and AZ is also present in NM. We present the first genetic analysis of clinical isolates from NM. Travel and demographic information was available for a number of patients, which included individuals from NM and the Southwestern US Four Corners region. Multi-gene phylogenetic analyses revealed the presence of both C. posadasii and C. immitis. While NM is predicted to be within the endemic range for C. posadasii, our results expand the known range of C. immitis, often considered to be the “California species”. Five of eight infections for which patient ethnicity existed occurred in Native Americans, and two occurred in African Americans. Several isolates came from the northwestern part of NM—outside the predicted “highly-endemic” region. Our study suggests Native Americans represent an unrecognized at-risk group, and it provides a foundation for better defining the geographic distribution of the Coccidioides species and for preventing exposure among populations at risk. In the course of this study, we developed a reliable PCR-based method to distinguish species targeting regions of the mitochondrial genome.
1. Introduction
Coccidioidomycosis, commonly known as Valley Fever, is caused by the soil-dwelling fungi Coccidioides immitis and C. posadasii. As is common for fungi that infect humans and animals, species of Coccidioides are dimorphic in terms of their life cycle, growing saprobically as multicellular filaments on non-living organic matter and, upon entry into a host lung, growing pathogenically in a yeast-like spherule stage [1]. The endospores formed by spherules can disseminate within a host but are not transmissible to new hosts. Outside a living host, species of Coccidioides form arthroconidia (asexual spores) that can become airborne via soil disturbance and can be inhaled by potential hosts [2]. Although coccidioidomycosis presents in about 40% of patients as a pulmonary infection, a chronic and disseminated form of coccidioidomycosis, often resulting in lifelong treatment, occurs in roughly 5% of patients [3,4].
Although arthroconidia can be detected in soil and in dust, the specific niches of Coccidioides species are unknown [5]. The disease has been characterized as endemic to arid environments that include the Southwestern United States, Central and South America and Mexico [6]. In the United States, highly endemic hotspots have been documented in California and Arizona [7,8,9]. Although less prevalent, Valley Fever is present in Nevada, Colorado, Texas, New Mexico, Utah and Washington state [10,11,12,13].
Molecular and phenotypic analyses have resulted in Coccidioides being divided into two species: C. immitis, known primarily from California, and C. posadasii, recognized originally as the non–California group [6]. While clinical diagnosis and treatment of coccidioidomycosis have not depended on identifying isolates to species, medical and scientific communities are becoming more cognizant of the potential need to recognize previously undetected phenotypic, morphological, ecological, and genetic differences between species. Multi-locus genetic analyses and whole-genome studies have attempted to map the distribution of these species. To date, however, the isolates studied have been mainly limited to California, Arizona, Central America, Mexico, and Texas [14,15], and the genetics of isolates from New Mexico have not been examined. The study reported here employed genetic analyses of 18 isolates collected from patients diagnosed with coccidioidomycosis from diverse locations across New Mexico and the Four Corners region. Although Southern New Mexico has been recognized as part of the endemic region for Coccidioides [12,16], several of the isolates examined here were from Northern and Central New Mexico, suggesting a broad range for Coccidioides that includes the Four Corners region. Also noteworthy is the fact that isolates of both C. immitis and C. posadasii were obtained from patients in New Mexico with C. immitis being obtained from a patient who resides in San Juan County, NM.
The risk for coccidioidomycosis has been reported as much higher among some ethnic groups, particularly African Americans and Filipinos [17]. In these ethnic groups, the risk for disseminated coccidioidomycosis is ten-fold that of the general population [18]. Health records from patients in our study suggest the possibility that Native American Indians represent an additional risk group for disseminated disease.
2. Materials and Methods
2.1. Sample Collection and Patient Information
Eighteen human clinical specimens from seventeen patients diagnosed with coccidioidomycosis obtained between 2013 and 2017 were submitted to the New Mexico Department of Health (NMDOH) Scientific Laboratory Division (SLD). Coccidioidomycosis is a reportable condition in New Mexico. The NMDOH collects information on all confirmed and probable cases (case status definitions follow CSTE standards) who reside in New Mexico at the time of diagnosis to look for potential risk factors via chart review and data extraction. When possible, patient information was noted with specific interest in type of infection, residency, travel history, occupation, race/ethnicity, gender, age, and medical history (Table 1).

Table 1.
Isolate and patient information.
2.2. Molecular Methods
DNA was extracted at the NMDOH SLD from Coccidioides isolates using the PrepMan® Ultra Reagent method (Applied Biosystems, Foster City, CA) and stored at −80 °C until further processing. The DNA extractions were transferred to the Natvig Laboratory at the University of New Mexico where a 1/10 DNA dilution of each preparation was used for PCR amplification of three AFToL (Assembling the Fungal Tree of Life)-designated nuclear gene regions in addition to a serine proteinase gene region diagnostic for species (Table 2). For the serine proteinase, MCM7, and RPB1 genes, primers were designed to amplify and sequence regions that we determined from comparisons of sequences in GenBank to be diagnostic in distinguishing between the two Coccidioides species. Serine proteinase, specifically, was chosen as a target based on results presented by Koufopanou et al. [19]. ITS amplification and sequencing involved the entire ITS1-5.8S rRNA-ITS2 region.

Table 2.
Primers used in gene amplification.
We designed a primer pair, with one primer anchored in a sequence unique to C. posadasii, to amplify a mitochondrial intron sequence from C. posadasii but not C. immitis. A second primer set was designed to amplify a portion of the first cox1 exon along with an upstream intergenic region in both Coccidioides species but with the potential to differentiate Coccidioides from other Onygenales (Table 2).
All PCR reactions began with an initial step at 95 °C for 5 min. This was followed by 35 cycles of 94 °C for 30 s, a gene-specific annealing temperature (Table 2) for 30 s, then 72 °C for 45 s with a final extension at 72 °C for 7 min. PCR products were cleaned with ExoSAP-IT (Thermo Fisher Scientific, Waltham, MA, USA) before DNA sequencing using BigDye v3.1 (Applied Biosystems, Foster City, CA, USA) chain termination with the Big Dye STeP protocol [20]. Forward and reverse sequences were assembled and edited with Sequencher 5.1 (Gene Codes, Ann Arbor, MI, USA). Sequences were deposited in GenBank under accessions MH748760–MH748777 for serine proteinase; MH748742–MH748759 for MCM7; MH748724–MH748741 for RPB1; and MH725244–MH725261 for ITS (Table 3).

Table 3.
GenBank accession numbers for sequences employed in phylogenetic analysis.
2.3. Phylogenetic Analysis
Sequences for the four gene regions were aligned individually using Clustal Omega version 1.2.45 [21]. For outgroup sequences, each alignment included the appropriate homologous gene region from Aspergillus steynii from the GenBank accessions (Table 3). This species was chosen as an outgroup because it had a clear homolog for each of the four gene regions examined. The serine proteinase in particular, appears to undergo rapid evolution and perhaps gene loss, and as a result, it was difficult to identify clear orthologs in many other species of Eurotiomycetes.
Tree-building analyses employed maximum likelihood analysis with PHYLIP (version 3.695) DNAMLK and parsimony analysis with PHYLIP DNAPARS [22]. In each case, tree building employed 1000 bootstrap datasets. Analyses were done on each of the four gene alignments separately, as well as on a concatenated alignment of all four genes. The concatenated alignment has been deposited at TreeBase (Submission ID 24737).
2.4. Ethics Statement
All patient data analyzed for this study were anonymized.
3. Results
All four gene regions examined were capable of distinguishing between the two Coccidioides species, based on comparisons with sequences reported in Genbank. Three of the isolates were revealed to be C. immitis, while the remaining 15 were C. posadasii. Isolates NM3006, NM9443, and NM9737 were clustered together as a C. immitis clade in single-locus trees and in the concatenated four-gene phylogeny (Figure 1 and Figure S1). Both NM9443 and NM9737 came from a single patient in Utah whose type of infection was unknown (Table 1). Isolate NM3006 was from a 60-year-old male from San Juan County, New Mexico, with disco-vertebral osteomyelitis due to C. immitis. Human coccidioidomycosis due to C. posadasii was represented by twelve cases of pulmonary infections (one also had a facial lesion), one of osteomyelitis, and one knee infection. The type of infection associated with the remaining patient was unknown (Table 1). Five of eight infections for which patient ethnicity was known occurred in Native Americans, while two of the eight occurred in African Americans (Table 1).
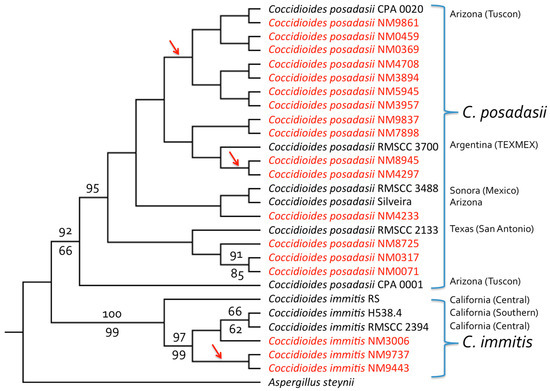
Figure 1.
Four-gene phylogeny (DNAMLK) for clinical Coccidioides isolates from New Mexico and the Four Corners region. The tree was derived from a concatenated alignment of partial sequences from four gene regions: Serine protease, MCM7, RPB1, and ITS; trees for individual genes are shown in Figure S1 (with all four genes agreeing in terms of species separation). Both C. posadasii and C. immitis isolates were present among those obtained from patients in New Mexico (shown in red). As expected, based on previous analyses of isolates from California, Arizona and Texas, most NM isolates were from C. posadasii (C. immitis being known primarily from CA). One patient infected with C. immitis (isolates NM9443 and NM9737) was a resident of the Four Corners region of Utah, while another (isolate NM3006) was from the Four Corners region of NM with no apparent travel history to California. GenBank accession numbers are given in Table 3. Bootstrap values (percentage of 1000 replicates) greater than 60% are shown for maximum likelihood analysis above the branches and for parsimony analysis below the branches. The tree was rooted with Aspergillus steynii. Maximum likelihood and parsimony analyses performed without sequences from A. steynii, and employing midpoint rooting, separated C. immitis and C. posadasii clades with 100% bootstrap support and placed the root between the two species (results not shown). Arrows indicate branches leading to isolates with identical sequences.
In addition to the four nuclear genes examined, we explored the possibility that mitochondrial DNA (mtDNA) regions might be useful in distinguishing between C. immitis and C. posadasii. In comparisons of the sequences available in GenBank, it appeared that the first intron of the cytochrome c oxidase I (cox1) gene possesses a large insertion/deletion that separates the two species. This allowed the design of PCR primers that resulted in amplification of a sequence in all 15 C. posadasii isolates but not isolates of C. immitis (Figure 2).
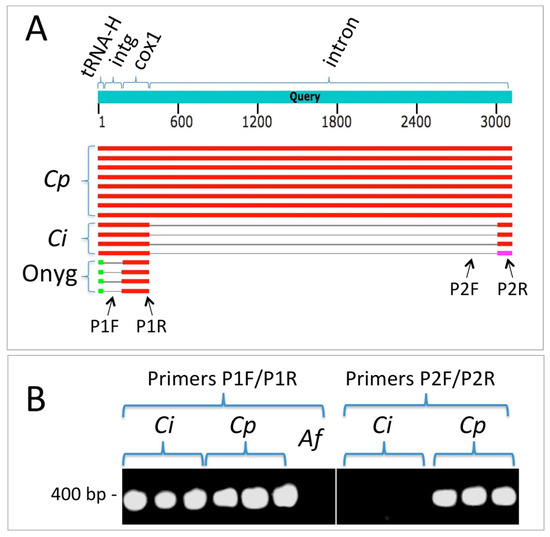
Figure 2.
An mtDNA region can be employed to distinguish between C. immitis and C. posadasii using PCR. (A) Summary results from a BLASTN search employing the cox1 mtDNA region from C. posadasii strain C735 delta SOWgp (GenBank: ACFW01000039). Cp = hits against C. posadasii sequences, Ci = hits against C. immitis, and Onyg = hits against other Onygenales (Microsporum canis CBS 113480, Trichophyton interdigitale M8436, Trichophyton mentagrophytes TIMM 2789, and Trichophyton interdigitale H6). Note that the gap in the intron of C. immitis relative to C. posadasii represents a large indel. The intergenic region (intg) between the histidine tRNA and cox1 is conserved between the two Coccidioides species but not between the latter and other Onygenales. The sequence corresponding to primer P1F is present in both Coccidioides species but not in other genera for which sequences are currently available in Genbank. The P1R sequence is contained within the first cox1 exon and is conserved in both species. The P2F primer sequence is entirely absent from C. immitis. The P2R primer sequence is wholly contained within the C. posadasii intron but spans the predicted exon–intron border of C. immitis. (B) Primer pair P1F/P1R amplifies a fragment from both C. immitis and C. posadasii (Aspergillus fumigatus, Af, included as negative control). Primer pair P2F/P2R amplifies a fragment from C. posadasii only (although this fragment was amplified in all 15 C. posadasii strains, results are shown for only three strains.
4. Discussion
The isolates employed in this study came from individuals across New Mexico, with several isolates coming from the northwestern part of the State (San Juan County, NM, USA; Table 1). This is a surprising result given that species of Coccidioides are expected to occur primarily in the southern portions of New Mexico, where the environment is similar to the Sonoran Desert regions of Arizona and California. Although 14 of the 15 patients who were infected with C. posadasii were residents of New Mexico, two reported recent travel to Arizona (Table 1). New Mexico sees far fewer cases of coccidioidomycosis than its neighbor Arizona (approximately 47 cases/yr compared to 9680 cases/yr from 2008–2014) [23]. There are three non-mutually-exclusive possible reasons for this: (1) physicians in New Mexico do not have a thorough understanding of the disease for diagnosis and treatment, (2) Coccidioides is most common in less populated areas of the state, and/or (3) the low human population density across most portions of the state means that there are fewer targets for infection and less human-caused soil disturbance on a wide scale (a known factor in generating the airborne spores that cause infections). From a Knowledge, Attitudes, and Practices Survey of New Mexican physicians in 2010, 72% were not confident in their ability to diagnose and 70% were not confident in their ability to treat coccidioidomycosis. There is a clear need for an increased understanding of disease ecology, including the environmental conditions related both to where the fungus grows and factors that aid in dispersion to better inform our healthcare professionals of the distribution in New Mexico. An increased awareness will help avoid complications from delayed diagnosis and inappropriate treatment that can result in severe disease progression and even death. This point is driven home by a news report of several recent severe cases of human coccidioidomycosis in Southern New Mexico, all of which resulted in delayed diagnosis that prevented timely treatment [24].
Native Americans and African Americans represent only 11% and 2.5% of the population of New Mexico, respectively [25], so it was surprising to see five of eight infections for which patient ethnicity was known occurred in Native Americans, and two of the eight occurred in African Americans (Table 1). The increased risk for coccidioidomycosis among African Americans is well known [26]. The risk among Native Americans is not documented in the literature. Although one study [27] found Southwest Native Americans in regions of high Coccidoides endemism (Lower Sonoran desert) to have high rates of positive response to coccidioidin skin tests, that study did not find that rates were higher than for non-Native Americans in the same region. A separate study found a possible correlation between diabetes and coccidioidomycosis-associated death among Native Americans [28]. We acknowledge that any or all of multiple factors could contribute to increased risk of coccidioidomycosis. These include living in remote rural areas with high airborne dust loads, working in agriculture or construction, as well as comorbidities such as diabetes and other health conditions. Nonetheless, our results suggest a need for health professionals in New Mexico and the Four Corners region to be aware of potential at-risk groups, as well as a need for a better understanding of locations of Coccidioides endemism.
Coccidioidomycosis incidence is on the rise in highly endemic areas such as Arizona and California and in more sparsely populated reporting regions including New Mexico, Nevada, and Utah [11]. One reason for this increase may be that clinicians are becoming more aware of the disease. Other hypotheses include changes in testing practices, increased travel or relocation to endemic areas, and/or growth of the “at-risk” immunosuppressed population (although coccidioidomycosis can infect healthy individuals). Climatic factors, such as temperature and moisture, in addition to increases in human activities such as construction that produce dust from soil disturbance, can result in increased spore dispersal [29]. Our genetic analysis of isolates collected from patients diagnosed with coccidioidomycosis in New Mexico provides a foundation for future exploration of distribution, incidence, and susceptibility of patients in New Mexico and the American Southwest Four Corners region.
Because C. immitis has been considered to be the “California species” and the vast majority of C. immitis infections occur in California, the presence of C. immitis among our isolates was unexpected. The reported range for C. immitis does, however, include locations in Washington state and Northeastern Utah [13,30]. Acknowledging that the C. immitis isolates we examined could reflect infections acquired as a result of travel outside the Four-Corners region, it is entirely possible that the range of C. immitis includes Southern Utah and/or Northern New Mexico. Related to the question of species distributions, we note that although hybridization between C. immitis and C. posadasii has been reported [31,32], the fact that all four nuclear genes and the mitochondrial region examined for our isolates agreed in terms of species separations would suggest that any introgression of genes across species would be minimal for the isolates we examined.
Our observation that C. immitis was present among isolates obtained from patients in New Mexico argues that it is important for health professionals and researchers to have rapid methods to distinguish between the two species. This is true in part because, while differences in distribution of the two species have become increasingly clear in the past decade, the ecological niche difference between the species is unknown [33,34]. Although physicians do not currently rely on speciation for diagnosis and treatment, this may well change in the future with the increasing discovery of genetic and phenotypic differences between the species. For example, C. immitis has a tendency to grow faster than C. posadasii on high-salt media [6], which suggests there may be other growth differences in physiology that affect virulence and ecology. Rapid methods to distinguish between the two Coccidioides species such as the PCR-based method we employed here with mtDNA, along with similar methods that can be used to detect species of Coccidioides in environmental DNA samples, should prove valuable in future studies. Given the reported hybridization between C. immitis and C. posadasii cited above [31,32], we acknowledge that species assignments made based on a single gene region should be viewed as tentative.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2309-608X/5/3/74/s1. Figure S1. Phylogenetic trees for individual gene regions: Serine protease, MCM7, RPB1, and ITS.
Author Contributions
P.S.H., M.I.H. (equal contributors) and D.O.N. conceived and completed the molecular analyses. P.S.H. and D.O.N. acquired funding. P.L. obtained isolates and DNA. S.M. analyzed patient histories. All authors participated in writing the manuscript.
Funding
This work was funded in part by a University of New Mexico (UNM) Graduate and Professional Student Association High Priority Grant to P.S.H. Additional funding was provided by the UNM Biology Department’s Graduate Research Allocations Committee Research Award and by the University of New Mexico’s Sevilleta LTER Summer Graduate Student Fellowship program (P.S.H.) (National Science Foundation awards DEB 1655499 and DEB 1440478). We thank the UNM Department of Biology’s Molecular Biology Facility for help with sequencing and analysis, supported by the UNM Center for Evolutionary & Theoretical Immunology (CETI) under National Institutes of Health grant P30GM110907. Data analysis was aided by computing resources of the Center for Advanced Research Computing, supported in part by the National Science Foundation.
Acknowledgments
The authors would like to thank the New Mexico Department of Health (NMDOH) State Laboratory Division, General Microbiology Section, for providing sample DNA, and Pamela Morden (NMDOH) and Terry Torres-Cruz (Penn State University) for helpful comments and insights.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nguyen, C.; Barker, B.M.; Hoover, S.; Nix, D.E.; Ampel, N.M.; Frelinger, J.A.; Orbach, M.J.; Galgiani, J.N. Recent advances in our understanding of the environmental, epidemiological, immunological, and clinical dimensions of coccidioidomycosis. Clin. Microbiol. Rev. 2013, 26, 505–525. [Google Scholar] [CrossRef] [PubMed]
- Pappagianis, D. Epidemiology of coccidioidomycosis. In Coccidioidomycosis. Current Topics in Infectious Disease; Stevens, D.A., Ed.; Springer: Boston, MA, USA, 1980; pp. 63–85. [Google Scholar]
- Galgiani, J.; Ampel, N.M.; Blair, J.; Catanzaro, A. Coccidioidomycosis. Clin. Infect. Dis. 2005, 4, 1217–1223. [Google Scholar] [CrossRef] [PubMed]
- Hector, R.F.; Laniado-Laborin, R. Coccidioidomycosis: A fungal disease of the Americas. PLoS Med. 2005, 2, e2. [Google Scholar] [CrossRef] [PubMed]
- Baptista-Rosas, R.C.; Hinojosa, A.; Riquelme, M. Ecological niche modeling of Coccidioides spp. in Western North American deserts. Ann. N. Y. Acad. Sci. 2007, 1111, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Koenig, G.L.; White, T.J.; Taylor, J.W. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 2002, 94, 73–84. [Google Scholar] [CrossRef]
- Barker, B.M.; Jewell, K.A.; Kroken, S.; Orbach, M.J. The population biology of Coccidioides: Epidemiologic implications for disease outbreaks. Ann. N. Y. Acad. Sci. 2007, 1111, 147–163. [Google Scholar] [CrossRef] [PubMed]
- Edwards, P.Q.; Palmer, C.E. Prevalence of sensitivity to coccidioidin, with special reference to specific and nonspecific reactions to coccidioidin and to histoplasmin. Dis. Chest 1957, 31, 35–60. [Google Scholar] [CrossRef]
- Ampel, N.M.; Mosley, D.G.; England, B.; Vertz, P.D.; Komatsu, K.; Hajjeh, R.A. Coccidioidomycosis in Arizona: Increase in incidence from 1990 to 1995. Clin. Infect. Dis. 1998, 27, 1528–1530. [Google Scholar] [CrossRef]
- Petersen, L.R.; Marshall, S.L.; Barton–Dickson, C.; Hajjeh, R.A.; Lindsley, M.D.; Warnock, D.W.; Panackal, A.A.; Shaffer, J.B.; Haddad, M.B.; Fisher, F.S.; et al. Coccidioidomycosis among workers at an archaeological site, northeastern Utah. Emerg. Infect. Dis. 2004, 10, 637–642. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Coccidioidomycosis—United States, 1991–1992. Morb. Mortal Wkly. Rep. 2011, 62, 217–221. [Google Scholar]
- Kirkland, T.N.; Fierer, J. Coccidioidomycosis: A reemerging infectious disease. Emerg. Infect. Dis. 1996, 3, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Litvintseva, A.P.; Marsden-Haug, N.; Hurst, S.; Hill, H.; Gade, L.; Driebe, E.M.; Ralston, C.; Roe, C.; Barker, B.M.; Goldoft, M.; et al. Valley fever: Finding new places for an old disease: Coccidioides immitis found in Washington State soil associated with recent human infection. Clin. Infect. Dis. 2014, 60, e1–e3. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Rannala, B.; Chaturvedi, V.; Taylor, J.W. Disease surveillance in recombining pathogens: Multilocus genotypes identify sources of human Coccidioides infections. Proc. Natl. Acad. Sci. USA 2002, 99, 9067–9071. [Google Scholar] [CrossRef] [PubMed]
- Engelthaler, D.M.; Roe, C.C.; Hepp, C.M.; Teixeira, M.; Driebe, E.M.; Schupp, J.M.; Gade, L.; Waddell, V.; Komatsu, K.; Arathoon, E.; et al. Local population structure and patterns of western hemisphere dispersal for Coccidioides spp., the fungal cause of valley fever. mBio 2016, 7, e00550-16. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Benedict, K.; Park, B.J.; Thompson, G.R., III. Coccidioidomycosis: Epidemiology. Clin. Epidemiol. 2013, 5, 185–197. [Google Scholar] [PubMed]
- Louie, L.; Ng, S.; Hajjeh, R.; Johnson, R.; Vugia, D.; Werner, S.B.; Talbot, R.; Klitz, W. Influence of host genetics on the severity of coccidioidomycosis. Emerg. Infect. Dis. 1999, 5, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Pappagianis, D. Epidemiology of coccidioidomycosis. Curr. Top. Med. Mycol. 1988, 2, 199–238. [Google Scholar] [PubMed]
- Koufopanou, V.; Burt, A.; Taylor, J.W. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. USA 1997, 94, 5478–5482. [Google Scholar] [CrossRef] [PubMed]
- Platt, A.R.; Woodhall, R.W.; George, A.L., Jr. Improved DNA sequencing quality and efficiency using an optimized fast cycle sequencing protocol. BioTechniques 2007, 43, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.G.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 1989, 5, 164–166. [Google Scholar]
- Shriber, J.; Conlon, K.C.; Benedict, K.; McCotter, O.Z.; Bell, J.E. Assessment of vulnerability to coccidioidomycosis in Arizona and California. Int. J. Environ. Res. Public Health 2017, 14, 680. [Google Scholar] [CrossRef] [PubMed]
- Gumprecht, B. Beware the spore: Fungus in New Mexico soils causes serious illness. Las Cruces Sun-News. 12 March 2019. Available online: https://www.lcsun-news.com/story/life/wellness/2019/03/11/soil-fungus-serious-illness-new-mexico/2711787002/ (accessed on 26 July 2019).
- US Census Bureau Quick Facts: New Mexico. Available online: https://www.census.gov/quickfacts/NM (accessed on 28 Dec 2018).
- Centers for Disease Control and Prevention (CDC). Summary of Notifiable Infectious Diseases and Conditions—United States, 2014. Morb. Mortal Wkly. Rep. 2016, 63, 1–152. [Google Scholar] [CrossRef] [PubMed]
- Sievers, M.L. Coccidioidomycosis among Southwestern American Indians. Am. Rev. Respir. Dis. 1964, 90, 920–926. [Google Scholar] [PubMed]
- Sievers, M.L.; Nelson, R.G.; Knowler, W.C.; Bennett, P.H. Impact of NIDDM on mortality and causes of death in Pima Indians. Diabetes Care 1992, 15, 1541–1549. [Google Scholar] [CrossRef] [PubMed]
- Park, B.J.; Sigel, K.; Vaz, V.; Komatsu, K.; McRill, C.; Phelan, M.; Colman, T.; Comrie, A.C.; Warnock, D.W.; Galgiani, J.N.; et al. An epidemic of coccidioidomycosis in Arizona associated with climatic changes, 1998–2001. J. Infect. Dis. 2005, 191, 1981–1987. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.M.; Carlson, E.L.; Fisher, F.S.; Pappagianis, D. Demonstration of Coccidioides immitis and Coccidioides posadasii DNA in soil samples collected from Dinosaur National Monument, Utah. Med. Mycol. 2014, 52, 610–617. [Google Scholar] [CrossRef]
- Neafsey, D.E.; Barker, B.M.; Sharpton, T.J.; Stajich, J.E.; Park, D.J.; Whiston, E.; Hung, C.-Y.; McMahan, C.; White, J.; Sykes, S.; et al. Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res. 2010, 20, 938–946. [Google Scholar] [CrossRef]
- Teixeira, M.M.; Barker, B.M. Use of population genetics to assess the ecology, evolution, and population structure of Coccidioides. Emerg. Infect. Dis. 2016, 22, 1022–1030. [Google Scholar] [CrossRef]
- Barker, B.M.; Tabor, J.A.; Shubitz, L.F.; Perrill, R.; Orbach, M.J. Detection and phylogenetic analysis of Coccidioides posadasii in Arizona soil samples. Fungal Ecol. 2012, 5, 163–176. [Google Scholar] [CrossRef]
- Taylor, J.W.; Barker, B.M. The endozoan, small-mammal reservoir hypothesis and the life cycle of Coccidioides species. Med. Mycol. 2019, 57, S16–S20. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).