Preoperative Molecular Subtype Classification Prediction of Ovarian Cancer Based on Multi-Parametric Magnetic Resonance Imaging Multi-Sequence Feature Fusion Network
Abstract
:1. Introduction
2. Materials and Methods
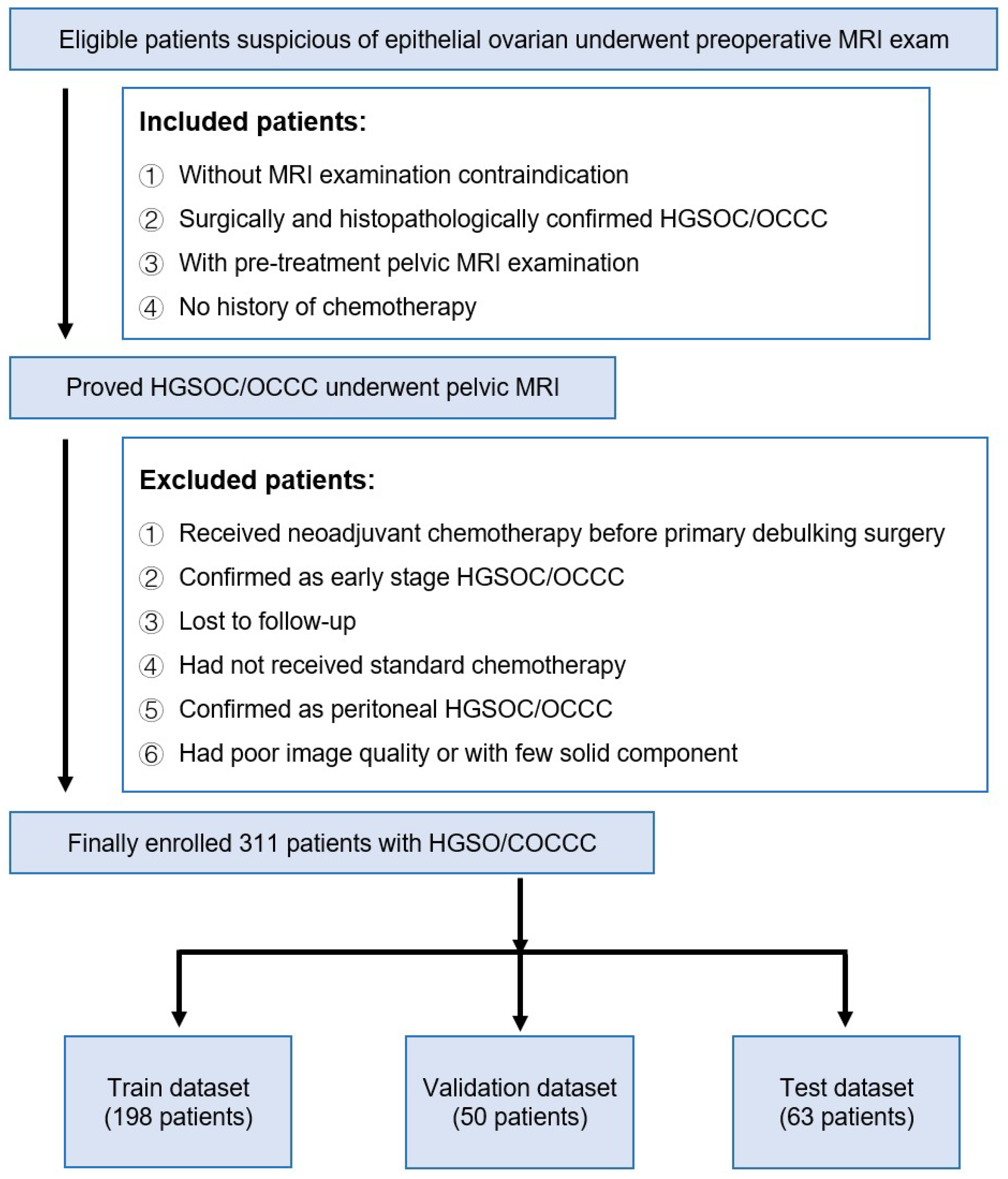
2.1. Data Sources
2.2. MRI Image Preprocessing
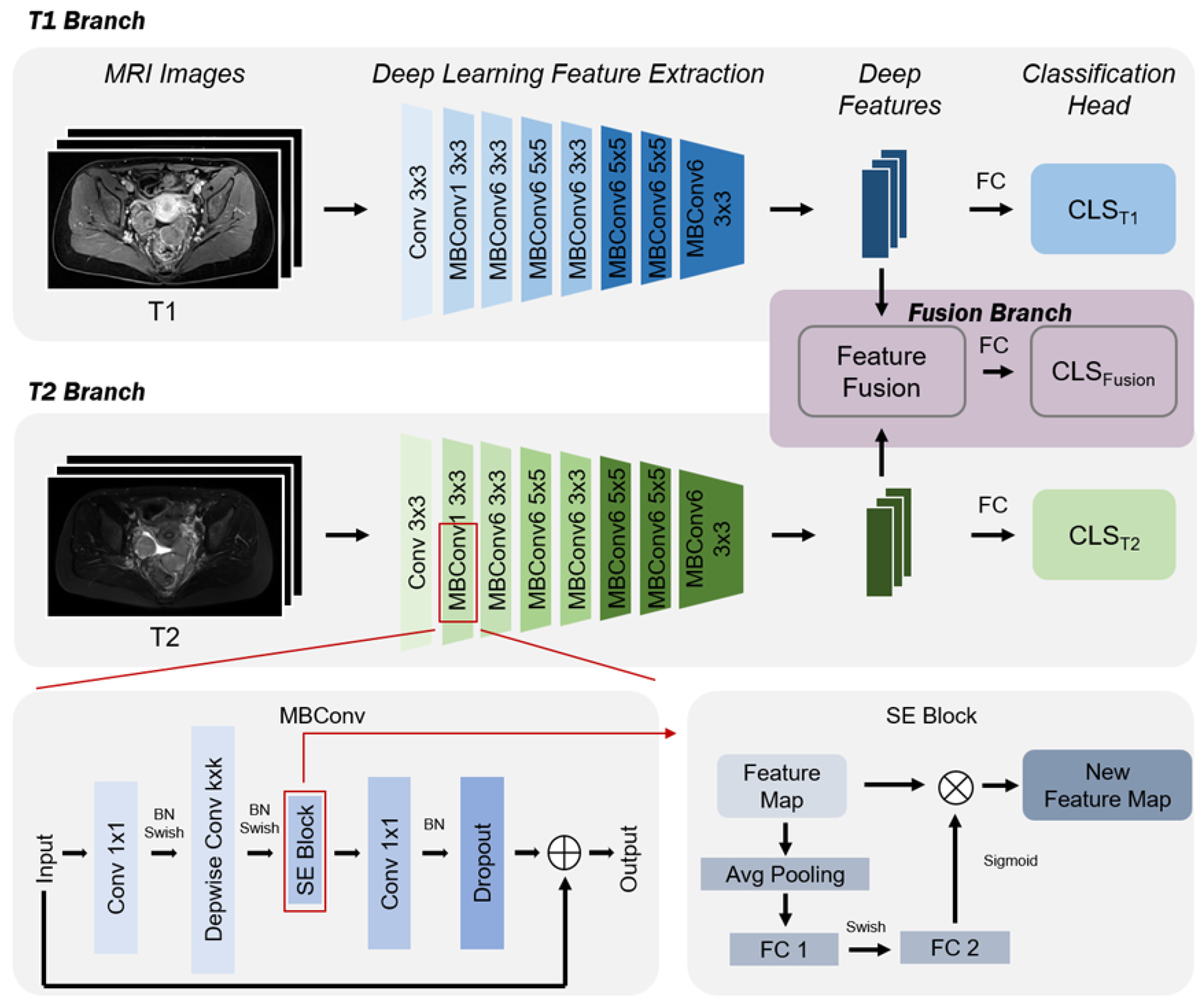
2.3. Ovarian Cancer Classification Prediction Model Based on mpMRI and EfficientNet Multi-Sequence Feature Fusion
2.4. Deep Feature Extraction Based on EfficientNet
2.5. Multi-Sequence Feature Fusion
2.6. Ovarian Cancer Subtype Prediction
2.7. Training and Implementation Details
2.8. Metrics
- •
- AUC (area under the curve): This is commonly used with the ROC (receiver operating characteristic) curve, referred to as ROC-AUC. The ROC curve is generated by plotting the true positive rate (TPR) against the false positive rate (FPR) for all possible classification thresholds. The AUC value is the area under this curve, ranging from 0 to 1. A higher AUC value indicates better model classification performance;
- •
- AP (average precision): This measures the average performance of the model’s precision (precision) across different thresholds. It is the area under the precision–recall curve, particularly suitable for evaluating imbalanced datasets. A higher AP indicates better model performance;
- •
- F1-Score (F1): This is the harmonic mean of precision (precision) and recall (recall). It is a number between 0 and 1 used to measure the model’s precision and robustness. A higher F1 score indicates a better balance between the model’s precision and recall;
- •
- ACC (accuracy): The most intuitive performance metric, indicating the proportion of correctly classified samples out of the total number of samples. A high accuracy means that the model can correctly classify more samples;
- •
- SEN (sensitivity) or recall: This is the true positive rate (TPR), measuring the model’s ability to correctly identify positive cases. A higher sensitivity means the model is more accurate in identifying positive cases;
- •
- SPEC (specificity): This is the true negative rate, measuring the model’s ability to correctly identify negative cases. A higher specificity means the model is more accurate in identifying negative cases.
3. Results
3.1. Impact of Feature Fusion on Results
3.2. Impact of Baseline Network Architecture on Results
3.3. Impact of Hyperparameters on Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA A Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef]
- Manuel, A.V.; Inés, P.M.; Oleg, B.; Andy, R.; Aleksandra, G.M.; Jatinderpal, K.; Ranjit, M.; Ian, J.; Usha, M.; Alexey, Z. A quantitative performance study of two automatic methods for the diagnosis of ovarian cancer. Biomed. Signal Process. Control 2018, 46, 86–93. [Google Scholar]
- Kurman, R.J.; Shih, I.M. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer—shifting the paradigm. Hum. Pathol. 2011, 42, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Fairman, A.; Tan, J.; Quinn, M. Women with low-grade abnormalities on Pap smear should be referred for colposcopy. Aust. N. Z. J. Obstet. Gynaecol. 2004, 44, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Wang, H.; Wu, C.; Liu, X.; Qu, L.; Shi, Y. Prediction model of hemorrhage transformation in patient with acute ischemic stroke based on multiparametric MRI radiomics and machine learning. Brain Sci. 2022, 12, 858. [Google Scholar] [CrossRef] [PubMed]
- Polanec, S.H.; Pinker-Domenig, K.; Brader, P.; Georg, D.; Shariat, S.; Spick, C.; Susani, M.; Helbich, T.H.; Baltzer, P.A. Multiparametric MRI of the prostate at 3 T: Limited value of 3D 1 H-MR spectroscopy as a fourth parameter. World J. Urol. 2016, 34, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Loffroy, R.; Chevallier, O.; Moulin, M.; Favelier, S.; Genson, P.Y.; Pottecher, P.; Crehange, G.; Cochet, A.; Cormier, L. Current role of multiparametric magnetic resonance imaging for prostate cancer. Quant. Imaging Med. Surg. 2015, 5, 754. [Google Scholar]
- Kim, J.H.; Choi, S.H.; Ryoo, I.; Yun, T.J.; Kim, T.M.; Lee, S.H.; Park, C.K.; Kim, J.H.; Sohn, C.H.; Park, S.H.; et al. Prognosis prediction of measurable enhancing lesion after completion of standard concomitant chemoradiotherapy and adjuvant temozolomide in glioblastoma patients: Application of dynamic susceptibility contrast perfusion and diffusion-weighted imaging. PLoS ONE 2014, 9, e113587. [Google Scholar] [CrossRef] [PubMed]
- Gondo, T.; Hricak, H.; Sala, E.; Zheng, J.; Moskowitz, C.S.; Bernstein, M.; Eastham, J.A.; Vargas, H.A. Multiparametric 3T MRI for the prediction of pathological downgrading after radical prostatectomy in patients with biopsy-proven Gleason score 3+ 4 prostate cancer. Eur. Radiol. 2014, 24, 3161–3170. [Google Scholar] [CrossRef] [PubMed]
- Turkbey, B.; Mani, H.; Aras, O.; Ho, J.; Hoang, A.; Rastinehad, A.R.; Agarwal, H.; Shah, V.; Bernardo, M.; Pang, Y.; et al. Prostate cancer: Can multiparametric MR imaging help identify patients who are candidates for active surveillance? Radiology 2013, 268, 144–152. [Google Scholar] [CrossRef]
- Neto, J.A.O.; Parente, D.B. Multiparametric magnetic resonance imaging of the prostate. Magn. Reson. Imaging Clin. 2013, 21, 409–426. [Google Scholar] [CrossRef]
- Wu, M.; Yan, C.; Liu, H.; Liu, Q. Automatic classification of ovarian cancer types from cytological images using deep convolutional neural networks. Biosci. Rep. 2018, 38, BSR20180289. [Google Scholar] [CrossRef] [PubMed]
- Shibusawa, M.; Nakayama, R.; Okanami, Y.; Kashikura, Y.; Imai, N.; Nakamura, T.; Kimura, H.; Yamashita, M.; Hanamura, N.; Ogawa, T. The usefulness of a computer-aided diagnosis scheme for improving the performance of clinicians to diagnose non-mass lesions on breast ultrasonographic images. J. Med. Ultrason. 2016, 43, 387–394. [Google Scholar] [CrossRef]
- Chen, S.J.; Chang, C.Y.; Chang, K.Y.; Tzeng, J.E.; Chen, Y.T.; Lin, C.W.; Hsu, W.C.; Wei, C.K. Classification of the thyroid nodules based on characteristic sonographic textural feature and correlated histopathology using hierarchical support vector machines. Ultrasound Med. Biol. 2010, 36, 2018–2026. [Google Scholar] [CrossRef]
- Chang, C.Y.; Liu, H.Y.; Tseng, C.H.; Shih, S.R. Computer-aided diagnosis for thyroid graves’disease in ultrasound images. Biomed. Eng. Appl. Basis Commun. 2010, 22, 91–99. [Google Scholar] [CrossRef]
- Acharya, U.R.; Sree, S.V.; Swapna, G.; Gupta, S.; Molinari, F.; Garberoglio, R.; Witkowska, A.; Suri, J.S. Effect of complex wavelet transform filter on thyroid tumor classification in three-dimensional ultrasound. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2013, 227, 284–292. [Google Scholar] [CrossRef]
- Guo, Q.; Qu, L.; Zhu, J.; Li, H.; Wu, Y.; Wang, S.; Yu, M.; Wu, J.; Wen, H.; Ju, X.; et al. Predicting Lymph Node Metastasis From Primary Cervical Squamous Cell Carcinoma Based on Deep Learning in Histopathologic Images. Mod. Pathol. 2023, 36, 100316. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.P.; Hadjiiski, L.M.; Samala, R.K. Computer-aided diagnosis in the era of deep learning. Med. Phys. 2020, 47, e218–e227. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Liu, S.; Wang, M.; Song, Z. Transmef: A transformer-based multi-exposure image fusion framework using self-supervised multi-task learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtually, 22 February–1 March 2022; Volume 36, pp. 2126–2134. [Google Scholar]
- Qu, L.; Luo, X.; Liu, S.; Wang, M.; Song, Z. Dgmil: Distribution guided multiple instance learning for whole slide image classification. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 24–34. [Google Scholar]
- Qu, L.; Liu, S.; Liu, X.; Wang, M.; Song, Z. Towards label-efficient automatic diagnosis and analysis: A comprehensive survey of advanced deep learning-based weakly-supervised, semi-supervised and self-supervised techniques in histopathological image analysis. Phys. Med. Biol. 2022, 67, 20TR01. [Google Scholar] [CrossRef]
- Qu, L.; Wang, M.; Song, Z. Bi-directional weakly supervised knowledge distillation for whole slide image classification. Adv. Neural Inf. Process. Syst. 2022, 35, 15368–15381. [Google Scholar]
- Luo, X.; Qu, L.; Guo, Q.; Song, Z.; Wang, M. Negative instance guided self-distillation framework for whole slide image analysis. IEEE J. Biomed. Health Inform. 2023, 28, 964–975. [Google Scholar] [CrossRef]
- Sun, Z.; Qu, L.; Luo, J.; Song, Z.; Wang, M. Label correlation transformer for automated chest X-ray diagnosis with reliable interpretability. Radiol. Medica 2023, 128, 726–733. [Google Scholar] [CrossRef]
- Qu, L.; Yang, Z.; Duan, M.; Ma, Y.; Wang, S.; Wang, M.; Song, Z. Boosting whole slide image classification from the perspectives of distribution, correlation and magnification. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2–3 October 2023; pp. 21463–21473. [Google Scholar]
- Qu, L.; Ma, Y.; Luo, X.; Wang, M.; Song, Z. Rethinking multiple instance learning for whole slide image classification: A good instance classifier is all you need. arXiv 2023, arXiv:2307.02249. [Google Scholar]
- Liu, X.; Qu, L.; Xie, Z.; Zhao, J.; Shi, Y.; Song, Z. Towards more precise automatic analysis: A comprehensive survey of deep learning-based multi-organ segmentation. arXiv 2023, arXiv:2303.00232. [Google Scholar]
- Liu, S.; Yin, S.; Qu, L.; Wang, M.; Song, Z. A Structure-aware Framework of Unsupervised Cross-Modality Domain Adaptation via Frequency and Spatial Knowledge Distillation. IEEE Trans. Med. Imaging 2023, 42, 3919–3931. [Google Scholar] [CrossRef]
- Park, H.; Yun, J.; Lee, S.M.; Hwang, H.J.; Seo, J.B.; Jung, Y.J.; Hwang, J.; Lee, S.H.; Lee, S.W.; Kim, N. Deep learning–based approach to predict pulmonary function at chest CT. Radiology 2023, 307, e221488. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, L.; Yu, M.; Wu, R.; Steffens, D.C.; Potter, G.G.; Liu, M. Hybrid representation learning for cognitive diagnosis in late-life depression over 5 years with structural MRI. Med. Image Anal. 2024, 94, 103135. [Google Scholar] [CrossRef]
- Suzuki, K. Computational Intelligence in Biomedical Imaging; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Theodoridis, S.; Koutroumbas, K. Pattern Recognition; Elsevier: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Kott, O.; Linsley, D.; Amin, A.; Karagounis, A.; Jeffers, C.; Golijanin, D.; Serre, T.; Gershman, B. Development of a deep learning algorithm for the histopathologic diagnosis and Gleason grading of prostate cancer biopsies: A pilot study. Eur. Urol. Focus 2021, 7, 347–351. [Google Scholar] [CrossRef]
- Ismael, S.A.A.; Mohammed, A.; Hefny, H. An enhanced deep learning approach for brain cancer MRI images classification using residual networks. Artif. Intell. Med. 2020, 102, 101779. [Google Scholar] [CrossRef]
- Booma, P.; Vinesh, T.; Julius, T.S.H. Max Pooling Technique to Detect and Classify Medical Image for Ovarian Cancer Diagnosis. Test Eng. Manag. J. 2020, 82, 8423–8442. [Google Scholar]
- Bruce, W.; Kirby, R.C.; Karissa, T.; Oleg, N.; Molly, A.B.; Manish, P.; Vikas, S.; Kevin, W.E.; Paul, J.C. 3D texture analysis for classification of second harmonic generation images of human ovarian cancer. Sci. Rep. 2016, 6, 35734. [Google Scholar]
- Wang, C.; Lee, Y.; Chang, C.; Lin, Y.; Liou, Y.; Hsu, P.; Chang, C.; Sai, A.; Wang, C.; Chao, T. A Weakly Supervised Deep Learning Method for Guiding Ovarian Cancer Treatment and Identifying an Effective Biomarker. Cancers 2022, 14, 1651. [Google Scholar] [CrossRef] [PubMed]
- Saida, T.; Mori, K.; Hoshiai, S.; Sakai, M.; Urushibara, A.; Ishiguro, T.; Minami, M.; Satoh, T.; Nakajima, T. Diagnosing Ovarian Cancer on MRI: A Preliminary Study Comparing Deep Learning and Radiologist Assessments. Cancers 2022, 14, 987. [Google Scholar] [CrossRef] [PubMed]
- Ziyambe, B.; Yahya, A.; Mushiri, T.; Tariq, M.U.; Abbas, Q.; Babar, M.; Albathan, M.; Asim, M.; Hussain, A.; Jabbar, S. A Deep Learning Framework for the Prediction and Diagnosis of Ovarian Cancer in Pre- and Post-Menopausal Women. Diagnostics 2023, 13, 1703. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.; Sawyer, T.W.; Thurston, N.; Barton, J.; Ditzler, G. Ovarian Cancer Detection Using Optical Coherence Tomography and Convolutional Neural Networks. Neural Comput. Appl. 2022, 34, 8977–8987. [Google Scholar] [CrossRef]
- Gao, Y.; Zeng, S.; Xu, X.; Li, H.; Yao, S.; Song, K.; Li, X.; Chen, L.; Tang, J.; Xing, H.; et al. Deep Learning-Enabled Pelvic Ultrasound Images for Accurate Diagnosis of Ovarian Cancere in China: A Retrospective, Multicentre, Disgnostic Study. Digit. Health 2022, 4, 179–187. [Google Scholar]
- Jung, Y.; Kim, T.; Han, M.R.; Kim, S.; Kim, G.; Lee, S.; Choi, Y. Ovarian Tumor Diagnosis Using Deep Convolutional Neural Networks and a Denoising Convolutional Autoencoder. Sci. Rep. 2022, 12, 17024. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Ren, P.; Xiao, Y.; Chang, X.; Huang, P.Y.; Li, Z.; Chen, X.; Wang, X. A comprehensive survey of neural architecture search: Challenges and solutions. ACM Comput. Surv. (CSUR) 2021, 54, 1–34. [Google Scholar] [CrossRef]
- He, T.; Zhang, Z.; Zhang, H.; Zhang, Z.; Xie, J.; Li, M. Bag of Tricks for Image Classification with Convolutional Neural Networks. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; IEEE Computer Society: New York, NY, USA, 2019; pp. 558–567. [Google Scholar]
- Zhang, N.; Yang, G.; Gao, Z.; Xu, C.; Zhang, Y.; Shi, R.; Keegan, J.; Xu, L.; Zhang, H.; Fan, Z.; et al. Deep Learning for Diagnosis of Chronic Myocardial Infarction on Nonenhanced Cardiac Cine MRI. Radiology 2019, 291, 606–617. [Google Scholar] [CrossRef]
- Lithens, G.; Sanchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen, C.v.d.K.; Bult, P.; Ginneken, B.v.; van der Laak, J. Deep Learning as a Tool for Increased Accuracy and Efficiency of Histopathological Diagnosis. Sci. Rep. 2016, 6, 1–11. [Google Scholar]

| Stage | Operator | Resolution | Channels | Layers |
|---|---|---|---|---|
| 1 | Conv3 × 3 | 224 × 224 | 32 | 1 |
| 2 | MBConv1, k3 × 3 | 112 × 112 | 16 | 2 |
| 3 | MBConv6, k3 × 3 | 112 × 112 | 24 | 3 |
| 4 | MBConv6, k5 × 5 | 56 × 56 | 48 | 3 |
| 5 | MBConv6, k3 × 3 | 28 × 28 | 88 | 4 |
| 6 | MBConv6, k5 × 5 | 14 × 14 | 120 | 4 |
| 7 | MBConv6, k5 × 5 | 14 × 14 | 208 | 5 |
| 8 | MBConv6, k3 × 3 | 7 × 7 | 352 | 2 |
| Hyperparameters | Description | Value |
|---|---|---|
| Learning Rate | Learning rate in model training | 0.001 |
| Batch Size | Number of samples per training batch | 32 |
| Number of Epochs | The total number of rounds of model training | 100 |
| Optimizer | Optimization algorithm used to train the model | AdamW |
| L2 Regularization | Only apply to the weight norm scale factors | 0.0001 |
| learning rate decay | Learning rate decay method with the number of training rounds | Warmup_Cosine(25) |
| Weight initialization | Initialization strategy for model weights | ImageNet Pretraining |
| Upsampling strategy | Dealing with imbalanced class distribution problems | Weighted Random Sampling |
| Data Augmentation Rate | Probability of applying data augmentation | 0.3 |
| AUC | AP | F1-Score | ACC | SEN | SPEC | |
|---|---|---|---|---|---|---|
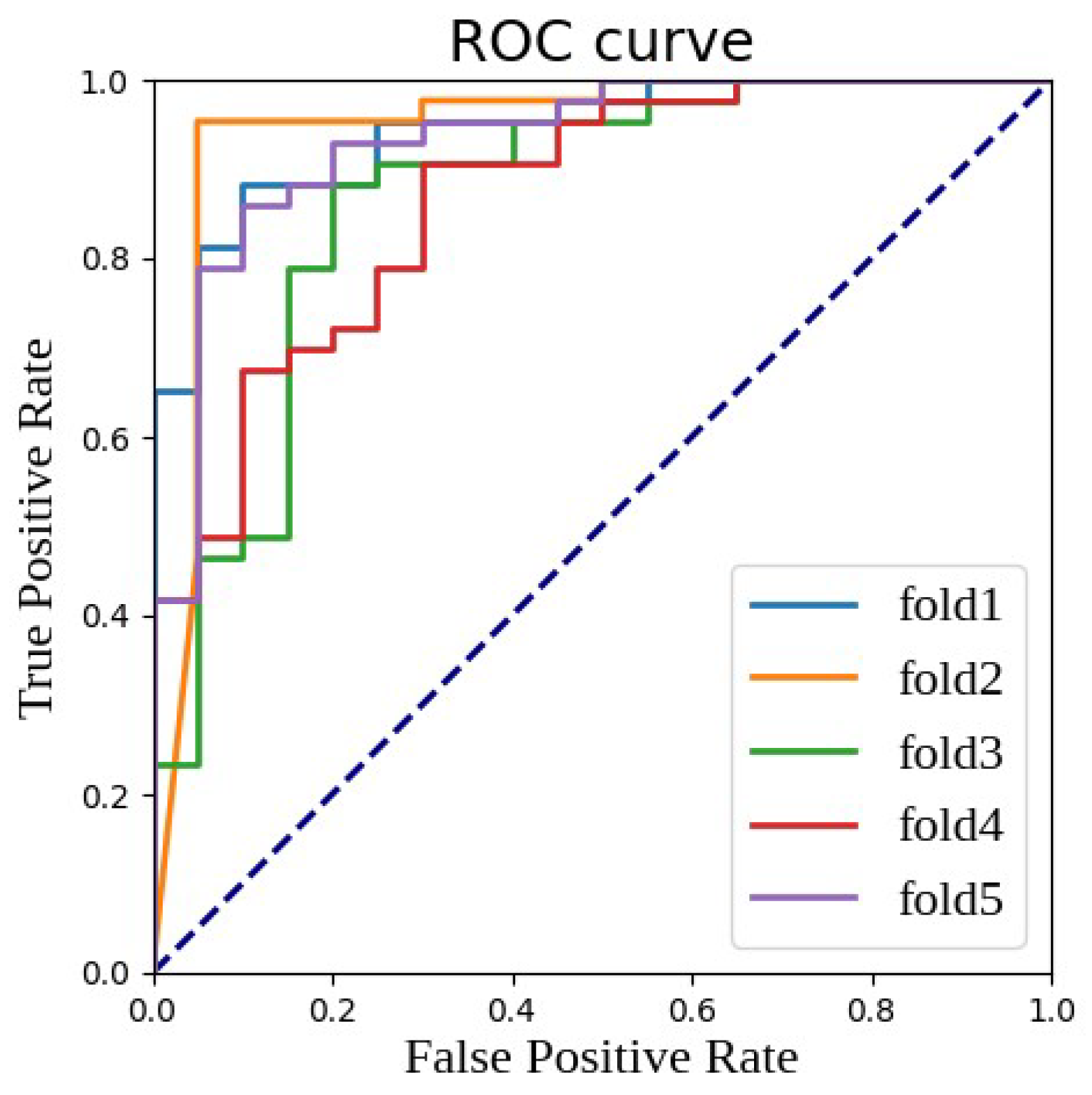
| Fold 1 | 0.9442 | 0.9742 | 0.8916 | 0.8571 | 0.8605 | 0.8500 |
| Fold 2 | 0.9413 | 0.9580 | 0.9302 | 0.9048 | 0.9302 | 0.8500 |
| Fold 3 | 0.8814 | 0.9288 | 0.8989 | 0.8571 | 0.9302 | 0.7000 |
| Fold 4 | 0.9116 | 0.9560 | 0.8889 | 0.8571 | 0.8372 | 0.9000 |
| Fold 5 | 0.9023 | 0.9393 | 0.8571 | 0.8254 | 0.7674 | 0.9500 |
| AUC | AP | F1-Score | ACC | SEN | SPEC |
|---|---|---|---|---|---|
| 0.9162 ± 0.0226 | 0.9513 ± 0.0176 | 0.8933 ± 0.0261 | 0.8603 ± 0.0284 | 0.8651 ± 0.0686 | 0.850 ± 0.0935 |
| Branch | AUC | AP | F1-Score | ACC | SEN | SPEC |
|---|---|---|---|---|---|---|
| T1 Branch | 0.7823 | 0.8615 | 0.8065 | 0.7524 | 0.7628 | 0.73 |
| T1 Branch | 0.8958 | 0.9472 | 0.8629 | 0.8286 | 0.8047 | 0.88 |
| Fusion Branch | 0.9162 | 0.9513 | 0.8933 | 0.8603 | 0.8651 | 0.85 |
| Baseline Network | AUC | AP | F1-Score | ACC | SEN | SPEC |
|---|---|---|---|---|---|---|
| EfficientNet-B0 | 0.8884 | 0.9216 | 0.8941 | 0.8571 | 0.8837 | 0.80 |
| EfficientNet-B1 | 0.7663 | 0.8382 | 0.8913 | 0.8413 | 0.9535 | 0.63 |
| EfficientNet-B2 | 0.9162 | 0.9513 | 0.8933 | 0.8603 | 0.8651 | 0.85 |
| EfficientNet-B3 | 0.8215 | 0.9004 | 0.75 | 0.7143 | 0.6279 | 0.85 |
| EfficientNet-B4 | 0.8291 | 0.909 | 0.8764 | 0.8254 | 0.907 | 0.65 |
| Description | EfficientNet-B2 | |||||
|---|---|---|---|---|---|---|
| Learning Rate | 0.001 | 0.0001 | 0.001 | 0.001 | 0.001 | 0.001 |
| Fusion Method | Concatenate | Concatenate | Add | Concatenate | Concatenate | Concatenate |
| Upsample | Yes | Yes | Yes | No | Yes | Yes |
| Crop Method | CenterCrop | CenterCrop | CenterCrop | CenterCrop | RandomCrop | Resize |
| AUC | 0.9162 | 0.9093 | 0.9107 | 0.9009 | 0.6337 | 0.8756 |
| AP | 0.9513 | 0.9489 | 0.9521 | 0.9566 | 0.7874 | 0.9373 |
| F1-Score | 0.8933 | 0.8924 | 0.8571 | 0.8991 | 0.6389 | 0.881 |
| ACC | 0.8603 | 0.8467 | 0.8254 | 0.853 | 0.5873 | 0.8413 |
| SEN | 0.8651 | 0.8902 | 0.7674 | 0.9302 | 0.5349 | 0.8605 |
| SPEC | 0.85 | 0.78 | 0.88 | 0.73 | 0.70 | 0.80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, Y.; Wang, T.; Qu, L.; Li, H.; Guo, Q.; Wang, H.; Liu, X.; Wu, X.; Song, Z. Preoperative Molecular Subtype Classification Prediction of Ovarian Cancer Based on Multi-Parametric Magnetic Resonance Imaging Multi-Sequence Feature Fusion Network. Bioengineering 2024, 11, 472. https://doi.org/10.3390/bioengineering11050472
Du Y, Wang T, Qu L, Li H, Guo Q, Wang H, Liu X, Wu X, Song Z. Preoperative Molecular Subtype Classification Prediction of Ovarian Cancer Based on Multi-Parametric Magnetic Resonance Imaging Multi-Sequence Feature Fusion Network. Bioengineering. 2024; 11(5):472. https://doi.org/10.3390/bioengineering11050472
Chicago/Turabian StyleDu, Yijiang, Tingting Wang, Linhao Qu, Haiming Li, Qinhao Guo, Haoran Wang, Xinyuan Liu, Xiaohua Wu, and Zhijian Song. 2024. "Preoperative Molecular Subtype Classification Prediction of Ovarian Cancer Based on Multi-Parametric Magnetic Resonance Imaging Multi-Sequence Feature Fusion Network" Bioengineering 11, no. 5: 472. https://doi.org/10.3390/bioengineering11050472





