Coordination Compounds of Nickel(II) with 3,5–Dibromo–Salicylaldehyde: Structure and Interaction with Biomolecules
Abstract
:1. Introduction
2. Results
2.1. Synthesis and Characterization
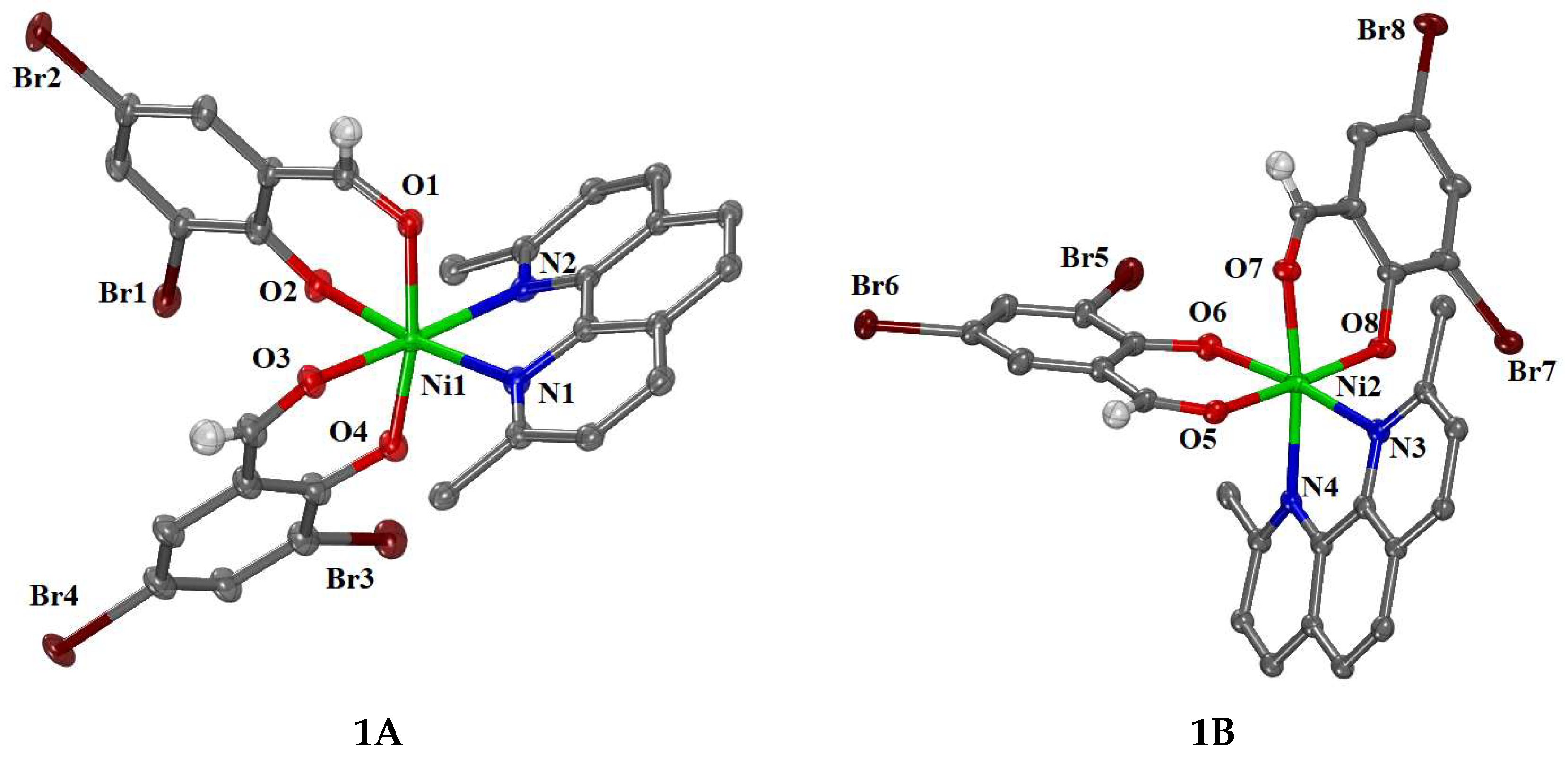
2.2. Structure of the Complexes
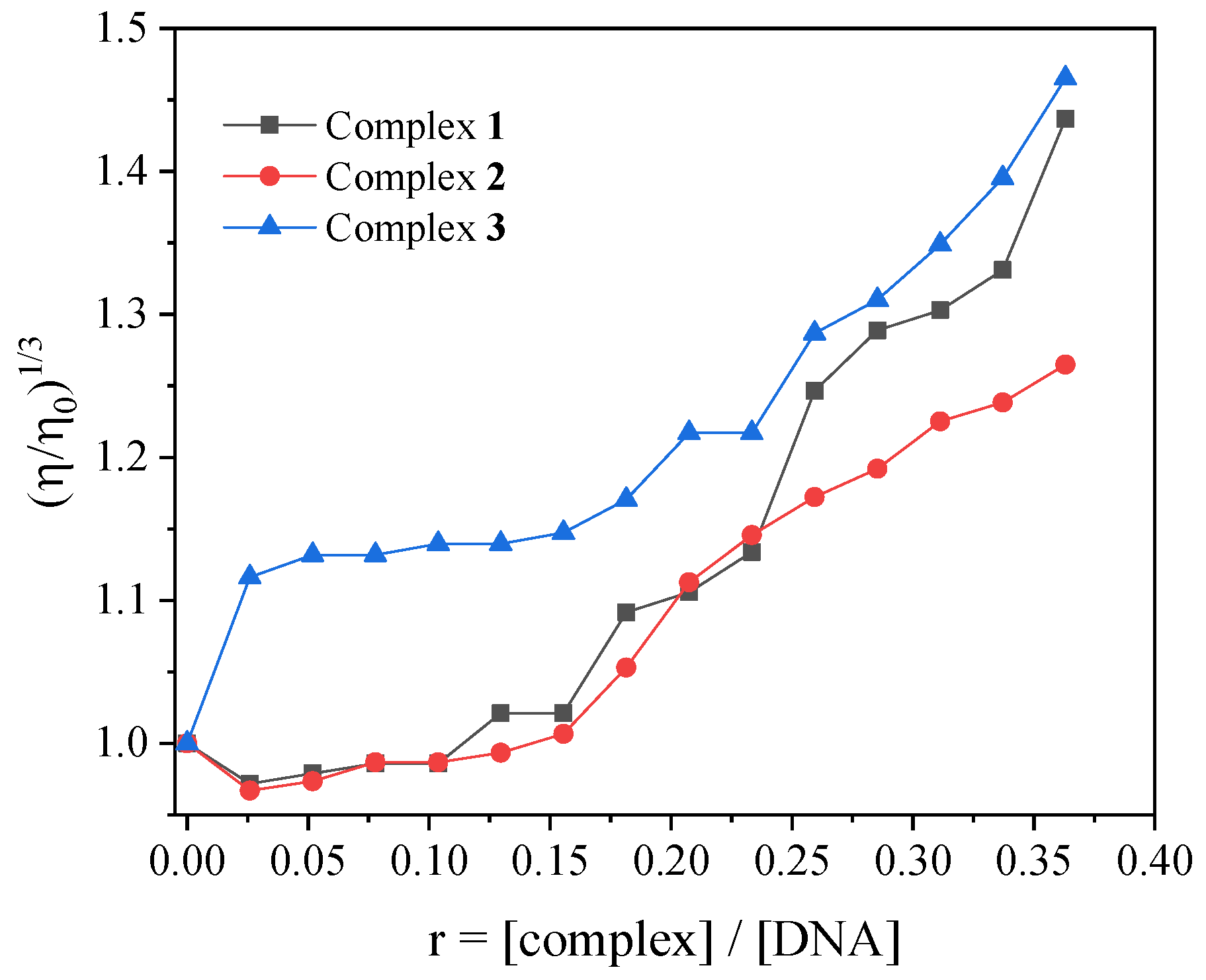
2.3. Interaction of the Complexes with CT DNA
2.4. Interaction of the Complexes with Albumins
3. Materials and Methods
3.1. Materials–Instrumentation—Physical Measurements
3.2. Synthesis of the Complexes
3.3. Single-Crystal X-ray Crystallography
3.4. Study of the Biological Profile of the Complexes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Claudel, M.; Schwarte, J.V.; Fromm, K.M. New Antimicrobial Strategies Based on Metal Complexes. Chemistry 2020, 2, 849–899. [Google Scholar] [CrossRef]
- Gasser, G. Metal Complexes and Medicine: A Successful Combination. Chimia 2015, 69, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Pauls, G.; Becker, T.; Rahfeld, P.; Gretscher, R.R.; Paetz, C.; Pasteels, J.; von Reuss, S.H.; Burse, A.; Boland, W. Two Defensive Lines in Juvenile Leaf Beetles; Esters of 3-Nitropropionic Acid in the Hemolymph and Aposematic Warning. J. Chem. Ecol. 2016, 42, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Devi, J.; Dubey, A.; Kumar, D.; Jindal, D.K.; Asija, S.; Sharma, A. Co(II), Ni(II), Cu(II) and Zn(II) Complexes of Schiff Base Ligands: Synthesis, Characterization, DFT, in Vitro Antimicrobial Activity and Molecular Docking Studies. Res. Chem. Intermed. 2023, 49, 939–965. [Google Scholar] [CrossRef]
- Kargar, H.; Ardakani, A.A.; Tahir, M.N.; Ashfaq, M.; Munawar, K.S. Synthesis, Spectral Characterization, Crystal Structure Determination and Antimicrobial Activity of Ni(II), Cu(II) and Zn(II) Complexes with the Schiff Base Ligand Derived from 3,5-Dibromosalicylaldehyde. J. Mol. Struct. 2021, 1229, 129842. [Google Scholar] [CrossRef]
- Ghasemi, L.; Abedi, A.; Abbasi, A.; Kucerakova, M.; Dusek, M.; Behzad, M. Subtle Structural Variations in New Mixed-Ligand Cu(II) Complexes with NN’O Type Unsymmetrical Schiff Bases: Molecular Docking against SARS-CoV-2 and Its Omicron Variant Main Proteases. Inorg. Chem. Commun. 2024, 159, 111795. [Google Scholar] [CrossRef]
- Zianna, A.; Geromichalos, G.D.; Pekou, A.; Hatzidimitriou, A.G.; Coutouli-Argyropoulou, E.; Lalia-Kantouri, M.; Pantazaki, A.A.; Psomas, G. A Palladium(II) Complex with the Schiff Base 4-Chloro-2-(N-Ethyliminomethyl)-Phenol: Synthesis, Structural Characterization, and in Vitro and in Silico Biological Activity Studies. J. Inorg. Biochem. 2019, 199, 110792. [Google Scholar] [CrossRef] [PubMed]
- Kargar, H.; Centore, R.; Fallah-Mehrjardi, M.; Santagata, E.; Munawar, K.S. Dihalosalicylaldehyde Based Molybdenum(VI) Complexes: Synthesis, Spectral Characterization, Crystal Structures, and Catalytic Activities for the Selective Oxidation of Benzylic Alcohols. Inorg. Chim. Acta 2024, 566, 122015. [Google Scholar] [CrossRef]
- Karmakar, M.; Kumar, P.; Jiyaur Rahaman, S.; Chattopadhyay, S. An Overview on the Synthesis, Structure, and Application of Vanadyl Complexes with Hydrazonic Acid Ligands Based on Salicylaldehyde or Its Derivatives. Inorg. Chim. Acta 2024, 565, 121969. [Google Scholar] [CrossRef]
- Zhuo, S.; Wu, W.; Liu, Y.; Zou, L.; Wu, Y.; Botha, L.; Kumar, A.; Afzal, M.; Alarifi, A. 3,5-Dibromosalicylaldehyde Nicotinoylhydrazone and 4,4′-Bipyridine Appended New Zn(II) Coordination Polymer: Secnidazole Sensing and Rhodamine B Photocatalytic Degradation Properties. J. Mol. Struct. 2022, 1264, 133304. [Google Scholar] [CrossRef]
- Gui, M.; Wu, W.; Liu, Y.; Wu, Y.; Singh, S.; Kumar, A.; Afzal, M.; Alarifi, A.; Shi, C. Sensing and Photocatalytic Properties of a New 1D Zn(II)-Based Coordination Polymer Derived from the 3,5-Dibromosalicylaldehyde Nicotinoylhydrazone Ligand. Polyhedron 2022, 222, 115900. [Google Scholar] [CrossRef]
- Rani, M.; Devi, J.; Kumar, B.; Arora, T.; Taxak, B. Tridentate Xanthene-Based Hydrazone Ligands and Their Mononuclear Transition Metal Complexes: Synthesis, Anti-Malarial, Antimicrobial and Molecular Docking Studies. Res. Chem. Intermed. 2024, 50, 1409–1434. [Google Scholar] [CrossRef]
- Zahirović, A.; Hadžalić, S.; Višnjevac, A.; Fočak, M.; Tüzün, B.; Žilić, D.; Roca, S.; Jurec, J.; Topčagić, A.; Osmanković, I. Vanadium(IV) Complexes of Salicylaldehyde-Based Furoic Acid Hydrazones: Synthesis, BSA Binding and in Vivo Antidiabetic Potential. J. Inorg. Biochem. 2023, 244, 112232. [Google Scholar] [CrossRef]
- Adhikari, H.S.; Garai, A.; Khanal, C.; Yadav, P.N. Synthesis and Comprehensive Characterization with Anticancer Activity Assessment of Salicylaldehyde and 2-Acetylphenol Based Chitosan Thiosemicarbazones and Their Copper(II) Complexes. Carbohydr. Polym. Technol. Appl. 2024, 7, 100469. [Google Scholar] [CrossRef]
- Scaccaglia, M.; Pinelli, S.; Manini, L.; Ghezzi, B.; Nicastro, M.; Heinrich, J.; Kulak, N.; Mozzoni, P.; Pelosi, G.; Bisceglie, F. Gold(III) Complexes with Thiosemicarbazone Ligands: Insights into Their Cytotoxic Effects on Lung Cancer Cells. J. Inorg. Biochem. 2024, 251, 112438. [Google Scholar] [CrossRef]
- Andriichuk, Y.M.; Lyapunov, A.Y.; Hotynchan, A.H.; Savchenko, D.V.; Karavan, V.V.; Turash, M.M.; Khalavka, Y.B. Synergetic Radical-Scavenging Effect in the Complex of Copper(II) with the Thiosemicarbazone of Salicylaldehyde. J. Coord. Chem. 2023, 76, 1763–1775. [Google Scholar] [CrossRef]
- Elo, H.; Kuure, M.; Pelttari, E. Correlation of the Antimicrobial Activity of Salicylaldehydes with Broadening of the NMR Signal of the Hydroxyl Proton. Possible Involvement of Proton Exchange Processes in the Antimicrobial Activity. Eur. J. Med. Chem. 2015, 92, 750–753. [Google Scholar] [CrossRef] [PubMed]
- Pelttari, E.; Lehtinen, M.; Elo, H. Substituted Salicylaldehydes as Potential Antimicrobial Drugs: Minimal Inhibitory and Microbicidal Concentrations. Z. Naturforsch.—Sect. C J. Biosci. 2011, 66, 571–580. [Google Scholar] [CrossRef]
- Kordestani, N.; Rudbari, H.A.; Fernandes, A.R.; Raposo, L.R.; Baptista, P.V.; Ferreira, D.; Bruno, G.; Bella, G.; Scopelliti, R.; Braun, J.D.; et al. Antiproliferative Activities of Diimine-Based Mixed Ligand Copper(II) Complexes. ACS Comb. Sci. 2020, 22, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Varadwaj, P.R.; Varadwaj, A.; Marques, H.M. Halogen Bonding: A Halogen-Centered Noncovalent Interaction Yet to Be Understood. Inorganics 2019, 7, 40. [Google Scholar] [CrossRef]
- Feng, D.; Wang, B.; Wang, L.; Abraham, N.; Tao, K.; Huang, L.; Shi, W.; Dong, Y.; Qu, Y. Pre-Ischemia Melatonin Treatment Alleviated Acute Neuronal Injury after Ischemic Stroke by Inhibiting Endoplasmic Reticulum Stress-Dependent Autophagy via PERK and IRE1 Signalings. J. Pineal Res. 2017, 62, e12395. [Google Scholar] [CrossRef]
- Thapa, K.; Khan, H.; Singh, T.G.; Kaur, A. Traumatic Brain Injury: Mechanistic Insight on Pathophysiology and Potential Therapeutic Targets. J. Mol. Neurosci. 2021, 71, 1725–1742. [Google Scholar] [CrossRef] [PubMed]
- Vitomirov, T.; Dimiza, F.; Matić, I.Z.; Stanojković, T.; Pirković, A.; Živković, L.; Spremo-Potparević, B.; Novaković, I.; Anđelković, K.; Milčić, M.; et al. Copper(II) Complexes with 4-(Diethylamino)Salicylaldehyde and α-Diimines: Anticancer, Antioxidant, Antigenotoxic Effects and Interaction with DNA and Albumins. J. Inorg. Biochem. 2022, 235, 111942. [Google Scholar] [CrossRef]
- Ntanatsidis, S.; Perontsis, S.; Konstantopoulou, S.; Kalogiannis, S.; Hatzidimitriou, A.G.; Papadopoulos, A.N.; Psomas, G. Manganese(II) Complexes of Substituted Salicylaldehydes and α-Diimines: Synthesis, Characterization and Biological Activity. J. Inorg. Biochem. 2022, 227, 111693. [Google Scholar] [CrossRef]
- Papadopoulos, Z.; Doulopoulou, E.; Zianna, A.; Hatzidimitriou, A.G.; Psomas, G. Copper(II) Complexes of 5–Fluoro–Salicylaldehyde: Synthesis, Characterization, Antioxidant Properties, Interaction with DNA and Serum Albumins. Molecules 2022, 27, 8929. [Google Scholar] [CrossRef]
- Zianna, A.; Vradi, E.; Hatzidimitriou, A.G.; Kalogiannis, S.; Psomas, G. Zinc(II) Complexes of 3-Bromo-5-Chloro-Salicylaldehyde: Characterization and Biological Activity. Dalton Trans. 2022, 51, 17629–17641. [Google Scholar] [CrossRef]
- Zianna, A.; Geromichalos, G.; Psoma, E.; Kalogiannis, S.; Hatzidimitriou, A.G.; Psomas, G. Structure and in Vitro and in Silico Biological Activity of Zinc(II) Complexes with 3,5–Dichloro–Salicylaldehyde. J. Inorg. Biochem. 2022, 229, 111727. [Google Scholar] [CrossRef]
- Zianna, A.; Geromichalou, E.; Geromichalos, G.; Fiotaki, A.M.; Hatzidimitriou, A.G.; Kalogiannis, S.; Psomas, G. Zinc(II) Complexes of 3,5–Dibromo–Salicylaldehyde and α–Diimines: Synthesis, Characterization and in Vitro and in Silico Biological Profile. J. Inorg. Biochem. 2022, 226, 111659. [Google Scholar] [CrossRef]
- Christidou, A.; Zavalani, K.; Hatzidimitriou, A.G.; Psomas, G. Copper(II) Complexes with 3,5–Dihalogeno–Salicylaldehydes: Synthesis, Structure and Interaction with DNA and Albumins. J. Inorg. Biochem. 2023, 238, 112049. [Google Scholar] [CrossRef] [PubMed]
- Aryaeifar, M.; Rudbari, H.A.; Moreno-Pineda, E.; Cuevas-Vicario, J.V.; Paul, S.; Schulze, M.; Wernsdorfer, W.; Lloret, F.; Moini, N.; Blacque, O. Synthesis, Characterization and Magnetic Properties of Halogenated Tetranuclear Cubane-like Nickel(II) Complexes. New J. Chem. 2024, 48, 3603–3613. [Google Scholar] [CrossRef]
- Zianna, A.; Geromichalos, G.; Fiotaki, A.M.; Hatzidimitriou, A.G.; Kalogiannis, S.; Psomas, G. Palladium(II) Complexes of Substituted Salicylaldehydes: Synthesis, Characterization and Investigation of Their Biological Profile. Pharmaceuticals 2022, 15, 886. [Google Scholar] [CrossRef]
- Fontecilla-Camps, J.C. Nickel and the Origin and Early Evolution of Life. Metallomics 2022, 14, 16. [Google Scholar] [CrossRef]
- Meyer, F.; Kozlowski, H. Nickel. Compr. Coord. Chem. II 2004, 6, 247–554. [Google Scholar] [CrossRef]
- Kumar, S.; Trivedi, A.V. A Review on Role of Nickel in the Biological System. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 719–727. [Google Scholar] [CrossRef]
- Prueitt, R.L.; Li, W.; Chang, Y.C.; Boffetta, P.; Goodman, J.E. Systematic Review of the Potential Respiratory Carcinogenicity of Metallic Nickel in Humans. Crit. Rev. Toxicol. 2020, 50, 605–639. [Google Scholar] [CrossRef]
- Buttice, C. Nickel. In The SAGE Encyclopedia of Cancer and Society; Colditz, G.A., Ed.; SAGE Publications: Thousand Oaks, CA, USA, 2015; Volume 2, pp. 828–831. [Google Scholar]
- Zhao, J.; Shi, X.; Castranova, V.; Ding, M. Occupational Toxicology of Nickel and Nickel Compounds. J. Environ. Pathol. Toxicol. Oncol. 2009, 28, 177–208. [Google Scholar] [CrossRef]
- Barceloux, D.G. Nickel. J. Toxicol. Clin. Toxicol. 1999, 37, 239–258. [Google Scholar] [CrossRef]
- Dixon, N.E.; Gazzola, C.; Blakeley, R.L.; Zerner, B. Jack Bean Urease (EC 3.5.1.5). A Metalloenzyme. A Simple Biological Role for Nickel? J. Am. Chem. Soc. 1975, 97, 4131–4133. [Google Scholar] [CrossRef]
- Company, A.; McDonald, A.R. Bio-Relevant Chemistry of Nickel. In Comprehensive Coordination Chemistry III; Constable Edwin, C., Parkin, G., Que, L., Jr., Eds.; Elsevier: Hoboken, NJ, USA, 2021; Volume 8, pp. 846–877. ISBN 9780081026885. [Google Scholar]
- Glazer, E.C.; Casini, A. Metals in Medicine. Eur. J. Inorg. Chem. 2022, 2022, e202101093. [Google Scholar] [CrossRef]
- Orvig, C.; Abrams, M.J. Medicinal Inorganic Chemistry: Introduction. Chem. Rev. 1999, 99, 2202–2203. [Google Scholar] [CrossRef]
- Bisceglie, F.; Pinelli, S.; Alinovi, R.; Goldoni, M.; Mutti, A.; Camerini, A.; Piola, L.; Tarasconi, P.; Pelosi, G. Cinnamaldehyde and Cuminaldehyde Thiosemicarbazones and Their Copper(II) and Nickel(II) Complexes: A Study to Understand Their Biological Activity. J. Inorg. Biochem. 2014, 140, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Betanzos-Lara, S.; Gómez-Ruiz, C.; Barrón-Sosa, L.R.; Gracia-Mora, I.; Flores-Álamo, M.; Barba-Behrens, N. Cytotoxic Copper(II), Cobalt(II), Zinc(II), and Nickel(II) Coordination Compounds of Clotrimazole. J. Inorg. Biochem. 2012, 114, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Haribabu, J.; Jeyalakshmi, K.; Arun, Y.; Bhuvanesh, N.S.P.; Perumal, P.T.; Karvembu, R. Synthesis, DNA/Protein Binding, Molecular Docking, DNA Cleavage and in Vitro Anticancer Activity of Nickel(II) Bis(Thiosemicarbazone) Complexes. RSC Adv. 2015, 5, 46031–46049. [Google Scholar] [CrossRef]
- Kisa, D.; Korkmaz, N.; Taslimi, P.; Tuzun, B.; Tekin, Ş.; Karadag, A.; Sen, F. Bioactivity and Molecular Docking Studies of Some Nickel Complexes: New Analogues for the Treatment of Alzheimer, Glaucoma and Epileptic Diseases. Bioorg Chem. 2020, 101, 104066. [Google Scholar] [CrossRef] [PubMed]
- Alomar, K.; Landreau, A.; Allain, M.; Bouet, G.; Larcher, G. Synthesis, Structure and Antifungal Activity of Thiophene-2,3- Dicarboxaldehyde Bis(Thiosemicarbazone) and Nickel(II), Copper(II) and Cadmium(II) Complexes: Unsymmetrical Coordination Mode of Nickel Complex. J. Inorg. Biochem. 2013, 126, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Shawish, H.B.; Wong, W.Y.; Wong, Y.L.; Loh, S.W.; Looi, C.Y.; Hassandarvish, P.; Phan, A.Y.L.; Wong, W.F.; Wang, H.; Paterson, I.C.; et al. Nickel(II) Complex of Polyhydroxybenzaldehyde N4-Thiosemicarbazone Exhibits Anti-Inflammatory Activity by Inhibiting NF-ΚB Transactivation. PLoS ONE 2014, 9, e100933. [Google Scholar] [CrossRef] [PubMed]
- Skyrianou, K.C.; Efthimiadou, E.K.; Psycharis, V.; Terzis, A.; Kessissoglou, D.P.; Psomas, G. Nickel-Quinolones Interaction. Part 1—Nickel(II) Complexes with the Antibacterial Drug Sparfloxacin: Structure and Biological Properties. J. Inorg. Biochem. 2009, 103, 1617–1625. [Google Scholar] [CrossRef]
- Xu, B.B.; Pei, X.S.; Guan, Q.Y.; Shi, X.; Zhao, G.L. Synthesis, Crystal Structure, and Biological Activity of a Nickel(II) Complex Constructed by 2-Phenyl-4-Selenazole Carboxylic Acid and 1,10-Phenanthroline. J. Coord. Chem. 2013, 66, 2605–2614. [Google Scholar] [CrossRef]
- Totta, X.; Papadopoulou, A.A.; Hatzidimitriou, A.G.; Papadopoulos, A.; Psomas, G. Synthesis, Structure and Biological Activity of Nickel(II) Complexes with Mefenamato and Nitrogen-Donor Ligands. J. Inorg. Biochem. 2015, 145, 79–93. [Google Scholar] [CrossRef]
- Perontsis, S.; Hatzidimitriou, A.G.; Papadopoulos, A.N.; Psomas, G. Nickel-Diflunisal Complexes: Synthesis, Characterization, in Vitro Antioxidant Activity and Interaction with DNA and Albumins. J. Inorg. Biochem. 2016, 162, 9–21. [Google Scholar] [CrossRef]
- Lazou, M.; Hatzidimitriou, A.G.; Papadopoulos, A.N.; Psomas, G. Transition Metal(II) Complexes with the Non–Steroidal Anti–Inflammatory Drug Oxaprozin: Characterization and Biological Profile. J. Inorg. Biochem. 2023, 243, 112196. [Google Scholar] [CrossRef]
- Barmpa, A.; Geromichalos, G.D.; Hatzidimitriou, A.G.; Psomas, G. Nickel(II)–Meclofenamate Complexes: Structure, in Vitro and in Silico DNA– and Albumin–Binding Studies, Antioxidant and Anticholinergic Activity. J. Inorg. Biochem. 2021, 222, 111507. [Google Scholar] [CrossRef] [PubMed]
- Zianna, A.; Psomas, G.; Hatzidimitriou, A.; Lalia-Kantouri, M. Ni(II) Complexes with 2,2-Dipyridylamine and Salicylaldehydes: Synthesis, Crystal Structure and Interaction with Calf-Thymus DNA and Albumins. J. Inorg. Biochem. 2016, 163, 131–142. [Google Scholar] [CrossRef]
- Zianna, A.; Psomas, G.; Hatzidimitriou, A.; Lalia-Kantouri, M. Synthesis, Structural, Thermal Characterization and Interaction with Calf-Thymus DNA and Albumins of Cationic Ni(II) Complexes with 2,2′-Dipyridylamine and Salicylaldehydes. Polyhedron 2017, 124, 104–116. [Google Scholar] [CrossRef]
- Gkritzali, M.; Georgila, M.; Hatzidimitriou, A.G.; Kalogiannis, S.; Psomas, G. Neutral and Cationic Nickel(II) Complexes with Substituted Salicylaldehydes: Characterization, Antibacterial Activity, and Interaction with Biomacromolecules. J. Inorg. Biochem. 2023, 247, 112339. [Google Scholar] [CrossRef] [PubMed]
- Geary, W.J. The Use of Conductivity Measurements in Organic Solvents for the Characterisation of Coordination Compounds. Coord. Chem. Rev. 1971, 7, 81–122. [Google Scholar] [CrossRef]
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds: Part B: Applications in Coordination, Organometallic, and Bioinorganic Chemistry; John Wiley and Sons: Hoboken, NJ, USA, 2008; ISBN 9780471744931. [Google Scholar]
- Totta, X.; Hatzidimitriou, A.G.; Papadopoulos, A.N.; Psomas, G. Nickel(II)-Naproxen Mixed-Ligand Complexes: Synthesis, Structure, Antioxidant Activity and Interaction with Albumins and Calf-Thymus DNA. New J. Chem. 2017, 41, 4478–4492. [Google Scholar] [CrossRef]
- Stamou, P.; Hatzidimitriou, A.G.; Psomas, G. Manganese(II) Complexes with 5–Nitro–2–Hydroxy–Benzaldehyde or Substituted 2–Hydroxy–Phenones: Structure and Interaction with Bovine Serum Albumin and Calf–Thymus DNA. J. Inorg. Biochem. 2022, 235, 111923. [Google Scholar] [CrossRef] [PubMed]
- Zianna, A.; Psomas, G.; Hatzidimitriou, A.; Coutouli-Argyropoulou, E.; Lalia-Kantouri, M. Zinc Complexes of Salicylaldehydes: Synthesis, Characterization and DNA-Binding Properties. J. Inorg. Biochem. 2013, 127, 116–126. [Google Scholar] [CrossRef]
- Zianna, A.; Ristović, M.Š.; Psomas, G.; Hatzidimitriou, A.; Coutouli-Argyropoulou, E.; Lalia-Kantouri, M. Cadmium(II) Complexes of 5-Bromo-Salicylaldehyde and α-Diimines: Synthesis, Structure and Interaction with Calf-Thymus DNA and Albumins. Polyhedron 2016, 107, 136–147. [Google Scholar] [CrossRef]
- Zianna, A.; Sumar Ristovic, M.; Psomas, G.; Hatzidimitriou, A.; Coutouli-Argyropoulou, E.; Lalia-Kantouri, M. Cadmium(II) Complexes of 5-Nitro-Salicylaldehyde and α-Diimines: Synthesis, Structure and Interaction with Calf-Thymus DNA. J. Coord. Chem. 2015, 68, 4444–4463. [Google Scholar] [CrossRef]
- Papadopoulos, C.D.; Lalia-Kantouri, M.; Jaud, J.; Hatzidimitriou, A.G. Substitution Effect on New Co(II) Addition Compounds with Salicylaldehydes and the Nitrogenous Bases Phen or Neoc: Crystal and Molecular Structures of [CoII(5-NO2-Salicylaldehyde)2(Phen)], [CoII(5-CH3-Salicylaldehyde)2(Neoc)] and [CoII(5-Cl-Salicylaldehyde)2(Neoc)]. Inorg. Chim. Acta 2007, 360, 3581–3589. [Google Scholar] [CrossRef]
- Papadopoulos, C.D.; Hatzidimitriou, A.G.; Voutsas, G.P.; Lalia-Kantouri, M. Synthesis and Characterization of New Addition Compounds of Bis(Substituted-Salicylaldehydo)Cobalt(II) with 2,2′-Bipyridine (Bipy). Crystal and Molecular Structures of [CoII(3-Methoxy-Salicylaldehyde)2(Bipy)]·CH3OH (1) and [CoII(Bipy)3]Br2·0.5(5-Chloro-SalicylaldehydeH)·1.5CH3OH (5). Polyhedron 2007, 26, 1077–1086. [Google Scholar] [CrossRef]
- Lalia-Kantouri, M.; Gdaniec, M.; Czapik, A.; Chrissafis, K.; Ferenc, W.; Sarzynski, J.; Papadopoulos, C.D. Thermoanalytical, Magnetic and Structural Study of Co(II) Complexes with Substituted Salicylaldehydes and Neocuproine. J. Therm. Anal. Calorim. 2012, 109, 131–139. [Google Scholar] [CrossRef]
- Papadopoulos, C.; Cristóvão, B.; Ferenc, W.; Hatzidimitriou, A.; Ciprioti, S.V.; Risoluti, R.; Lalia-Kantouri, M. Thermoanalytical, Magnetic and Structural Investigation of Neutral Co(II) Complexes with 2,2′-Dipyridylamine and Salicylaldehydes. J. Therm. Anal. Calorim. 2016, 123, 717–729. [Google Scholar] [CrossRef]
- Pages, B.J.; Ang, D.L.; Wright, E.P.; Aldrich-Wright, J.R. Metal Complex Interactions with DNA. Dalton Trans. 2015, 44, 3505–3526. [Google Scholar] [CrossRef] [PubMed]
- Duchackova, L.; Steinmetz, V.; Lemaire, J.; Roithova, J. Comparison of 2,2′-Bipyridine and 2,2′-Bipyridyl-N,N′-Dioxide as Ligands in Zinc Complexes. Inorg. Chem. 2010, 49, 8897–8903. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.D.; Paul, A. Reversible Small Molecule–Nucleic Acid Interactions. In Nucleic Acids in Chemistry and Biology; Blackburn, G.M., Egli, M., Gait, M.J., Watts, J.K., Eds.; The Royal Society of Chemistry: London, UK, 2022; pp. 477–521. [Google Scholar]
- Wolfe, A.; Shimer, G.H.; Meehan, T. Polycyclic Aromatic Hydrocarbons Physically Intercalate into Duplex Regions of Denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef] [PubMed]
- Dimitrakopoulou, A.; Dendrinou-Samara, C.; Pantazaki, A.A.; Alexiou, M.; Nordlander, E.; Kessissoglou, D.P. Synthesis, Structure and Interactions with DNA of Novel Tetranuclear, [Mn4(II/II/II/IV)] Mixed Valence Complexes. J. Inorg. Biochem. 2008, 102, 618–628. [Google Scholar] [CrossRef]
- Gkisiou, C.; Malis, G.; Hatzidimitriou, A.G.; Psomas, G. Erbium(III) Coordination Compounds with Substituted Salicylaldehydes: Characterization and Biological Profile. J. Inorg. Biochem. 2023, 242, 112161. [Google Scholar] [CrossRef]
- Garcia-Gimenez, J.L.; Gonzalez-Alvarez, M.; Liu-Gonzalez, M.; Macías, B.; Borras, J.; Alzuet, G. Toward the Development of Metal-Based Synthetic Nucleases: DNA Binding and Oxidative DNA Cleavage of a Mixed Copper(II) Complex with N-(9H-Purin-6-yl)Benzenesulfonamide and 1,10-Phenantroline. Antitumor Activity in Human Caco-2 Cells and Jurkat T Lymphocytes. Evaluation of P53 and Bcl-2 Proteins in the Apoptotic Mechanism. J. Inorg. Biochem. 2009, 103, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: Berlin/Heidelberg, Germany, 2006; ISBN 0387312781. [Google Scholar]
- Heller, D.P.; Greenstock, C.L. Fluorescence Lifetime Analysis of DNA Intercalated Ethidium Bromide and Quenching by Free Dye. Biophys. Chem. 1994, 50, 305–312. [Google Scholar] [CrossRef] [PubMed]
- He, X.M.; Carter, D.C. Atomic Structure and Chemistry of Human Serum Albumin. Nature 1992, 358, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.E.; Christ, D.D. Chapter 33. Plasma Protein Binding of Drugs. Annu. Rep. Med. Chem. 1996, 31, 327–336. [Google Scholar] [CrossRef]
- Zhao, G.; Lin, H.; Zhu, S.; Sun, H.; Chen, Y. Dinuclear Palladium(II) Complexes Containing Two Monofunctional [Pd(En)(Pyridine)Cl]+ Units Bridged by Se or S. Synthesis, Characterization, Cytotoxicity and Kinetic Studies of DNA-Binding. J. Inorg. Biochem. 1998, 70, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Stella, L.; Capodilupo, A.L.; Bietti, M. A Reassessment of the Association between Azulene and [60]Fullerene. Possible Pitfalls in the Determination of Binding Constants through Fluorescence Spectroscopy. Chem. Commun. 2008, 4744–4746. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.S.; Yuan, W.B.; Wang, H.Y.; Zhang, Q.; Liu, M.; Yu, K.B. Synthesis, Crystal Structure and Interaction with DNA and HSA of (N,N′-Dibenzylethane-1,2-Diamine) Transition Metal Complexes. J. Inorg. Biochem. 2008, 102, 2026–2034. [Google Scholar] [CrossRef] [PubMed]
- Selaković, S.; Rodić, M.V.; Novaković, I.; Matić, I.Z.; Stanojković, T.; Pirković, A.; Živković, L.; Spremo-Potparević, B.; Milčić, M.; Medaković, V.; et al. Cu(II) Complexes with a Salicylaldehyde Derivative and α-Diimines as Co-Ligands: Synthesis, Characterization, Biological Activity. Experimental and Theoretical Approach. Dalton Trans. 2024, 53, 2770–2788. [Google Scholar] [CrossRef] [PubMed]
- Laitinen, O.H.; Hytönen, V.P.; Nordlund, H.R.; Kulomaa, M.S. Genetically Engineered Avidins and Streptavidins. Cell. Mol. Life Sci. 2006, 63, 2992–3017. [Google Scholar] [CrossRef]
- Marmur, J. A Procedure for the Isolation of Deoxyribonucleic Acid from Micro-Organisms. J. Mol. Biol. 1961, 3, 208–218. [Google Scholar] [CrossRef]
- Reichmann, M.E.; Rice, S.A.; Thomas, C.A.; Doty, P. A Further Examination of the Molecular Weight and Size of Desoxypentose Nucleic Acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]
- Bruker Analytical X-ray Systems, Inc. Apex2, Version 2 User Manual, M86–E01078; Bruker Analytical X-ray Systems, Inc.: Billerica, MA, USA, 2006. [Google Scholar]
- Siemens Industrial Automation, Inc. SADABS: Area–Detector Absorption Correction; Siemens Industrial Automation, Inc.: Madison, WI, USA, 1996. [Google Scholar]
- Palatinus, L.; Chapuis, G. SUPERFLIP—A Computer Program for the Solution of Crystal Structures by Charge Flipping in Arbitrary Dimensions. J. Appl. Crystallogr. 2007, 40, 786–790. [Google Scholar] [CrossRef]
- Betteridge, P.W.; Carruthers, J.R.; Cooper, R.I.; Prout, K.; Watkin, D.J. CRYSTALS Version 12: Software for Guided Crystal Structure Analysis. J. Appl. Crystallogr. 2003, 36, 1487. [Google Scholar] [CrossRef]


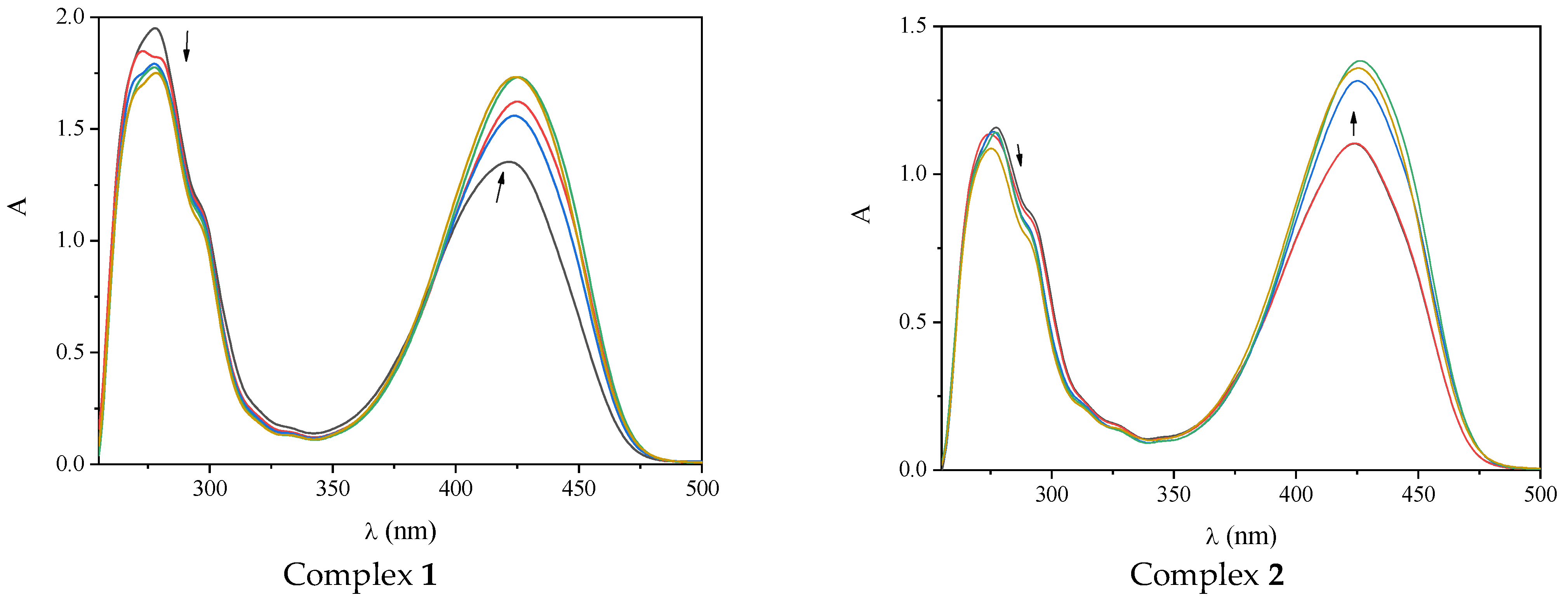


| Complex 1 | |
|---|---|
| Crystal data | |
| Chemical formula | C28H18Br4N2NiO4 |
| Mr | 824.79 |
| Crystal system | Monoclinic |
| Space group | P21/c |
| Temperature (K) | 295 |
| a (Å) | 16.604 (8) |
| b (Å) | 24.002 (13) |
| c (Å) | 14.571 (8) |
| β (°) | 103.381 (12) |
| V (Å3) | 5649 (5) |
| Z | 8 |
| Radiation type | Mo Kα |
| µ (mm−1) | 6.38 |
| Crystal size (mm) | 0.15 × 0.12 × 0.11 |
| Data collection | |
| Diffractometer | Bruker Kappa Apex2 |
| Absorption correction | Numerical Analytical Absorption (De Meulenaer and Tompa, 1965) |
| Tmin, Tmax | 0.46, 0.50 |
| No. of measured reflections | 58,802 |
| No. of independent reflections | 10,738 |
| No. of observed [I > 2.0σ(I)] reflections | 7018 |
| Rint | 0.042 |
| (sin θ/λ)max (Å−1) | 0.613 |
| Refinement | |
| R[F2 > 2σ(F2)] | 0.038 |
| wR(F2) | 0.085 |
| S | 1.00 |
| No. of reflections | 7018 |
| No. of parameters | 703 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.49, −0.98 |
| 1A | 1B | ||
|---|---|---|---|
| Bond | Length (Å) | Bond | Length (Å) |
| Ni1—O1 | 2.106(4) | Ni2—O5 | 2.092(4) |
| Ni1—O2 | 2.018(4) | Ni2—O6 | 2.033(4) |
| Ni1—O3 | 2.097(5) | Ni2—O7 | 2.060(4) |
| Ni1—O4 | 1.953(4) | Ni2—O8 | 2.037(4) |
| Ni1—N1 | 2.115(5) | Ni2—N3 | 2.134(5) |
| Ni1—N2 | 2.160(5) | Ni2—N4 | 2.103(5) |
| Bonds | Angle (°) | Bonds | Angle (°) |
| O1—Ni1—O2 | 86.99(16) | O5—Ni2—O6 | 89.41(17) |
| O1—Ni1—O4 | 170.44(17) | O5—Ni2—O8 | 173.40(18) |
| O2—Ni1—N1 | 173.05(19) | O6—Ni2—N3 | 172.78(17) |
| O3—Ni1—O4 | 87.93(19) | O7—Ni2—O8 | 88.58(17) |
| O3—Ni1—N2 | 172.04(17) | O7—Ni2—N4 | 171.09(17) |
| N1—Ni1—N2 | 78.3(2) | N3—Ni2—N4 | 78.54(19) |
| Compound | λmax (nm) ((ΔA/Aο, %) a (Δλ, nm) b) | Kb (M−1) |
|---|---|---|
| 3,5–diΒr–saloH [28] | 337 (<–50 c, elim d), 427(>+50 c, 0) | 3.71(±0.14) × 105 |
| Complex 1 | 277 (–10, +1); 422 (+35, +3) | 4.22(±0.86) × 105 |
| Complex 2 | 277 (–8, 0); 425(+25, +4) | 3.26(±0.75) × 105 |
| Complex 3 | 278(–6, 0); 422 (+30, +3) | 2.83(±0.70) × 105 |
| Compound | ΔΙ/Ι0(%) | Κsv (M−1) | Kq (M−1s−1) |
|---|---|---|---|
| 3,5–diΒr–saloH [28] | 46.6 | 3.95(±0.10) × 104 | 1.72(±0.04) × 1012 |
| Complex 1 | 54.8 | 4.11(±0.11) × 104 | 1.78(±0.05) × 1012 |
| Complex 2 | 51.8 | 8.56(±0.15) × 104 | 3.72(±0.06) × 1012 |
| Complex 3 | 58.7 | 5.33(±0.09) × 104 | 2.32(±0.04) × 1012 |
| Compound | Kq(BSA) (M−1s−1) | Κ(BSA) (M−1) | Kq(HSA) (M−1s−1) | Κ(HSA) (M−1) |
|---|---|---|---|---|
| 3,5–diΒr–saloH [28] | 1.63(±0.02) × 1014 | 5.76(±0.30) × 106 | 5.08(±0.15) × 1013 | 1.55(±0.10) × 106 |
| Complex 1 | 1.20(±0.10) × 1013 | 1.01(±0.10) × 105 | 8.72(±0.21) × 1012 | 2.23(±0.21) × 104 |
| Complex 2 | 3.43(±0.70) × 1013 | 8.21(±0.70) × 104 | 1.62(±0.15) × 1013 | 2.50(±0.15) × 104 |
| Complex 3 | 8.97(±0.50) × 1012 | 1.89(±0.50) × 105 | 6.00(±0.23) × 1012 | 9.13(±0.23) × 104 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Psarras, G.I.; Zianna, A.; Hatzidimitriou, A.G.; Psomas, G. Coordination Compounds of Nickel(II) with 3,5–Dibromo–Salicylaldehyde: Structure and Interaction with Biomolecules. Inorganics 2024, 12, 138. https://doi.org/10.3390/inorganics12050138
Psarras GI, Zianna A, Hatzidimitriou AG, Psomas G. Coordination Compounds of Nickel(II) with 3,5–Dibromo–Salicylaldehyde: Structure and Interaction with Biomolecules. Inorganics. 2024; 12(5):138. https://doi.org/10.3390/inorganics12050138
Chicago/Turabian StylePsarras, Georgios I., Ariadni Zianna, Antonios G. Hatzidimitriou, and George Psomas. 2024. "Coordination Compounds of Nickel(II) with 3,5–Dibromo–Salicylaldehyde: Structure and Interaction with Biomolecules" Inorganics 12, no. 5: 138. https://doi.org/10.3390/inorganics12050138






