Susceptibility Testing of Fungi to Antifungal Drugs
Abstract
:1. Introduction
2. Conventional Phenotypic Assays for Testing Fungal Susceptibility
2.1. Reference AFST Methods
2.2. Commercial AFST Methods
3. Nonconventional Phenotypic Assays for Testing Fungal Susceptibility
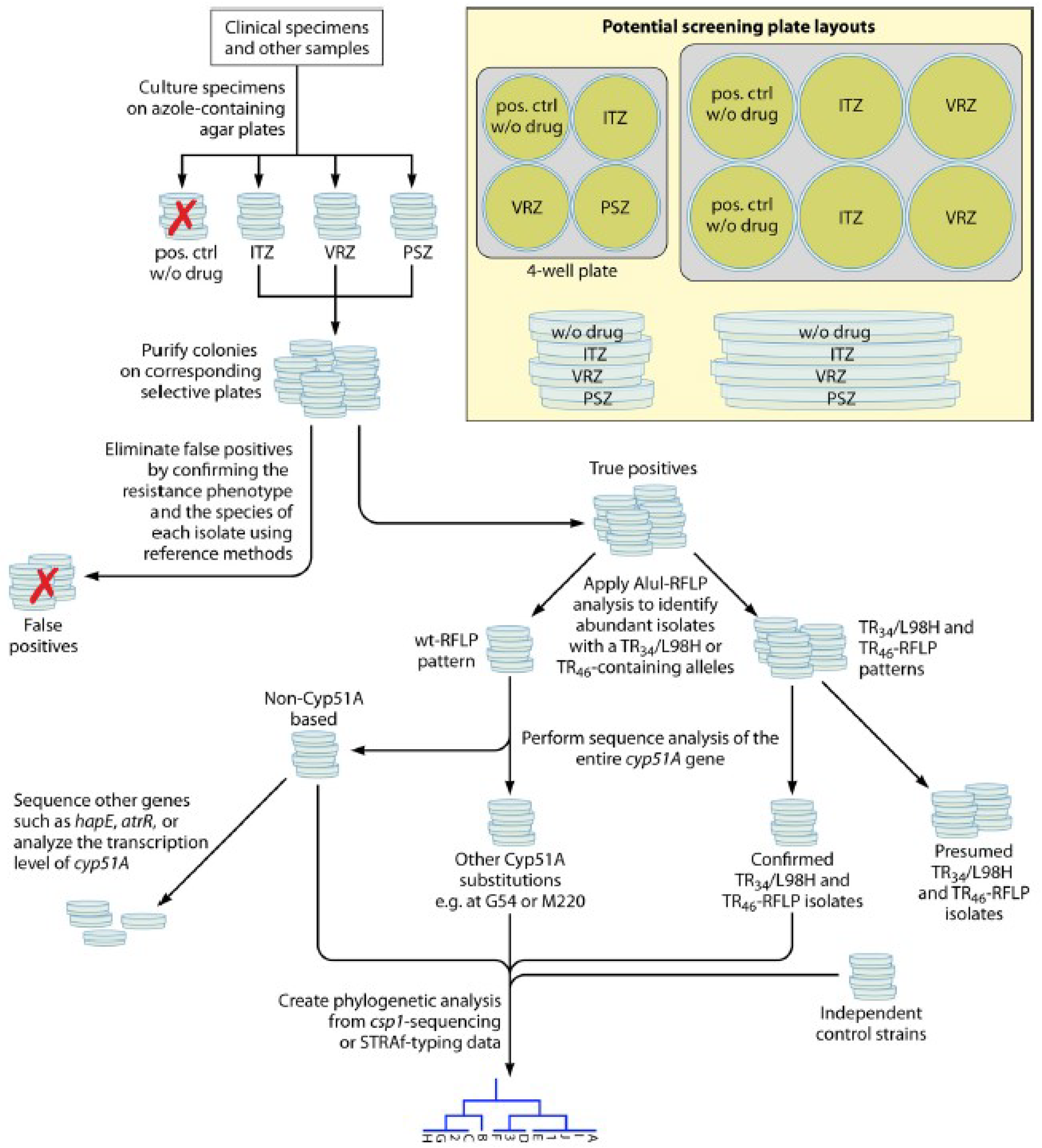
MALDI-TOF Mass Spectrometry-Based AFST Methods
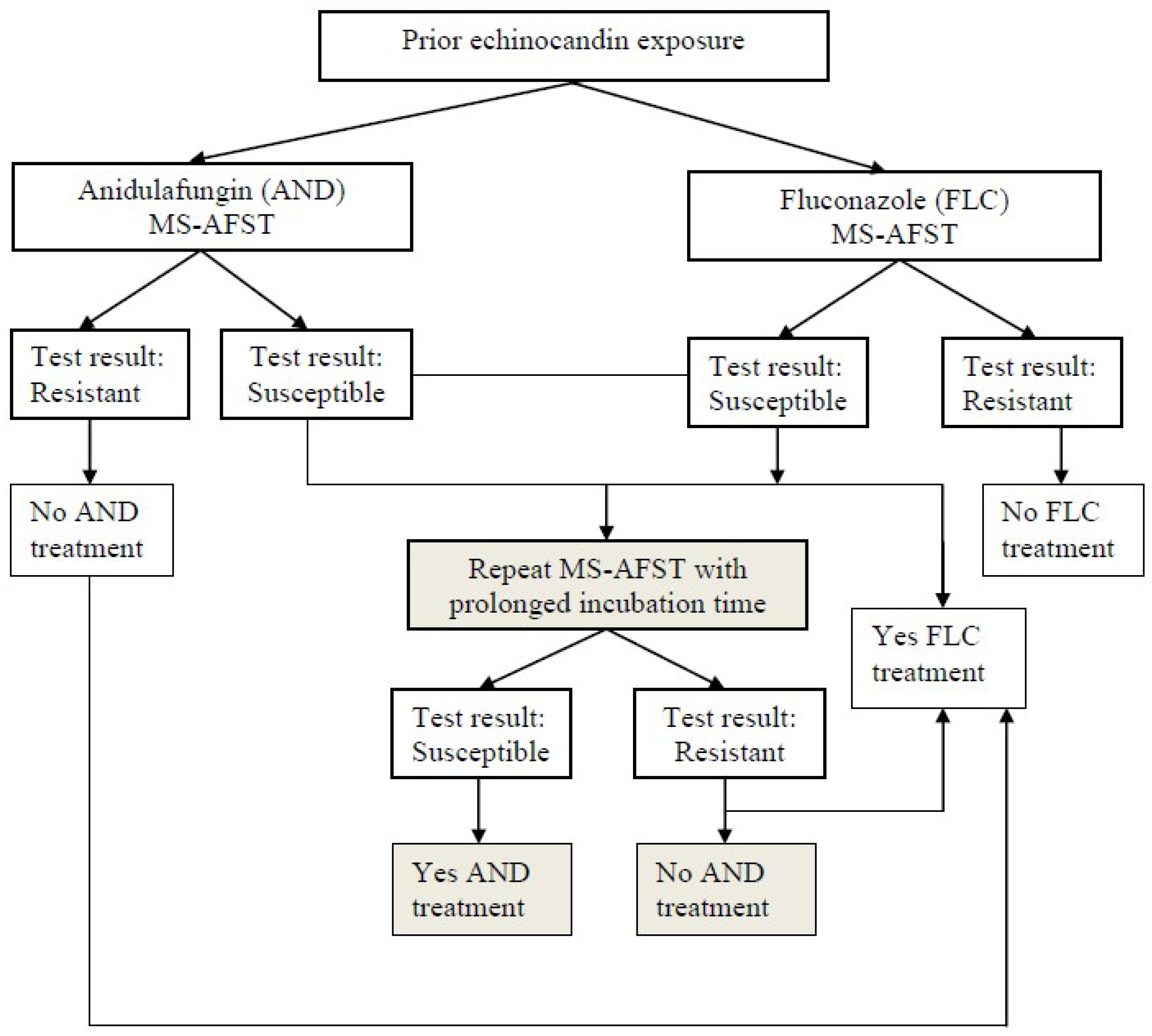
4. How to Better Use Phenotypic Fungal Susceptibility Results in the Clinic Setting
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 2018, 360, 739–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arendrup, M.C.; Patterson, T.F. Multidrug-resistant Candida: Epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 2017, 216, S445–S451. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Sharma, C.; Meis, J.F. Azole-resistant aspergillosis: Epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 2017, 216, S436–S444. [Google Scholar] [CrossRef] [PubMed]
- Pappas, P.G.; Kauffman, C.A.; Andes, D.R.; Clancy, C.J.; Marr, K.A.; Ostrosky-Zeichner, L.; Reboli, A.C.; Schuster, M.G.; Vazquez, J.A.; Walsh, T.J.; et al. Executive summary: Clinical practice guideline for the management of candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 62, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Patterson, T.F.; Thompson, G.R., 3rd; Denning, D.W.; Fishman, J.A.; Hadley, S.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Morrison, V.A.; Nguyen, M.H.; et al. Practice guidelines for the diagnosis and management of aspergillosis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 63, e1–e60. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.; Wright, G.D.; Cowen, L.E. Antifungal drugs: The current armamentarium and development of new agents. Microbiol. Spectr. 2016, 4. [Google Scholar]
- McCarthy, M.W.; Kontoyiannis, D.P.; Cornely, O.A.; Perfect, J.R.; Walsh, T.J. Novel agents and drug targets to meet the challenges of resistant fungi. J. Infect. Dis. 2017, 216, S474–S483. [Google Scholar] [CrossRef] [PubMed]
- Ostrosky-Zeichner, L.; Andes, D. The role of in vitro susceptibility testing in the management of Candida and Aspergillus. J. Infect. Dis. 2017, 216, S452–S457. [Google Scholar] [CrossRef] [PubMed]
- Wootton, M.; MacGowan, A.P.; Howe, R.A. Towards better antimicrobial susceptibility testing: Impact of the Journal of Antimicrobial Chemotherapy. J. Antimicrob. Chemother. 2017, 72, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Rex, J.H.; Pfaller, M.A. Has antifungal susceptibility testing come of age? Clin. Infect. Dis. 2002, 35, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. M27-A3: Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard—3rd ed.; CLSI: Wayne, PA, USA, 2008. [Google Scholar]
- Clinical and Laboratory Standards Institute. M38-A2: Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi; Approved Standard—2nd ed.; CLSI: Wayne, PA, USA, 2008. [Google Scholar]
- Arendrup, M.C.; Cuenca-Estrella, M.; Lass-Flörl, C.; Hope, W. EUCAST-AFST. EUCAST technical note on the EUCAST definitive document EDef 7.2: Method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts EDef 7.2 (EUCAST-AFST). Clin. Microbiol. Infect. 2012, 18, E246–E247. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C.; Guinea, J.; Cuenca-Estrella, M.; Meletiadis, J.; Mouton, J.W.; Lagrou, K.; Howard, S.J. Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST). In EUCAST Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Conidia Forming Moulds Version 9.3; EUCAST: Växjö, Sweden, 2015. [Google Scholar]
- Posteraro, B.; Sanguinetti, M. The future of fungal susceptibility testing. Future Microbiol. 2014, 9, 947–967. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; Torelli, R.; De Carolis, E.; Posteraro, P.; Sanguinetti, M. Antifungal susceptibility testing: Current role from the clinical laboratory perspective. Mediterr. J. Hematol. Infect. Dis. 2014, 6, e2014030. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Turnidge, J. The role of epidemiological cutoff values (ECVs/ECOFFs) in antifungal susceptibility testing and interpretation for uncommon yeasts and moulds. Rev. Iberoam. Micol. 2016, 33, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Espinel-Ingroff, A.; Boyken, L.; Hollis, R.J.; Kroeger, J.; Messer, S.A.; Tendolkar, S.; Diekema, D.J. Comparison of the broth microdilution (BMD) method of the European Committee on Antimicrobial Susceptibility Testing with the 24-hour CLSI BMD method for testing susceptibility of Candida species to fluconazole, posaconazole, and voriconazole by use of epidemiological cutoff values. J. Clin. Microbiol. 2011, 49, 845–850. [Google Scholar] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J. Clin. Microbiol. 2012, 50, 2846–2856. [Google Scholar] [CrossRef] [PubMed]
- EUCAST. Antifungal agents. Breakpoint Tables for Interpretation of MICs. 2018. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Clinical_breakpoints/Antifungal_breakpoints_v_9.0_180212.pdf (accessed on 13 July 2018).
- Clinical and Laboratory Standards Institute. M60: Performance Standards for Antifungal Susceptibility Testing of Yeasts; Supplement—1st ed.; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- Clinical and Laboratory Standards Institute. M59: Epidemiological Cutoff Values for Antifungal Susceptibility Testing of Yeasts; Supplement—2nd ed.; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Espinel-Ingroff, A.; Arendrup, M.C.; Pfaller, M.A.; Bonfietti, L.X.; Bustamante, B.; Canton, E.; Chryssanthou, E.; Cuenca-Estrella, M.; Dannaoui, E.; Fothergill, A.; et al. Interlaboratory variability of caspofungin MICs for Candida spp. Using CLSI and EUCAST methods: Should the clinical laboratory be testing this agent? Antimicrob. Agents Chemother. 2013, 57, 5836–5842. [Google Scholar] [CrossRef] [PubMed]
- Cuenca-Estrella, M.; Gomez-Lopez, A.; Alastruey-Izquierdo, A.; Bernal-Martinez, L.; Cuest, I.; Buitrago, M.J.; Rodriguez-Tudela, J.L. Comparison of the Vitek 2 antifungal susceptibility system with the clinical and laboratory standards institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) broth microdilution reference methods and with the Sensititre YeastOne and Etest techniques for in vitro detection of antifungal resistance in yeast isolates. J. Clin Microbiol. 2010, 48, 1782–1786. [Google Scholar] [PubMed]
- Pfaller, M.A.; Chaturvedi, V.; Diekema, D.J.; Ghannoum, M.A.; Holliday, N.M.; Killian, S.B.; Knapp, C.C.; Messer, S.A.; Miskou, A.; Ramani, R. Comparison of the Sensititre YeastOne colorimetric antifungal panel with CLSI microdilution for antifungal susceptibility testing of the echinocandins against Candida spp., using new clinical breakpoints and epidemiological cutoff values. Diagn. Microbiol. Infect. Dis. 2012, 73, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Castanheira, M.; Messer, S.A.; Rhomberg, P.R.; Jones, R.N. Comparison of EUCAST and CLSI broth microdilution methods for the susceptibility testing of 10 systemically active antifungal agents when tested against Candida spp. Diagn. Microbiol. Infect. Dis. 2014, 79, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Eschenauer, G.A.; Nguyen, M.H.; Shoham, S.; Vazquez, J.A.; Morris, A.J.; Pasculle, W.A.; Kubin, C.J.; Klinker, K.P.; Carver, P.L.; Hanson, K.E.; et al. Real-world experience with echinocandin MICs against Candida species in a multicenter study of hospitals that routinely perform susceptibility testing of bloodstream isolates. Antimicrob. Agents Chemother. 2014, 58, 1897–1906. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; Spanu, T.; Fiori, B.; De Maio, F.; De Carolis, E.; Giaquinto, A.; Prete, V.; De Angelis, G.; Torelli, R.; D’Inzeo, T.; et al. Antifungal susceptibility profiles of bloodstream yeast isolates by Sensititre YeastOne over nine years at a large Italian teaching hospital. Antimicrob. Agents Chemother. 2015, 59, 3944–3955. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Fan, X.; Chen, S.C.; Wang, H.; Sun, Z.Y.; Liao, K.; Chen, S.L.; Yan, Y.; Kang, M.; Hu, Z.D.; et al. Antifungal susceptibilities of Candida glabrata species complex, Candida krusei, Candida parapsilosis species complex and Candida tropicalis causing invasive candidiasis in China: 3 year national surveillance. J. Antimicrob. Chemother. 2015, 70, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Cornely, O.A.; Bassetti, M.; Calandra, T.; Garbino, J.; Kullberg, B.J.; Lortholary, O.; Meersseman, W.; Akova, M.; Arendrup, M.C.; Arikan-Akdagli, S.; et al. ESCMID* guideline for the diagnosis and management of Candida diseases 2012: Non-neutropenic adult patients. Clin. Microbiol. Infect. 2012, 18, 19e37. [Google Scholar]
- Ullmann, A.J.; Akova, M.; Herbrecht, R.; Viscoli, C.; Arendrup, M.C.; Arikan-Akdagli, S.; Bassetti, M.; Bille, J.; Calandra, T.; Castagnola, E.; et al. ESCMID* guideline for the diagnosis and management of Candida diseases 2012: Adults with haematological malignancies and after haematopoietic stem cell transplantation (HCT). Clin. Microbiol. Infect. 2012, 18, 53e67. [Google Scholar]
- Perlin, D.S. Echinocandin resistance in Candida. Clin. Infect. Dis. 2015, 61, S612–S617. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Messer, S.A.; Diekema, D.J.; Jones, R.N.; Castanheira, M. Use of micafungin as a surrogate marker to predict susceptibility and resistance to caspofungin among 3,764 clinical isolates of Candida by use of CLSI methods and interpretive criteria. J. Clin. Microbiol. 2014, 52, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J.; Jones, R.N.; Castanheira, M. Use of anidulafungin as a surrogate marker to predict susceptibility and resistance to caspofungin among 4,290 clinical isolates of Candida by using CLSI methods and interpretive criteria. J. Clin. Microbiol. 2014, 52, 3223–3229. [Google Scholar] [CrossRef] [PubMed]
- Cantón, E.; Pemán, J.; Hervás, D.; Iñiguez, C.; Navarro, D.; Echeverría, J.; Martínez-Alarcón, J.; Fontanals, D.; Gomila-Sard, B.; Buendía, B.; et al. Comparison of three statistical methods for establishing tentative wild-type population and epidemiological cutoff values for echinocandins, amphotericin B, flucytosine, and six Candida species as determined by the colorimetric Sensititre YeastOne method. J. Clin. Microbiol. 2012, 50, 3921–3926. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Alvarez-Fernandez, M.; Cantón, E.; Carver, P.L.; Chen, S.C.; Eschenauer, G.; Getsinger, D.L.; Gonzalez, G.M.; Govender, N.P.; Grancini, A.; et al. Multicenter study of epidemiological cutoff values and detection of resistance in Candida spp. to anidulafungin, caspofungin, and micafungin using the Sensititre YeastOne colorimetric method. Antimicrob. Agents Chemother. 2015, 59, 6725–6732. [Google Scholar] [CrossRef] [PubMed]
- Kritikos, A.; Neofytos, D.; Khanna, N.; Schreiber, P.W.; Boggian, K.; Bille, J.; Schrenzel, J.; Mühlethaler, K.; Zbinden, R.; Bruderer, T.; et al. Accuracy of Sensititre YeastOne echinocandins epidemiological cut-off values for identification of FKS mutant Candida albicans and Candida glabrata: A ten year national survey of the Fungal Infection Network of Switzerland (FUNGINOS). Clin. Microbiol. Infect. 2018. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Zimbeck, A.J.; Baddley, J.W.; Marr, K.A.; Andes, D.R.; Walsh, T.J.; Kauffman, C.A.; Kontoyiannis, D.P.; Ito, J.I.; Pappas, P.G.; et al. In vitro echinocandin susceptibility of Aspergillus isolates from patients enrolled in the Transplant-Associated Infection Surveillance Network. Antimicrob. Agents Chemother. 2011, 55, 3944–3946. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, R.E.; Souteropoulos, P.; Park, S.; Perlin, D.S. Characterization of Aspergillus fumigatus mutants with reduced susceptibility to caspofungin. Med. Mycol. 2005, 43, S299–S305. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Ortigosa, C.; Moore, C.; Denning, D.W.; Perlin, D.S. Emergence of echinocandin resistance due to a point mutation in the fks1 gene of Aspergillus fumigatus in a patient with chronic pulmonary aspergillosis. Antimicrob. Agents Chemother. 2017, 61, e01277-17. [Google Scholar]
- Siopi, M.; Pournaras, S.; Meletiadis, J. Comparative evaluation of Sensititre YeastOne and CLSI M38-A2 reference method for antifungal susceptibility testing of Aspergillus spp. against echinocandins. J. Clin. Microbiol. 2017, 55, 1714–1719. [Google Scholar] [CrossRef] [PubMed]
- Verweij, P.E.; Chowdhary, A.; Melchers, W.J.; Meis, J.F. Azole resistance in Aspergillus fumigatus: Can we retain the clinical use of mold-active antifungal azoles? Clin. Infect. Dis. 2016, 62, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Diekema, D.J.; Fothergill, A.; Johnson, E.; Pelaez, T.; Pfaller, M.A.; Rinaldi, M.G.; Canton, E.; Turnidge, J. Wild-type MIC distributions and epidemiological cutoff values for the triazoles and six Aspergillus spp. for the CLSI broth microdilution method (M38-A2 document). J. Clin. Microbiol. 2010, 48, 3251–3257. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Chowdhary, A.; Gonzalez, G.M.; Lass-Flörl, C.; Martin-Mazuelos, E.; Meis, J.; Peláez, T.; Pfaller, M.A.; Turnidge, J. Multicenter study of isavuconazole MIC distributions and epidemiological cutoff values for Aspergillus spp. for the CLSI M38-A2 broth microdilution method. Antimicrob. Agents Chemother. 2013, 57, 3823–3828. [Google Scholar] [CrossRef] [PubMed]
- Dudakova, A.; Spiess, B.; Tangwattanachuleeporn, M.; Sasse, C.; Buchheidt, D.; Weig, M.; Groß, U.; Bader, O. Molecular tools for the detection and deduction of azole antifungal drug resistance phenotypes in Aspergillus species. Clin. Microbiol. Rev. 2017, 30, 1065–1091. [Google Scholar] [CrossRef] [PubMed]
- Snelders, E.; van der Lee, H.A.; Kuijpers, J.; Rijs, A.J.; Varga, J.; Samson, R.A.; Mellado, E.; Donders, A.R.; Melchers, W.J.; Verweij, P.E. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 2008, 5, e219. [Google Scholar] [CrossRef] [PubMed]
- Mello, E.; Posteraro, B.; Vella, A.; De Carolis, E.; Torelli, R.; D’Inzeo, T.; Verweij, P.E.; Sanguinetti, M. Susceptibility testing of common and uncommon Aspergillus species against posaconazole and other mold-active antifungal azoles using the Sensititre method. Antimicrob. Agents Chemother. 2017, 61, e00168-17. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C.; Verweij, P.; Nielsen, H.V. Evaluation of MIC strip isavuconazole test for susceptibility testing of wild-type and non-wild-type Aspergillus fumigatus isolates. Antimicrob. Agents Chemother. 2017, 61, e01659-16. [Google Scholar] [CrossRef] [PubMed]
- Idelevich, E.A.; Groß, U.; Becker, K.; Bader, O. Comparative evaluation of different gradient diffusion tests for detection of azole resistance in Aspergillus fumigatus. Diagn. Microbiol. Infect. Dis. 2018, 91, 52–54. [Google Scholar] [CrossRef] [PubMed]
- Guinea, J.; Peláez, T.; Recio, S.; Torres-Narbona, M.; Bouza, E. In vitro antifungal activities of isavuconazole (BAL4815), voriconazole, and fluconazole against 1,007 isolates of zygomycete, Candida, Aspergillus, Fusarium, and Scedosporium species. Antimicrob. Agents Chemother. 2008, 52, 1396–1400. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C.; Meletiadis, J.; Mouton, J.W.; Guinea, J.; Cuenca-Estrella, M.; Lagrou, K.; Howard, S.J.; Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST). EUCAST technical note on isavuconazole breakpoints for Aspergillus, itraconazole breakpoints for Candida and updates for the antifungal susceptibility testing method documents. Clin. Microbiol. Infect. 2016, 22, 571.e1–571.e4. [Google Scholar]
- Bougnoux, M.E.; Dannaoui, E.; Accoceberry, I.; Angoulvant, A.; Bailly, E.; Botterel, F.; Chevrier, S.; Chouaki, T.; Cornet, M.; Dalle, F.; et al. Multicenter comparison of the Etest and EUCAST methods for antifungal susceptibility testing of Candida isolates to micafungin. Antimicrob. Agents Chemother. 2016, 60, 5088–5091. [Google Scholar] [CrossRef] [PubMed]
- Aigner, M.; Erbeznik, T.; Gschwentner, M.; Lass-Flörl, C. Etest and Sensititre YeastOne susceptibility testing of echinocandins against Candida species from a single center in Austria. Antimicrob. Agents Chemother. 2017, 61, e00512-17. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.E.; Kaleta, E.J.; Arora, A.; Wolk, D.M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: A fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 2013, 26, 547–603. [Google Scholar] [CrossRef] [PubMed]
- Patel, R. Matrix-assisted laser desorption ionization-time of flight mass spectrometry in clinical microbiology. Clin. Infect. Dis. 2013, 57, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.; Posteraro, B. Mass spectrometry applications in microbiology beyond microbe identification: Progress and potential. Expert Rev. Proteomics 2016, 13, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.E.; Rolain, J.M.; Raoult, D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; De Carolis, E.; Vella, A.; Sanguinetti, M. MALDI-TOF mass spectrometry in the clinical mycology laboratory: Identification of fungi and beyond. Expert Rev. Proteomics 2013, 10, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.; Posteraro, B. Identification of molds by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2017, 55, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Fiori, B.; D’Inzeo, T.; Giaquinto, A.; Menchinelli, G.; Liotti, F.M.; de Maio, F.; De Angelis, G.; Quaranta, G.; Nagel, D.; Tumbarello, M.; et al. Optimized use of the MALDI BioTyper system and the FilmArray BCID panel for direct identification of microbial pathogens from positive blood cultures. J. Clin. Microbiol. 2016, 54, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; Efremov, L.; Leoncini, E.; Amore, R.; Posteraro, P.; Ricciardi, W.; Sanguinetti, M. Are the conventional commercial yeast identification methods still helpful in the era of new clinical microbiology diagnostics? A meta-analysis of their accuracy. J. Clin. Microbiol. 2015, 53, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Arvanitis, M.; Anagnostou, T.; Fuchs, B.B.; Caliendo, A.M.; Mylonakis, E. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clin. Microbiol. Rev. 2014, 27, 490–526. [Google Scholar] [CrossRef] [PubMed]
- Sparbier, K.; Schubert, S.; Kostrzewa, M. MBT-ASTRA: A suitable tool for fast antibiotic susceptibility testing? Methods 2016, 104, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.; Posteraro, B. New approaches for antifungal susceptibility testing. Clin. Microbiol. Infect. 2017, 23, 931–934. [Google Scholar] [CrossRef] [PubMed]
- Marinach, C.; Alanio, A.; Palous, M.; Kwasek, S.; Fekkar, A.; Brossas, J.Y.; Brun, S.; Snounou, G.; Hennequin, C.; Sanglard, D.; et al. MALDI-TOF MS-based drug susceptibility testing of pathogens: The example of Candida albicans and fluconazole. Proteomics 2009, 9, 4627–4631. [Google Scholar] [CrossRef] [PubMed]
- De Carolis, E.; Vella, A.; Florio, A.R.; Posteraro, P.; Perlin, D.S.; Sanguinetti, M.; Posteraro, B. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for caspofungin susceptibility testing of Candida and Aspergillus species. J. Clin. Microbiol. 2012, 50, 2479e83. [Google Scholar] [CrossRef] [PubMed]
- Vella, A.; De Carolis, E.; Vaccaro, L.; Posteraro, P.; Perlin, D.S.; Kostrzewa, M.; Posteraro, B.; Sanguinetti, M. Rapid antifungal susceptibility testing by matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis. J. Clin. Microbiol. 2013, 51, 2964e9. [Google Scholar] [CrossRef] [PubMed]
- Vella, A.; De Carolis, E.; Mello, E.; Perlin, D.S.; Sanglard, D.; Sanguinetti, M.; Posteraro, B. Potential use of MALDI-ToF mass spectrometry for rapid detection of antifungal resistance in the human pathogen Candida glabrata. Sci. Rep. 2017, 7, 9099. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.E.; Sanglard, D.; Howard, S.J.; Rogers, P.D.; Perlin, D.S. Mechanisms of antifungal drug resistance. Cold Spring Harb. Perspect. Med. 2014, 5, a019752. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.; Posteraro, B.; Fiori, B.; Ranno, S.; Torelli, R.; Fadda, G. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob. Agents Chemother. 2005, 49, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Ischer, F.; Calabrese, D.; Posteraro, B.; Sanguinetti, M.; Fadda, G.; Rohde, B.; Bauser, C.; Bader, O.; Sanglard, D. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 2009, 5, e1000268. [Google Scholar] [CrossRef] [PubMed]
- Gitman, M.R.; McTaggart, L.; Spinato, J.; Poopalarajah, R.; Lister, E.; Husain, S.; Kus, J.V. Antifungal susceptibility testing of Aspergillus spp. by using a composite correlation index (CCI)-based matrix-assisted laser desorption ionization-time of flight mass spectrometry method appears to not offer benefit over traditional broth microdilution testing. J. Clin. Microbiol. 2017, 55, 2030–2034. [Google Scholar] [PubMed]
- Saracli, M.A.; Fothergill, A.W.; Sutton, D.A.; Wiederhold, N.P. Detection of triazole resistance among Candida species by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS). Med. Mycol. 2015, 53, 736e42. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef]
- Kontoyiannis, D.P. Antifungal resistance: An emerging reality and a global challenge. J. Infect. Dis. 2017, 216, S431–S435. [Google Scholar] [CrossRef] [PubMed]
- Farmakiotis, D.; Kontoyiannis, D.P. Epidemiology of antifungal resistance in human pathogenic yeasts: Current viewpoint and practical recommendations for management. Int. J. Antimicrob. Agents 2017, 50, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Zhao, Y.; Perez, W.B.; Lockhart, S.R.; Sobel, J.D.; Farmakiotis, D.; Kontoyiannis, D.P.; Sanglard, D.; Taj-Aldeen, S.J.; Alexander, B.D.; et al. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat. Commun. 2016, 7, 11128. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Verweij, P.E.; Ananda-Rajah, M.; Andes, D.; Arendrup, M.C.; Brüggemann, R.J.; Chowdhary, A.; Cornely, O.A.; Denning, D.W.; Groll, A.H.; Izumikawa, K.; et al. International expert opinion on the management of infection caused by azole-resistant Aspergillus fumigatus. Drug Resist. Updat. 2015, 21–22, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Ullmann, A.J.; Aguado, J.M.; Arikan-Akdagli, S.; Denning, D.W.; Groll, A.H.; Lagrou, K.; Lass-Flörl, C.; Lewis, R.E.; Munoz, P.; Verweij, P.E.; et al. Diagnosis and management of Aspergillus diseases: Executive summary of the 2017 ESCMID-ECMM-ERS guideline. Clin. Microbiol. Infect. 2018, 24, Se1–Se38. [Google Scholar] [CrossRef] [PubMed]
- Buil, J.B.; van der Lee, H.A.L.; Rijs, A.J.M.M.; Zoll, J.; Hovestadt, J.A.M.F.; Melchers, W.J.G.; Verweij, P.E. Single-center evaluation of an agar-based screening for azole resistance in Aspergillus fumigatus by using VIPcheck. Antimicrob. Agents Chemother. 2017, 61, e01250-17. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A. Antifungal drug resistance: Mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 2012, 125, S3–S13. [Google Scholar] [CrossRef] [PubMed]
- Rueda, C.; Puig-Asensio, M.; Guinea, J.; Almirante, B.; Cuenca-Estrella, M.; Zaragoza, O. CANDIPOP Project from GEIH-GEMICOMED (SEIMC) and REIPI. Evaluation of the possible influence of trailing and paradoxical effects on the clinical outcome of patients with candidemia. Clin. Microbiol. Infect. 2017, 23, 49.e1–49.e8. [Google Scholar] [CrossRef] [PubMed]
- Chong, G.M.; van der Beek, M.T.; von dem Borne, P.A.; Boelens, J.; Steel, E.; Kampinga, G.A.; Span, L.F.; Lagrou, K.; Maertens, J.A.; Dingemans, G.J.; et al. PCR-based detection of Aspergillus fumigatus Cyp51A mutations on bronchoalveolar lavage: A multicentre validation of the AsperGenius assay® in 201 patients with haematological disease suspected for invasive aspergillosis. J. Antimicrob. Chemother. 2016, 71, 3528–3535. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Garnaud, C.; Brenier-Pinchart, M.P.; Thiébaut-Bertrand, A.; Saint-Raymond, C.; Camara, B.; Hamidfar, R.; Cognet, O.; Maubon, D.; Cornet, M.; et al. Direct molecular diagnosis of aspergillosis and CYP51A profiling from respiratory samples of French patients. Front. Microbiol. 2016, 7, 1164. [Google Scholar] [CrossRef] [PubMed]
- Doern, C.D. The slow march toward rapid phenotypic antimicrobial susceptibility testing: Are we there yet? J. Clin. Microbiol. 2018, 56, e01999-17. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, X.; Zhao, W. Emerging microtechnologies and automated systems for rapid bacterial identification and antibiotic susceptibility testing. SLAS Technol. 2017, 22, 585–608. [Google Scholar] [CrossRef] [PubMed]
- Pancholi, P.; Carroll, K.C.; Buchan, B.W.; Chan, R.C.; Dhiman, N.; Ford, B.; Granato, P.A.; Harrington, A.T.; Hernandez, D.R.; Humphries, R.M.; et al. Multicenter evaluation of the Accelerate PhenoTest BC kit for rapid identification and phenotypic antimicrobial susceptibility testing using morphokinetic cellular analysis. J. Clin. Microbiol. 2018, 56, e01329-17. [Google Scholar] [CrossRef] [PubMed]
- Idelevich, E.A.; Sparbier, K.; Kostrzewa, M.; Becker, K. Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin. Microbiol. Infect. 2018, 24, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W.; Perlin, D.S.; Muldoon, E.G.; Colombo, A.L.; Chakrabarti, A.; Richardson, M.D.; Sorrell, T.C. Delivering on antimicrobial resistance agenda not possible without improving fungal diagnostic capabilities. Emerg. Infect. Dis. 2017, 23, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Lamoth, F.; Chung, S.J.; Damonti, L.; Alexander, B.D. Changing epidemiology of invasive mold infections in patients receiving azole prophylaxis. Clin. Infect. Dis. 2017, 64, 1619–1621. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanguinetti, M.; Posteraro, B. Susceptibility Testing of Fungi to Antifungal Drugs. J. Fungi 2018, 4, 110. https://doi.org/10.3390/jof4030110
Sanguinetti M, Posteraro B. Susceptibility Testing of Fungi to Antifungal Drugs. Journal of Fungi. 2018; 4(3):110. https://doi.org/10.3390/jof4030110
Chicago/Turabian StyleSanguinetti, Maurizio, and Brunella Posteraro. 2018. "Susceptibility Testing of Fungi to Antifungal Drugs" Journal of Fungi 4, no. 3: 110. https://doi.org/10.3390/jof4030110





