A Biotechnological Approach for the Production of Pharmaceutically Active Human Interferon-α from Raphanus sativus L. Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
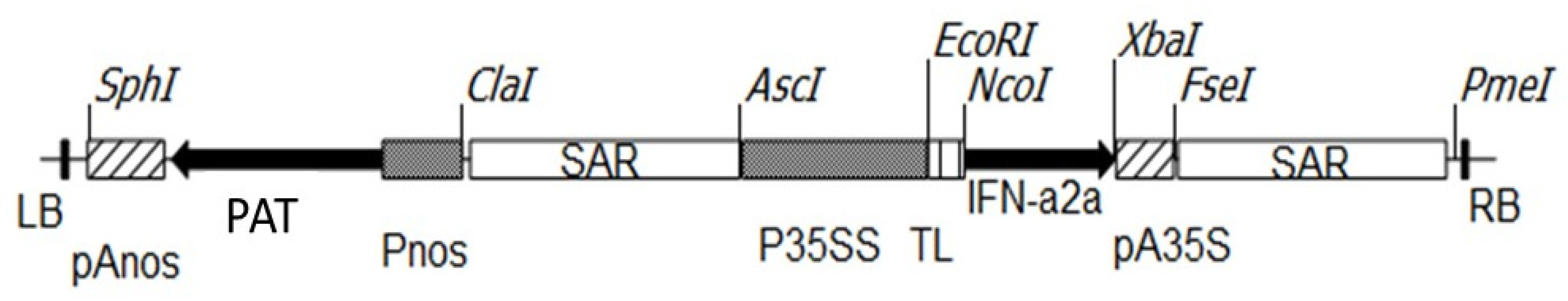
2.2. Plasmid Construct
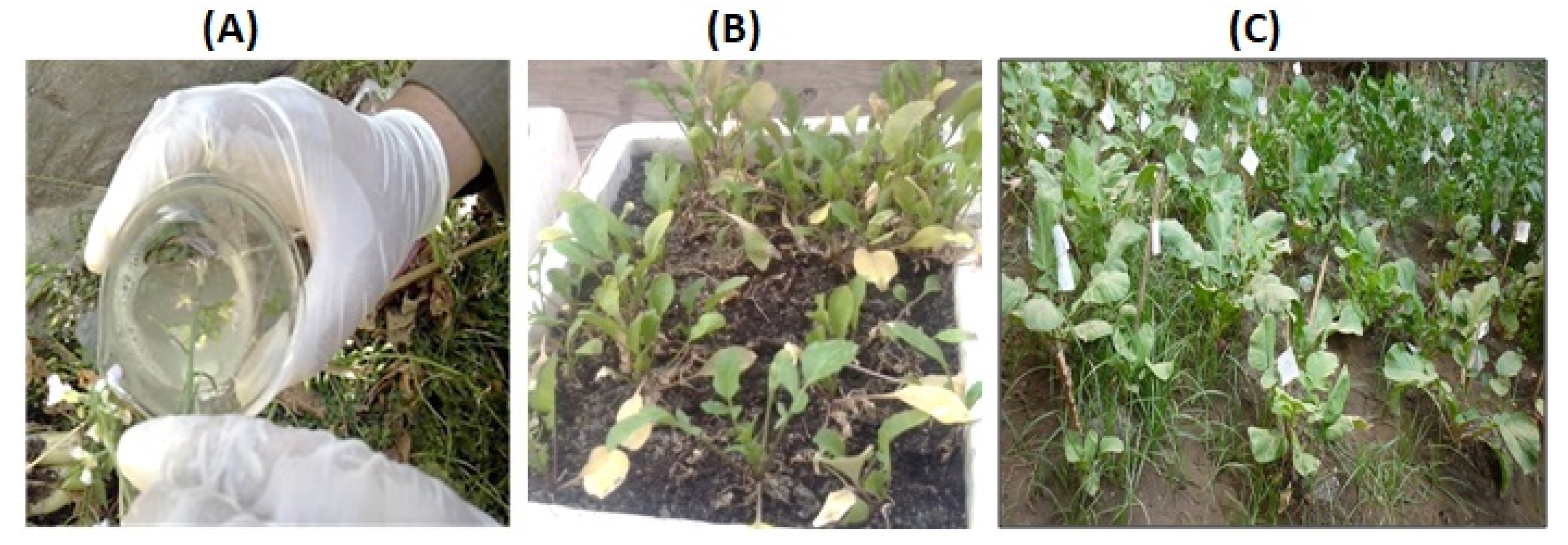
2.3. Transformation and Selection of Transgenic Raphanus sativus L.
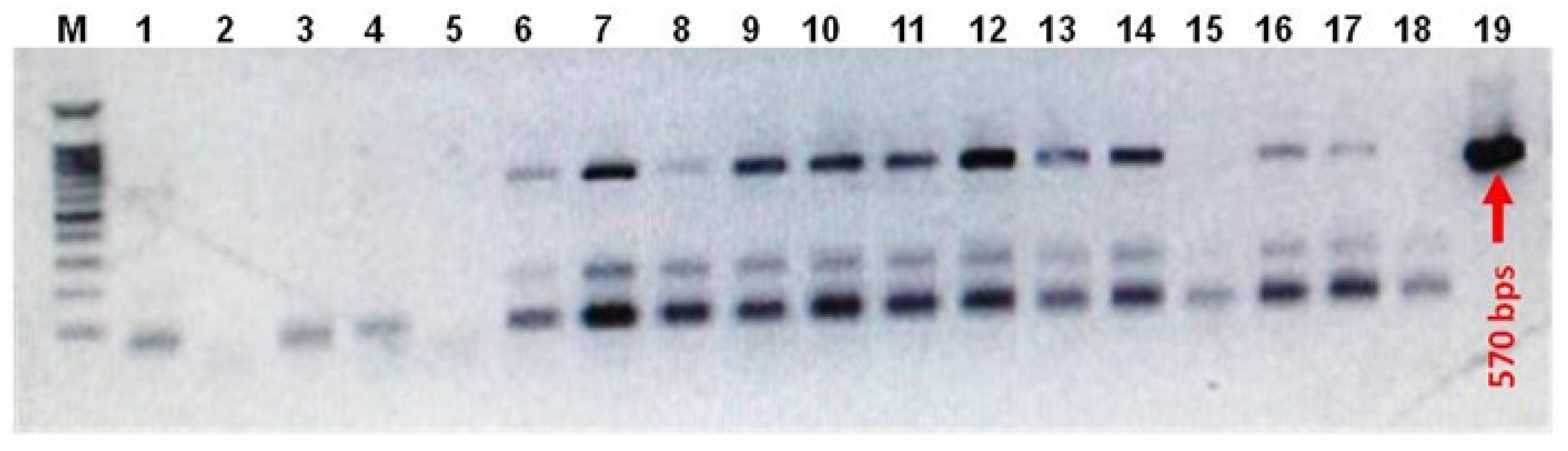
2.4. Real-Time RT-PCR
2.5. IFN-α2a Protein Extraction and Partial Purification
2.6. Enzyme-Linked Immunosorbent Assay (ELISA) for Detection of IFN-α2a Protein
2.7. Antiviral Activity of the Recombinant IFN-α2a
2.8. In Vitro CPE Assay of the Recombinant IFN-α2a on Hep-G2-Cells
2.9. Hep-G2-Cell Apoptosis Assay
2.10. Statistical Analysis
3. Results and Discussion
3.1. Generation and Selection of Transgenic Raphanus sativus L. plants
3.2. Analysis of IFN-α2a Expression in Transgenic White and Red Raphanus sativus L. plants
3.2.1. RT-PCR Analysis
3.2.2. Enzyme-Linked Immunosorbent Assay (ELISA) Detection of Human IFN-α2a and SDS-PAGE Analysis
3.3. Antiviral Activity of the Recombinant IFN-α2a
3.4. Antitumor Activity of the Recombinant IFN-α2a Isolated from Transgenic White and Red Raphanus sativus L. plants
3.4.1. Antitumor Effect of the Recombinant IFN-α2a on Hep-G2 Tumor Cell Line (In Vitro Assay)
3.4.2. Effect of Recombinant IFN-α2a on Hep-G2 Cell Apoptosis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sato, H.; Tsukamoto-Yasui, M.; Takado, Y.; Kawasaki, N.; Matsunaga, K.; Ueno, S.; Kanda, M.; Nishimura, M.; Karakawa, S.; Isokawa, M.; et al. Protein Deficiency-Induced Behavioral Abnormalities and Neurotransmitter Loss in Aged Mice Are Ameliorated by Essential Amino Acids. Front. Nutr. 2020, 7, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.K.; Drake, P.M.; Christou, P. The Production of Recombinant Pharmaceutical Proteins in Plants. Nat. Rev. Genet. 2003, 4, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Huang, Y.; Davis, A.; Yin, Z.; Mi, Q.; Menassa, R.; Brandle, J.E.; Jevnikar, A.M. Production of Biologically Active Human Interleukin-4 in Transgenic Tobacco and Potato. Plant Biotechnol. J. 2005, 3, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirono, H.; Morita, S.; Miki, Y.; Kurita, A.; Morita, S.; Koga, J.; Tanaka, K.; Masumura, T. Highly Efficient Production of Human Interferon-A by Transgenic Cultured Rice Cells. Plant Biotechnol. 2006, 23, 283–289. [Google Scholar] [CrossRef] [Green Version]
- Lowther, W.; Lorick, K.; Lawrence, S.D.; Yeow, W.S. Expression of Biologically Active Human Interferon Alpha 2 in Aloe Vera. Transgenic Res. 2012, 21, 1349–1357. [Google Scholar] [CrossRef]
- Nurjis, F.; Khan, M.S. Expression of Recombinant Interferon A-2a in Tobacco Chloroplasts Using Micro Projectile Bombardment. Afr. J. Biotechnol. 2011, 10, 17016–17022. [Google Scholar]
- Rotheim, P. The Biochemotechnology Revolution. Chemtech 1997, 27, 20–25. [Google Scholar]
- Sørensen, H.P.; Mortensen, K.K. Soluble Expression of Recombinant Proteins in the Cytoplasm of Escherichia coli. Microb. Cell Factories 2005, 4, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Gasdaska, J.R.; Spencer, D.; Dickey, L. Advantages of Therapeutic Protein Production in the Aquatic Plant Lemna. Bioprocess J. 2003, 2, 49–56. [Google Scholar] [CrossRef]
- Maeda, S.; McCandliss, R.; Gross, M.; Sloma, A.; Familletti, P.C.; Tabor, J.M.; Evinger, M.; Levy, W.P.; Pestka, S. Construction and Identification of Bacterial Plasmids Containing Nucleotide Sequence for Human Leukocyte Interferon. Proc. Natl. Acad. Sci. USA 1980, 77, 7010–7013. [Google Scholar] [CrossRef] [Green Version]
- Venkat, S.K.; Revathi, J.C.; Venkata, S.A.; Narayana, P.K.S.; Venkata, R. A Process for the Production of Human Interferon Alpha from Genetically Engineered Yeast. European Patent EP1272624, 20 September 2001. [Google Scholar]
- Digan, M.E.; Lair, S.V.; Brierley, R.A.; Siegel, R.S.; Williams, M.E.; Ellis, S.B.; Kellaris, P.A.; Provow, S.A.; Craig, W.S.; Velicelebi, G.; et al. Continuous Production of a Novel Lysozyme Via Secretion from the Yeast, Pichia pastoris. Bio/Technology 1989, 7, 160–164. [Google Scholar] [CrossRef]
- Yang, W. Bulking up Fuzeon. BioCentury 2002, 23, 10–18. [Google Scholar]
- Sahu, P.K.; Patel, T.S.; Sahu, P.; Singh, S.; Tirkey, P.; Sharma, D. Molecular Farming: A Biotechnological Approach in Agriculture for Production of Useful Metabolites. Int. J. Biotechnol. Biochem. 2014, 4, 23–30. [Google Scholar]
- Tarafdar, A.; Kamle, M.; Prakash, A.B.; Padaria, J.C. Transgenic Plants: Issues and Future Prospects. Biotechnol. Adv. 2014, 2, 1–47. [Google Scholar]
- Wu, H.Y.; Liu, K.H.; Wang, Y.C.; Wu, J.F.; Chiu, W.L.; Chen, C.Y.; Lai, E.M. Agrobest: An Efficient Agrobacterium-Mediated Transient Expression Method for Versatile Gene Function Analyses in Arabidopsis Seedlings. Plant Methods 2014, 10, 19. [Google Scholar] [CrossRef] [Green Version]
- Mehrizadeh, V.; Dorani, E.; Mohammadi, S.A.; Ghareyazie, B. Expression of Recombinant Human Ifn-Γ Protein in Soybean (Glycine max L.). Plant Cell Tissue Organ Cult. (PCTOC) 2021, 146, 127–136. [Google Scholar] [CrossRef]
- Ren, Q.; Wang, C.Y.; Song, Z.M.; Liu, D.; Yu, H.P.; Sheng, J. Expression of Human Interferon a 2b Gene in Ginseng Cells. Chem. Res. Chin. Univ. 2010, 26, 420–426. [Google Scholar]
- Razaghi, A.; Owens, L.; Heimann, K. Review of the Recombinant Human Interferon Gamma as an Immunotherapeutic: Impacts of Production Platforms and Glycosylation. J. Biotechnol. 2016, 240, 48–60. [Google Scholar] [CrossRef]
- Kebeish, R.; Niessen, M.; Thiruveedhi, K.; Bari, R.; Hirsch, H.J.; Rosenkranz, R.; Stäbler, N.; Schönfeld, B.; Kreuzaler, F.; Peterhänsel, C. Chloroplastic Photorespiratory Bypass Increases Photosynthesis and Biomass Production in Arabidopsis Thaliana. Nat. Biotechnol. 2007, 25, 593–599. [Google Scholar] [CrossRef]
- Herouet, C.; Esdaile, D.J.; Mallyon, B.A.; Debruyne, E.; Schulz, A.; Currier, T.; Hendrickx, K.; van der Klis, R.J.; Rouan, D. Safety Evaluation of the Phosphinothricin Acetyltransferase Proteins Encoded by the Pat and Bar Sequences That Confer Tolerance to Glufosinate-Ammonium Herbicide in Transgenic Plants. Regul. Toxicol. Pharmacol. 2005, 41, 134–149. [Google Scholar] [CrossRef]
- Banihani, S.A. Radish (Raphanus sativus) and Diabetes. Nutrients 2017, 9, 1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adolf, G.R.; Kalsner, I.; Ahorn, H.; Maurer-Fogy, I.; Cantell, K. Natural Human Interferon-A2 Is O-Glycosylated. Biochem. J. 1991, 276, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, G. Biological Basis for a Proper Clinical Application of Alpha Interferons. New Microbiol. 2008, 31, 305–318. [Google Scholar] [PubMed]
- Curtis, I.S.; Nam, H.G. Transgenic Radish (Raphanus sativus L. Longipinnatus Bailey) by Floral-Dip Method-Plant Development and Surfactant Are Important in Optimizing Transformation Efficiency. Transgenic Res. 2001, 10, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Kebeish, R.; Aboelmy, M.; El-Naggar, A.; El-Ayouty, Y.; Peterhansel, C. Simultaneous Overexpression of Cyanidase and Formate Dehydrogenase in Arabidopsis thaliana Chloroplasts Enhanced Cyanide Metabolism and Cyanide Tolerance. Environ. Exp. Bot. 2015, 110, 19–26. [Google Scholar] [CrossRef]
- Chomczynski, P.; Mackey, K. Substitution of Chloroform by Bromo-Chloropropane in the Single-Step Method of Rna Isolation. Anal. Biochem. 1995, 225, 163–164. [Google Scholar] [CrossRef]
- Niessen, M.; Thiruveedhi, K.; Rosenkranz, R.; Kebeish, R.; Hirsch, H.J.; Kreuzaler, F.; Peterhänsel, C. Mitochondrial Glycolate Oxidation Contributes to Photorespiration in Higher Plants. J. Exp. Bot. 2007, 58, 2709–2715. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bussereau, F.; Flamand, A.; Pese-Part, D. Reproducible Plaquing System for Rabies Virus in Cer Cells. J. Virol. Methods 1982, 4, 277–282. [Google Scholar] [CrossRef]
- Lansky, E.P.; Newman, R.A. Punica Granatum (Pomegranate) and Its Potential for Prevention and Treatment of Inflammation and Cancer. J. Ethnopharmacol. 2007, 109, 177–206. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Ozgur, O.; Karti, S.; Sonmez, M.; Yilmaz, M.; Karti, D.; Ozdemir, F.; Ovali, E. Effects of Interferon-Alfa-2a on Human Hepatoma Hepg2 Cells. Exp. Oncol. 2003, 25, 105–107. [Google Scholar]
- Barta, A.; Sommergruber, K.; Thompson, D.; Hartmuth, K.; Matzke, M.A.; Matzke, A.J. The Expression of a Nopaline Synthase—Human Growth Hormone Chimaeric Gene in Transformed Tobacco and Sunflower Callus Tissue. Plant Mol. Biol. 1986, 6, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Hiatt, A.; Caffferkey, R.; Bowdish, K. Production of Antibodies in Transgenic Plants. Nature 1989, 342, 76–78. [Google Scholar] [CrossRef] [PubMed]
- Burnett, M.J.; Burnett, A.C. Therapeutic Recombinant Protein Production in Plants: Challenges and Opportunities. Plants People Planet 2020, 2, 121–132. [Google Scholar] [CrossRef]
- Obembe, O.O.; Popoola, J.O.; Leelavathi, S.; Reddy, S.V. Advances in Plant Molecular Farming. Biotechnol. Adv. 2011, 29, 210–222. [Google Scholar] [CrossRef]
- Curtis, I.S. The Noble Radish: Past, Present and Future. Trends Plant Sci. 2003, 8, 305–307. [Google Scholar] [CrossRef]
- El-Ayouty, Y.; El-Manawy, I.; Nasih, S.; Hamdy, E.; Kebeish, R. Engineering Chlamydomonas Reinhardtii for Expression of Functionally Active Human Interferon-A. Mol. Biotechnol. 2019, 61, 134–144. [Google Scholar] [CrossRef]
- Babaeipour, V.; Shojaosadati, S.A.; Khalilzadeh, R.; Maghsoudi, N.; Farnoud, A.M. Enhancement of Human Γ-Interferon Production in Recombinant E. Coli Using Batch Cultivation. Appl. Biochem. Biotechnol. 2010, 160, 2366–2376. [Google Scholar] [CrossRef]
- Mizukami, T.; Komatsu, Y.; Hosoi, N.; Itoh, S.; Oka, T. Production of Active Human Interferon-A in E. coli I. Preferential Production by Lower Culture Temperature. Biotechnol. Lett. 1986, 8, 605–610. [Google Scholar] [CrossRef]
- Xu, J.; Dolan, M.C.; Medrano, G.; Cramer, C.L.; Weathers, P.J. Green Factory: Plants as Bioproduction Platforms for Recombinant Proteins. Biotechnol. Adv. 2012, 30, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Karg, S.R.; Kallio, P.T. The Production of Biopharmaceuticals in Plant Systems. Biotechnol. Adv. 2009, 27, 879–894. [Google Scholar] [CrossRef] [PubMed]
- Pestka, S.; Langer, J.A.; Zoon, K.C.; Samuel, C.E. Interferons and Their Actions. Annu. Rev. Biochem. 1987, 56, 727–777. [Google Scholar] [CrossRef] [PubMed]
- Nishiguchi, S.; Kuroki, T.; Nakatani, S.; Morimoto, H.; Takeda, T.; Nakajima, S.; Shiomi, S.; Seki, S.; Kobayashi, K.; Otani, S. Randomised Trial of Effects of Interferon-α on Incidence of Hepatocellular Carcinoma in Chronic Active Hepatitis C with Cirrhosis. Lancet 1995, 346, 1051–1055. [Google Scholar] [CrossRef]
- Masumura, T.; Morita, S.; Miki, Y.; Kurita, A.; Morita, S.; Shirono, H.; Koga, J.; Tanaka, K. Production of Biologically Active Human Interferon-A2 in Transgenic Rice. Plant Biotechnol. 2006, 23, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Gladilina, Y.A.; Sokolov, N.N.; Krasotkina, J.V. Cloning, Expression, and Purification of Helicobacter Pylori L-Asparaginase. Biochem. (Mosc.) Suppl. Ser. B Biomed. Chem. 2009, 3, 89–91. [Google Scholar] [CrossRef]
- Kebeish, R.; El-Sayed, A.; Fahmy, H.; Abdel-Ghany, A. Molecular Cloning, Biochemical Characterization, and Antitumor Properties of a Novel L-Asparaginase from Synechococcus elongatus Pcc6803. Biochemistry 2016, 81, 1173–1181. [Google Scholar] [CrossRef]
- Elshafei, A.M.; Hassan, M.M.; Abouzeid, M.A.E.; Mahmoud, D.A.; Elghonemy, D.H. Purification, Characterization and Antitumor Activity of L-Asparaginase from Penicillium brevicompactum 2 Nrc 829 3. Br. Microbiol. Res. J. 2011, 2, 158–174. [Google Scholar] [CrossRef]
- Enomoto, H.; Tao, L.; Eguchi, R.; Sato, A.; Honda, M.; Kaneko, S.; Iwata, Y.; Nishikawa, H.; Imanishi, H.; Iijima, H.; et al. The in Vivo Antitumor Effects of Type I-Interferon against Hepatocellular Carcinoma: The Suppression of Tumor Cell Growth and Angiogenesis. Sci. Rep. 2017, 7, 12189. [Google Scholar] [CrossRef] [Green Version]
- Melén, K.; Keskinen, P.; Lehtonen, A.; Julkunen, I. Interferon-Induced Gene Expression and Signaling in Human Hepatoma Cell Lines. J. Hepatol. 2000, 33, 764–772. [Google Scholar] [CrossRef]
- Hindi, N.N.; Saleh, M.I. Patient Characteristics associated with Peglyated Interferon Alfa-2a Induced Neutropenia in Chronic Hepatitis C Patients. Clin. Exp. Pharmacol. Physiol. 2018, 45, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Chuang, W.L.; Lee, C.M.; Yu, M.L.; Lu, S.N.; Wu, S.S.; Liao, L.Y.; Chen, C.L.; Kuo, H.T.; Chao, Y.C.; et al. Peginterferon Alfa-2a Plus Ribavirin for the Treatment of Dual Chronic Infection with Hepatitis B and C Viruses. Gastroenterology 2009, 136, 496–504.e3. [Google Scholar] [CrossRef] [PubMed]
- Ryff, J.-C. To Treat or Not to Treat? The Judicious Use of Interferon-A-2a for the Treatment of Chronic Hepatitis B. J. Hepatol. 1993, 17, S42–S46. [Google Scholar] [CrossRef]


| Sample Code | Reduction in VSV Titer (%) | |
|---|---|---|
| Direct | Indirect | |
| Peg-IFN | 7.8 ± 0.25 | 75.4 ± 6.1 |
| P-IFN-wRs | 6.7 ± 0.15 | 63.2 ± 3.2 |
| IFN-wRs-1 | 4.8 ± 0.20 | 50.8 ± 1.2 |
| IFN-wRs-2 | 5.5 ± 0.14 | 52.3 ± 1.6 |
| IFN-wRs-3 | 4.1 ± 0.21 | 48.2 ± 1.3 |
| P-IFN-rRs | 6.2 ± 0.18 | 60.2 ± 2.2 |
| IFN-rRs-1 | 4.4 ± 0.16 | 53.4 ± 1.2 |
| IFN-rRs-2 | 4.8 ± 0.11 | 55.6 ± 2.1 |
| IFN-rRs-3 | 3.8 ± 0.17 | 49. 8 ± 1.8 |
| WT-wRS | 0.8 ± 0.06 | 02.3 ± 0.1 |
| WT-rRS | 0.7 ± 0.05 | 02.0 ± 0.2 |
| Sample Name | IC50 (μgProtein) | SE |
|---|---|---|
| Peg-IFN | 6.2 | 0.62 |
| P-IFN-wRs | 7.4 | 0.77 |
| IFN-wRs-1 | 8.9 | 0.47 |
| IFN-wRs-2 | 8.5 | 0.55 |
| IFN-wRs-3 | 9.2 | 0.81 |
| P-IFN-rRs | 8.2 | 0.93 |
| IFN-rRs-1 | 10.3 | 0.65 |
| IFN-rRs-2 | 10.1 | 0.53 |
| IFN-rRs-3 | 11.7 | 0.68 |
| WT-wRS | 531.1 | 3.62 |
| WT-rRS | 468.5 | 4.23 |
| Sample Name | Normal Cells (%) | Early Apoptosis (%) | Late Apoptosis (%) | Necrotic Cells (%) |
|---|---|---|---|---|
| Control | 99.4 | 0.03 | 0.07 | 0.59 |
| Peg-IFN | 4.70 | 1.02 | 54.55 | 87.8 |
| P-IFN-wRs | 5.40 | 0.14 | 37.42 | 82.6 |
| IFN-wRs-1 | 8.47 | 0.07 | 26.4 | 63.8 |
| IFN-wRs-2 | 8.58 | 0.08 | 28.7 | 79.6 |
| IFN-wRs-3 | 7.82 | 0.06 | 25.2 | 57.1 |
| P-IFN-rRs | 5.10 | 0.12 | 39.52 | 81.2 |
| IFN-rRs-1 | 7.17 | 0.12 | 24.3 | 60.7 |
| IFN-rRs-2 | 7.51 | 0.18 | 26.7 | 76.2 |
| IFN-rRs-3 | 6.82 | 0.16 | 23.4 | 54.1 |
| WT-wRS | 96.7 | 0.72 | 2.21 | 0.62 |
| WT-rRS | 95.9 | 0.83 | 2.11 | 0.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kebeish, R.; Hamdy, E.; Al-Zoubi, O.; Habeeb, T.; Osailan, R.; El-Ayouty, Y. A Biotechnological Approach for the Production of Pharmaceutically Active Human Interferon-α from Raphanus sativus L. Plants. Bioengineering 2022, 9, 381. https://doi.org/10.3390/bioengineering9080381
Kebeish R, Hamdy E, Al-Zoubi O, Habeeb T, Osailan R, El-Ayouty Y. A Biotechnological Approach for the Production of Pharmaceutically Active Human Interferon-α from Raphanus sativus L. Plants. Bioengineering. 2022; 9(8):381. https://doi.org/10.3390/bioengineering9080381
Chicago/Turabian StyleKebeish, Rashad, Emad Hamdy, Omar Al-Zoubi, Talaat Habeeb, Raha Osailan, and Yassin El-Ayouty. 2022. "A Biotechnological Approach for the Production of Pharmaceutically Active Human Interferon-α from Raphanus sativus L. Plants" Bioengineering 9, no. 8: 381. https://doi.org/10.3390/bioengineering9080381






