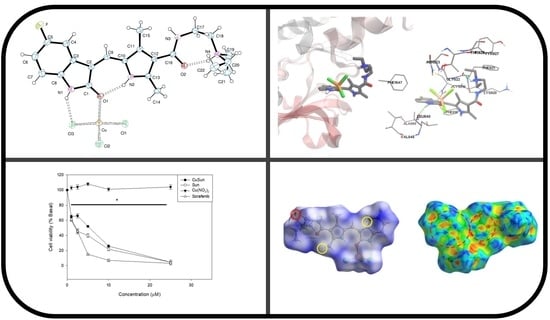
Synthesis and Characterization of Novel Copper(II)-Sunitinib Complex: Molecular Docking, DFT Studies, Hirshfeld Analysis and Cytotoxicity Studies
Abstract
:1. Introduction
2. Results and Discussion
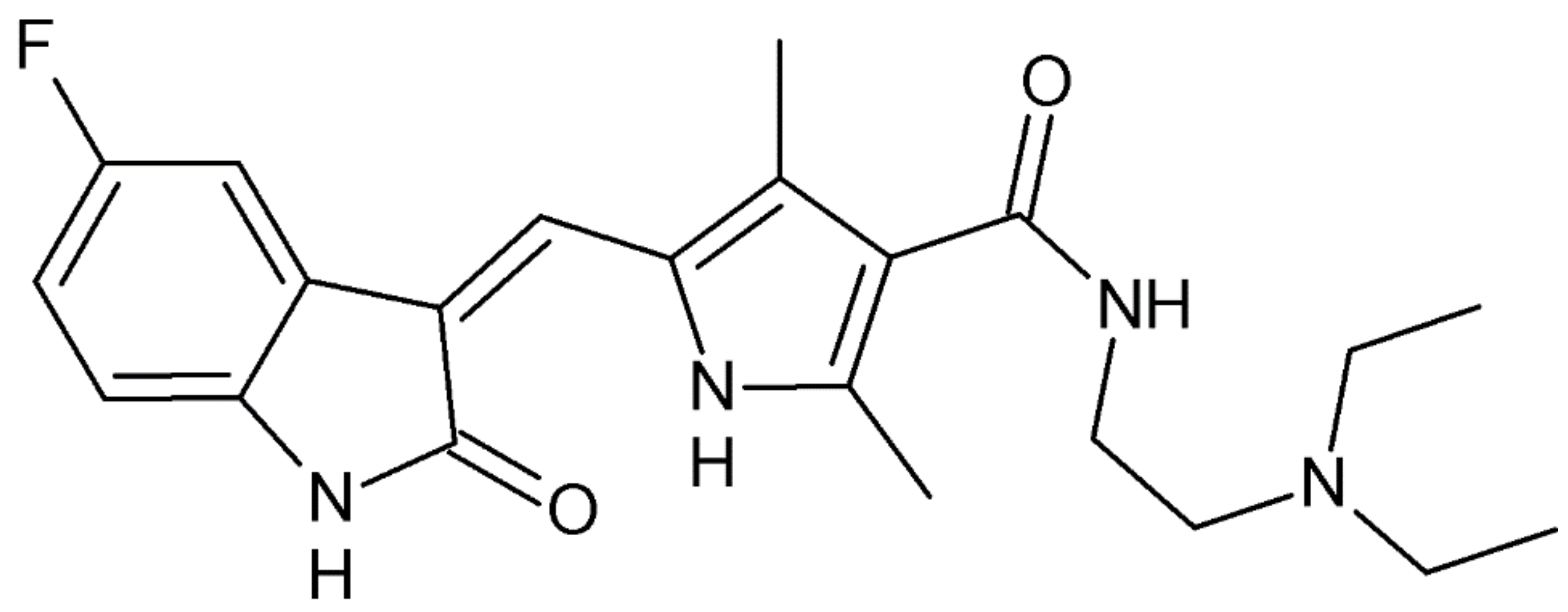
2.1. Synthesis and Characterization
2.2. Crystal Structure
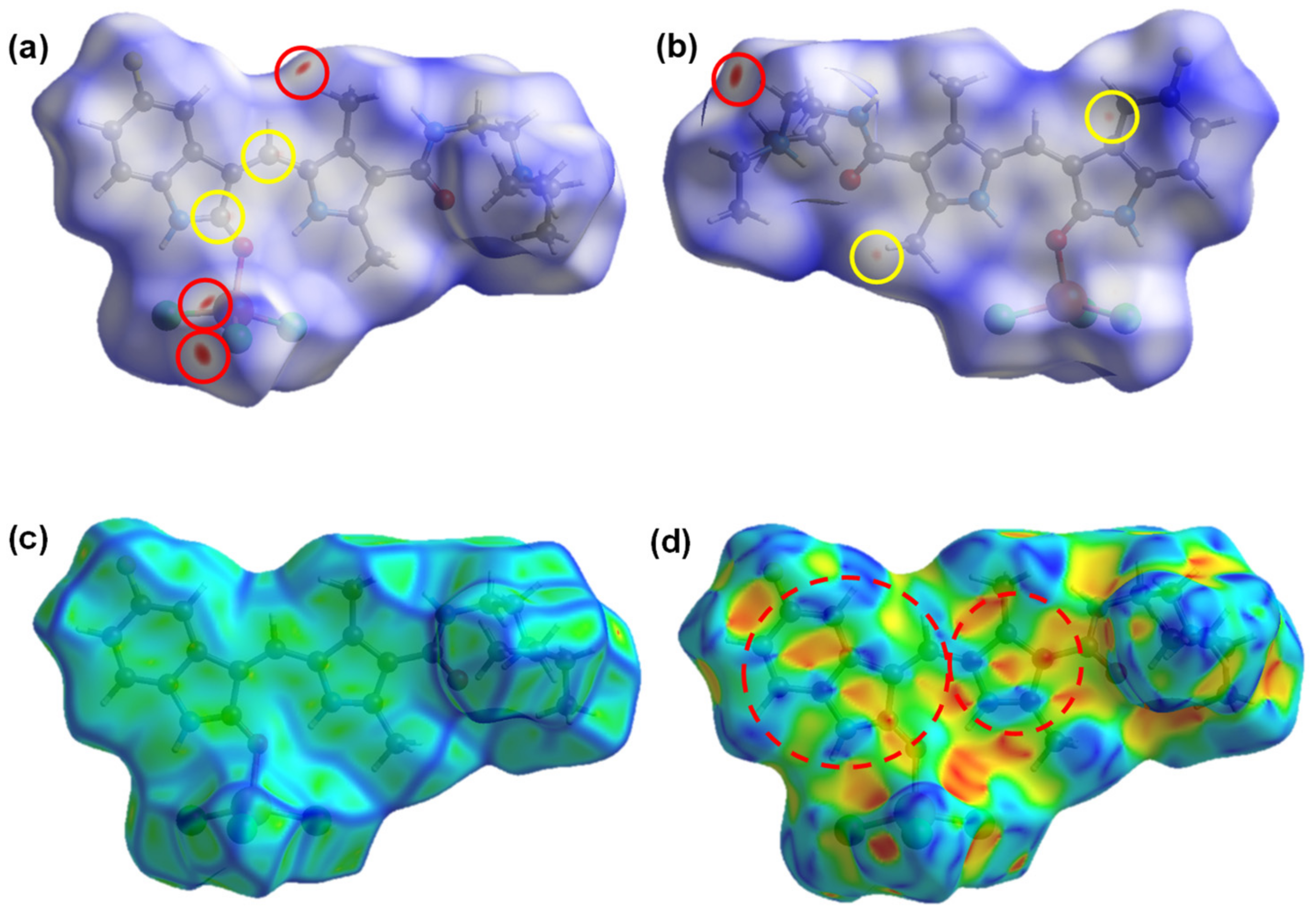
2.3. Hirshfeld Surface
2.4. Vibrational Spectroscopy
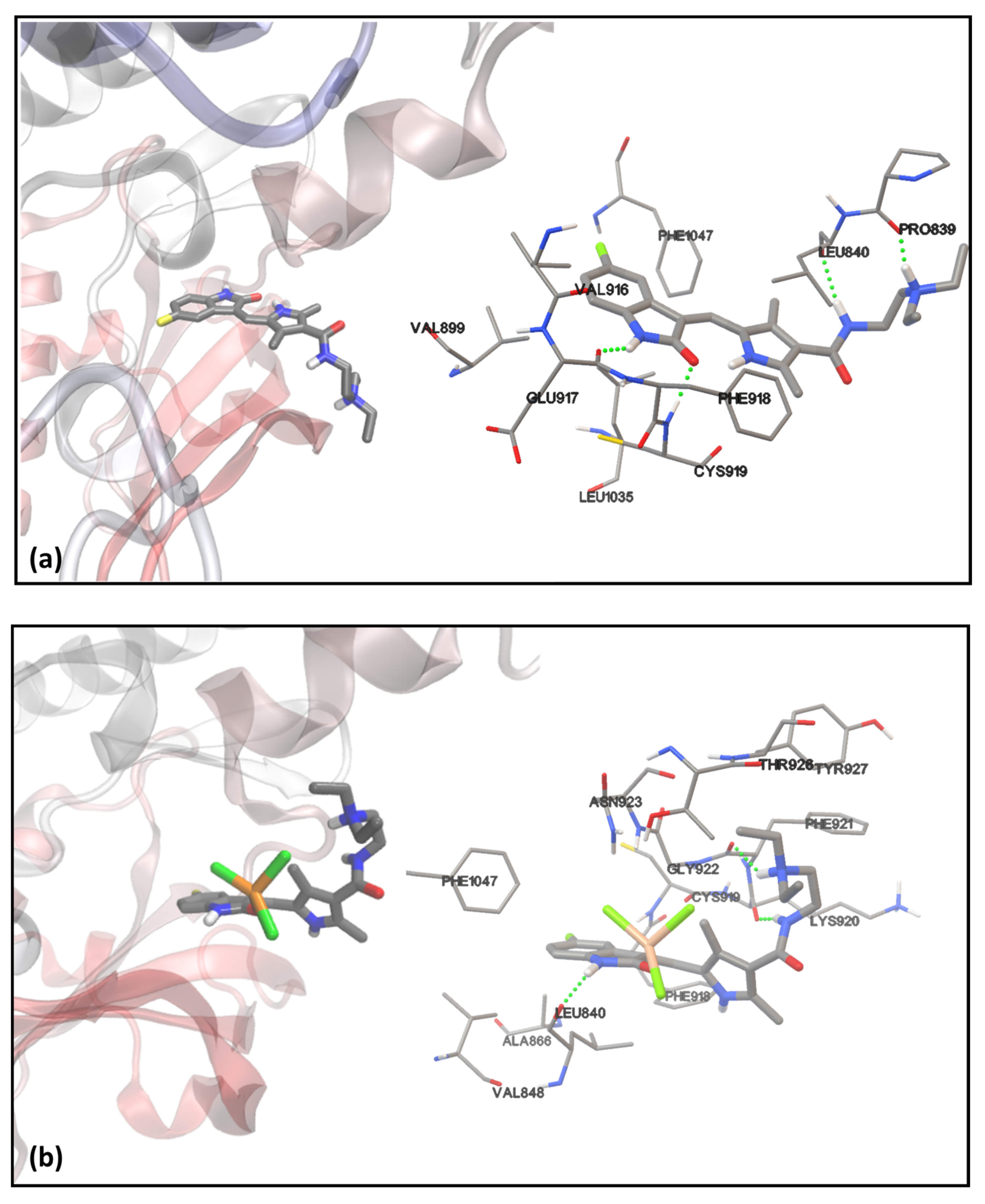
2.5. Molecular Docking with VEGFR2
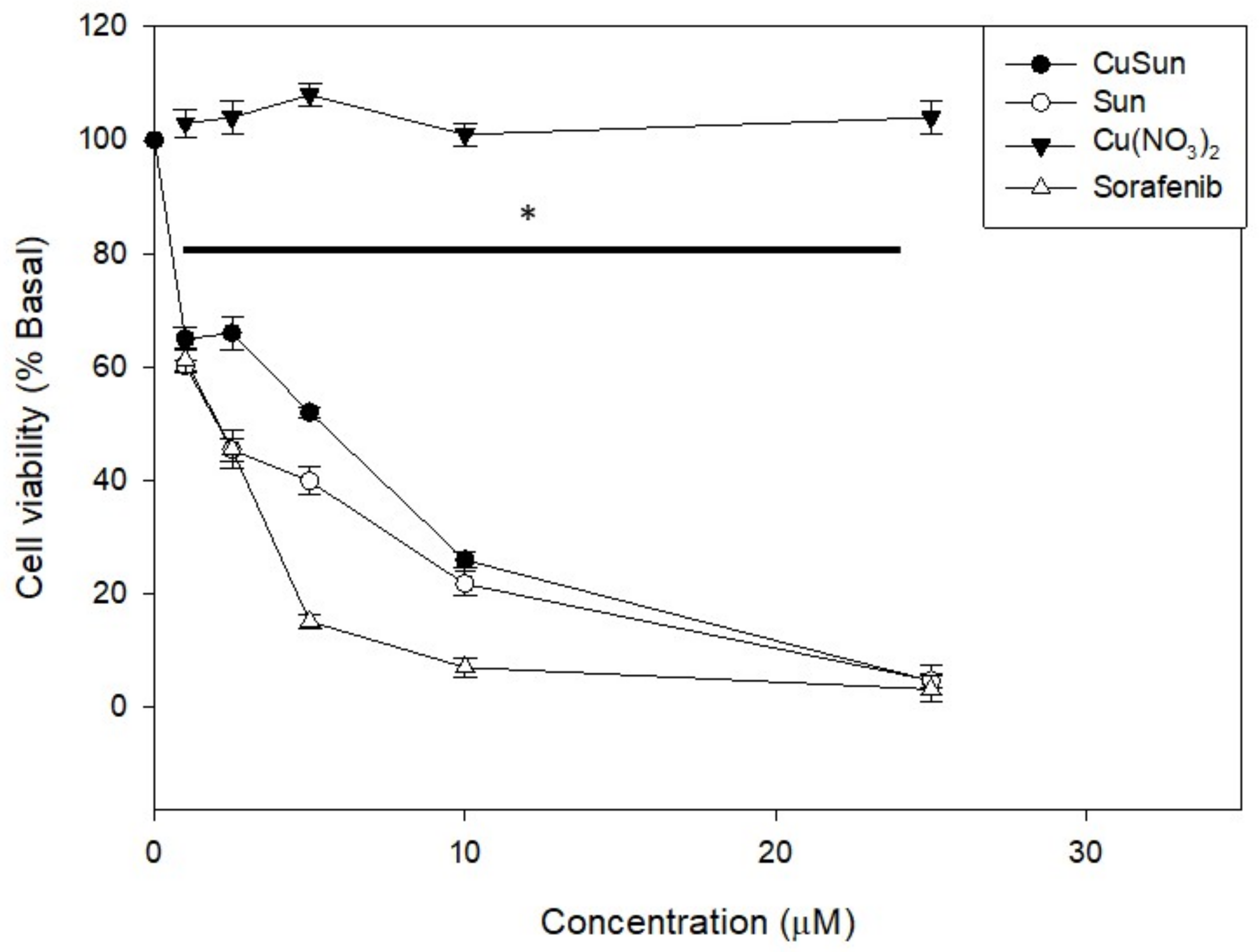
2.6. Cytotoxicity Study
3. Materials and Methods
3.1. Instruments, Reagents and Materials
3.2. X ray Diffraction Data
3.3. Computational Simulations
3.3.1. Hirshfeld Surface Analysis
3.3.2. Geometry Optimization of the CuSun Complex and FTIR Spectra Simulation
3.3.3. Molecular Docking with VEGFR2
3.4. Cell Culture Conditions and Viability Studies
3.4.1. Cell Line and Growth Conditions
3.4.2. Cell Viability Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerbel, R.; Folkman, J. Clinical translation of angiogenesis inhibitors. Nat. Rev. Cancer 2002, 2, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.; Haworth, L.; Sherry, R.M.; Hwu, P.; Schwartzentruber, D.J.; Topalian, S.L.; Steinberg, S.M.; Chen, H.X.; Rosenberg, S.A. A randomized trial of bevacizumab, an anti-vascular endothelial growth factor antibody, for metastatic renal cancer. N. Engl. J. Med. 2003, 349, 427–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrara, N.; Hillan, K.J.; Gerber, H.-P.; Novotny, W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat. Rev. Drug Discov. 2004, 3, 391–400. [Google Scholar] [CrossRef]
- Willett, C.G.; Boucher, Y.; Di Tomaso, E.; Duda, D.G.; Munn, L.L.; Tong, R.T.; Chung, D.C.; Sahani, D.V.; Kalva, S.P.; Kozin, S.V.; et al. Direct evidence that the VEGF-specific antibody bevacizumab has antivascular effects in human rectal cancer. Nat. Med. 2004, 10, 145–147. [Google Scholar] [CrossRef]
- Shaheen, R.M.; Tseng, W.W.; Davis, D.W.; Liu, W.; Reinmuth, N.; Vellagas, R.; Wieczorek, A.A.; Ogura, Y.; McConkey, D.J.; Drazan, K.E.; et al. Tyrosine kinase inhibition of multiple angiogenic growth factor receptors improves survival in mice bearing colon cancer liver metastases by inhibition of endothelial cell survival mechanisms. Cancer Res. 2001, 61, 1464–1468. [Google Scholar]
- Bergers, G.; Song, S.; Meyer-Morse, N.; Bergsland, E.; Hanahan, D. Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J. Clin. Investig. 2003, 111, 1287–1295. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, T.; Eisen, T. Kinase inhibition with BAY 43-9006 in renal cell carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2004, 10, 6388S–6392S. [Google Scholar] [CrossRef] [Green Version]
- Abrams, T.J.; Lee, L.B.; Murray, L.J.; Pryer, N.K.; Cherrington, J.M. SU11248 inhibits KIT and platelet-derived growth factor receptor beta in preclinical models of human small cell lung cancer. Mol. Cancer Ther. 2003, 2, 471–478. [Google Scholar]
- Mendel, D.B.; Laird, A.D.; Xin, X.; Louie, S.G.; Schreck, R.E.; Abrams, T.J.; Ngai, T.J.; Lee, L.B.; Murray, L.J.; Carver, J.; et al. In vivo Antitumor Activity of SU11248, a Novel Tyrosine Kinase Inhibitor Targeting Vascular Endotelial Growth Factor Receptors: Determination of a Parmacokinetic/Pharmacodynamic Relationship. Clin. Cancer Res. 2003, 9, 327–337. [Google Scholar]
- Carmeliet, P. VEGF as a key mediator of angiogenesis in cancer. Oncology 2005, 69 (Suppl. 3), 4–10. [Google Scholar] [CrossRef]
- Verheul, H.M.W.; Pinedo, H.M. The Role of Vascular Endothelial Growth Factor (VEGF) in Tumor Angiogenesis and Early Clinical Development of VEGFReceptor Kinase Inhibitors. Clin. Breast Cancer 2000, 1, S80–S84. [Google Scholar] [CrossRef]
- Lian, L.; Li, X.-L.; Xu, M.-D.; Li, X.-M.; Wu, M.-Y.; Zhang, Y.; Tao, M.; Li, W.; Shen, X.-M.; Zhou, C.; et al. VEGFR2 promotes tumorigenesis and metastasis in a pro-angiogenic-independent way in gastric cancer. BMC Cancer 2019, 19, 183. [Google Scholar] [CrossRef]
- Guo, S.; Colbert, L.S.; Fuller, M.; Zhang, Y.; Gonzalez-Perez, R.R. Vascular endothelial growth factor receptor-2 in breast cancer. Biochim. Biophys. Acta Rev. Cancer 2010, 1806, 108–121. [Google Scholar] [CrossRef] [Green Version]
- Committee for Medicinal Products for Human Use (CHMP). Sunitinib Accord Procedure No. EMEA/H/C/005419/0000. 2020. Available online: https://www.ema.europa.eu/en/documents/assessment-report/sunitinib-accord-epar-public-assessment-report_en.pdf (accessed on 21 December 2021).
- Ferrari, S.M.; Centanni, M.; Virili, C.; Miccoli, M.; Ferrari, P.; Ruffilli, I.; Ragusa, F.; Antonelli, A.; Fallahi, P. Sunitinib in the Treatment of Thyroid Cancer. Curr. Med. Chem. 2019, 26, 963–972. [Google Scholar] [CrossRef]
- Elgebaly, A.; Menshawy, A.; El Ashal, G.; Osama, O.; Ghanem, E.; Omar, A.; Negida, A. Sunitinib alone or in combination with chemotherapy for the treatment of advanced breast cancer: A systematic review and meta-analysis. Breast Dis. 2016, 36, 91–101. [Google Scholar] [CrossRef]
- Pokuri, V.K.; Tomaszewski, G.M.; Ait-Oudhia, S.; Groman, A.; Khushalani, N.I.; Lugade, A.A.; Thanavala, Y.; Ashton, E.A.; Grande, C.; Fetterly, G.J.; et al. Efficacy, Safety, and Potential Biomarkers of Sunitinib and Transarterial Chemoembolization (TACE) Combination in Advanced Hepatocellular Carcinoma (HCC): Phase II Trial. Am. J. Clin. Oncol. 2018, 41, 332–338. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, X.; Tang, Q.; Zhang, F.; Li, Y.; Feng, Z.; Zhu, J. Prognostic significance and potential therapeutic target of VEGFR2 in hepatocellular carcinoma. J. Clin. Pathol. 2011, 64, 343–348. [Google Scholar] [CrossRef]
- Lankheet, N.A.G.; Kloth, J.S.L.; Gadellaa-van Hooijdonk, C.G.M.; Cirkel, G.A.; Mathijssen, R.H.J.; Lolkema, M.P.J.K.; Schellens, J.H.M.; Voest, E.E.; Sleijfer, S.; de Jonge, M.J.A.; et al. Pharmacokinetically guided sunitinib dosing: A feasibility study in patients with advanced solid tumours. Br. J. Cancer 2014, 110, 2441–2449. [Google Scholar] [CrossRef] [Green Version]
- Demlová, R.; Turjap, M.; Peš, O.; Kostolanská, K.; Juřica, J. Therapeutic Drug Monitoring of Sunitinib in Gastrointestinal Stromal Tumors and Metastatic Renal Cell Carcinoma in Adults—A Review. Ther. Drug Monit. 2020, 42, 20–32. [Google Scholar] [CrossRef]
- Imbulgoda, A.; Heng, D.Y.C.; Kollmannsberger, C. Sunitinib in the Treatment of Advanced Solid Tumors. Small Mol. Oncol. 2014, 201, 165–184. [Google Scholar]
- Anthony, E.J.; Bolitho, E.M.; Bridgewater, H.E.; Carter, O.W.L.; Donnelly, J.M.; Imberti, C.; Lant, E.C.; Lermyte, F.; Needham, R.J.; Palau, M.; et al. Metallodrugs are unique: Opportunities and challenges of discovery and development. Chem. Sci. 2020, 11, 12888–12917. [Google Scholar] [CrossRef]
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The Next Generation of Platinum Drugs: Targeted Pt(II) Agents, Nanoparticle Delivery, and Pt(IV) Prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005, 4, 307–320. [Google Scholar] [CrossRef]
- Leon, I.; Cadavid-Vargas, J.; Di Virgilio, A.; Etcheverry, S. Vanadium, Ruthenium and Copper Compounds: A New Class of Nonplatinum Metallodrugs with Anticancer Activity. Curr. Med. Chem. 2017, 24, 112–148. [Google Scholar] [CrossRef]
- Gandin, V.; Ceresa, C.; Esposito, G.; Indraccolo, S.; Porchia, M.; Tisato, F.; Santini, C.; Pellei, M.; Marzano, C. Therapeutic potential of the phosphino Cu(I) complex (HydroCuP) in the treatment of solid tumors. Sci. Rep. 2017, 7, 13936. [Google Scholar] [CrossRef]
- Denoyer, D.; Clatworthy, S.A.S.; Cater, M.A. 16. Copper complexes in cancer therapy. In Metallo-Drugs: Development and Action of Anticancer Agents; Sigel, A., Sigel, H., Freisinger, E., Sigel, R.K.O., Eds.; De Gruyter: Berlin, Germany, 2018; pp. 469–506. [Google Scholar]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in Copper Complexes as Anticancer Agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef]
- Balsa, L.M.; Ferraresi-Curotto, V.; Lavecchia, M.J.; Echeverría, G.A.; Piro, O.E.; García-Tojal, J.; Pis-Diez, R.; González-Baró, A.C.; León, I.E. Anticancer activity of a new copper(ii) complex with a hydrazone ligand. Structural and spectroscopic characterization, computational simulations and cell mechanistic studies on 2D and 3D breast cancer cell models. Dalt. Trans. 2021, 50, 9812–9826. [Google Scholar] [CrossRef]
- Carcelli, M.; Tegoni, M.; Bartoli, J.; Marzano, C.; Pelosi, G.; Salvalaio, M.; Rogolino, D.; Gandin, V. In vitro and in vivo anticancer activity of tridentate thiosemicarbazone copper complexes: Unravelling an unexplored pharmacological target. Eur. J. Med. Chem. 2020, 194, 112266. [Google Scholar] [CrossRef]
- Shobha Devi, C.; Thulasiram, B.; Aerva, R.R.; Nagababu, P. Recent Advances in Copper Intercalators as Anticancer Agents. J. Fluoresc. 2018, 28, 1195–1205. [Google Scholar] [CrossRef]
- Shi, X.; Chen, Z.; Wang, Y.; Guo, Z.; Wang, X. Hypotoxic copper complexes with potent anti-metastatic and anti-angiogenic activities against cancer cells. Dalt. Trans. 2018, 47, 5049–5054. [Google Scholar] [CrossRef] [PubMed]
- Balsa, L.M.; Baran, E.J.; León, I.E. Copper complexes as antitumor agents: In vitro and in vivo evidences. Curr. Med. Chem. 2021. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Wani, W.A.; Saleem, K. Empirical Formulae to Molecular Structures of Metal Complexes by Molar Conductance. Synth. React. Inorg. Met. Nano Metal Chem. 2013, 43, 1162–1170. [Google Scholar] [CrossRef]
- Elleb, M.; Meullemeestre, J.; Schwing-Weill, M.J.; Vierling, F. Spectrophotometric study of copper(II) chloride complexes in propylene carbonate and in dimethyl sulfoxide. Inorg. Chem. 1982, 21, 1477–1483. [Google Scholar] [CrossRef]
- Meng, G.; Liu, C.; Qin, S.; Dong, M.; Wei, X.; Zheng, M.; Qin, L.; Wang, H.; He, X.; Zhang, Z. An improved synthesis of sunitinib malate via a solvent-free decarboxylation process. Res. Chem. Intermed. 2015, 41, 8941–8954. [Google Scholar] [CrossRef]
- Kassem, M.G.; Motiur Rahman, A.F.M.F.M.; Korashy, H.M. Chapter 9—Sunitinib Malate. Profiles Drug Subst. Excip. Relat. Methodol. 2012, 37, 363–388. [Google Scholar]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Clausen, H.F.; Chevallier, M.S.; Spackman, M.A.; Iversen, B.B. Three new co-crystals of hydroquinone: Crystal structures and Hirshfeld surface analysis of intermolecular interactions. New J. Chem. 2010, 34, 193–199. [Google Scholar] [CrossRef]
- Kashinski, D.O.; Chase, G.M.; Nelson, R.G.; Di Nallo, O.E.; Scales, A.N.; VanderLey, D.L.; Byrd, E.F.C. Harmonic Vibrational Frequencies: Approximate Global Scaling Factors for TPSS, M06, and M11 Functional Families Using Several Common Basis Sets. J. Phys. Chem. A 2017, 121, 2265–2273. [Google Scholar] [CrossRef]
- Mıhçıokur, Ö.; Özpozan, T. Molecular structure, vibrational spectroscopic analysis (IR & Raman), HOMO-LUMO and NBO analysis of anti-cancer drug sunitinib using DFT method. J. Mol. Struct. 2017, 1149, 27–41. [Google Scholar] [CrossRef]
- Alshetaili, A.S.; Anwer, M.K.; Alshahrani, S.M.; Alalaiwe, A.; Alsulays, B.B.; Ansari, M.J.; Imam, F.; Alshehri, S. Characteristics and anticancer properties of Sunitinib malate-loaded poly-lactic-co-glycolic acid nanoparticles against human colon cancer HT-29 cells lines. Trop. J. Pharm. Res. 2018, 17, 1263. [Google Scholar] [CrossRef]
- Karimi, M.H.; Mahdavinia, G.R.; Massoumi, B. pH-controlled sunitinib anticancer release from magnetic chitosan nanoparticles crosslinked with κ-carrageenan. Mater. Sci. Eng. C 2018, 91, 705–714. [Google Scholar] [CrossRef]
- McTigue, M.; Murray, B.W.; Chen, J.H.; Deng, Y.-L.; Solowiej, J.; Kania, R.S. Molecular conformations, interactions, and properties associated with drug efficiency and clinical performance among VEGFR TK inhibitors. Proc. Natl. Acad. Sci. USA 2012, 109, 18281–18289. [Google Scholar] [CrossRef] [Green Version]
- Faivre, S.; Demetri, G.; Sargent, W.; Raymond, E. Molecular basis for sunitinib efficacy and future clinical development. Nat. Rev. Drug Discov. 2007, 6, 734–745. [Google Scholar] [CrossRef]
- Modi, S.J.; Kulkarni, V.M. Vascular Endothelial Growth Factor Receptor (VEGFR-2)/KDR Inhibitors: Medicinal Chemistry Perspective. Med. Drug Discov. 2019, 2, 100009. [Google Scholar] [CrossRef]
- Garten, A.; Grohmann, T.; Kluckova, K.; Lavery, G.G.; Kiess, W.; Penke, M. Sorafenib-Induced Apoptosis in Hepatocellular Carcinoma is Reversed by SIRT1. Int. J. Mol. Sci. 2019, 20, 4048. [Google Scholar] [CrossRef] [Green Version]
- Konduri, S.K.M.; Satya, A.K.; Chowdary, N.V. Polymorphic Forms of Sunitinib Base. U.S. Patent 8,466,190, 18 June 2013. [Google Scholar]
- Rigaku Oxford Diffraction. CrysAlisPro Software System; Version 1.171.38.41; Rigaku Coorporation: Tokyo, Japan, 2015. [Google Scholar]
- Sheldrick, G.M. SHELXT Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Tan, S.L.; Jotani, M.M.; Tiekink, E.R.T. Utilizing Hirshfeld surface calculations, non-covalent interaction (NCI) plots and the calculation of interaction energies in the analysis of molecular packing. Acta Crystallogr. Sect. E Crystallogr. Commun. 2019, 75, 308–318. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16; Revision C.01; Gaussian, Inc.: Wallingford, UK, 2016. [Google Scholar]
- Yanai, T.; Tew, D.P.; Handy, N.C. A new hybrid exchange–correlation functional using the Coulomb-attenuating method (CAM-B3LYP). Chem. Phys. Lett. 2004, 393, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Schaftenaar, G.; Noordik, J.H. Molden: A pre- and post-processing program for molecular and electronic structures*. J. Comput. Aided. Mol. Des. 2000, 14, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besler, B.H.; Merz, K.M.; Kollman, P.A. Atomic charges derived from semiempirical methods. J. Comput. Chem. 1990, 11, 431–439. [Google Scholar] [CrossRef]
- Wang, Y.; Peng, C.; Wang, G.; Xu, Z.; Luo, Y.; Wang, J.; Zhu, W. Exploring binding mechanisms of VEGFR2 with three drugs lenvatinib, sorafenib, and sunitinib by molecular dynamics simulation and free energy calculation. Chem. Biol. Drug Des. 2019, 93, 934–948. [Google Scholar] [CrossRef]
- Kathiresan, S.; Mugesh, S.; Murugan, M.; Ahamed, F.; Annaraj, J. Mixed-ligand copper (II)-phenolate complexes: Structure and studies on DNA/protein binding profiles, DNA cleavage, molecular docking and cytotoxicity. RSC Adv. 2016, 6, 1810–1825. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
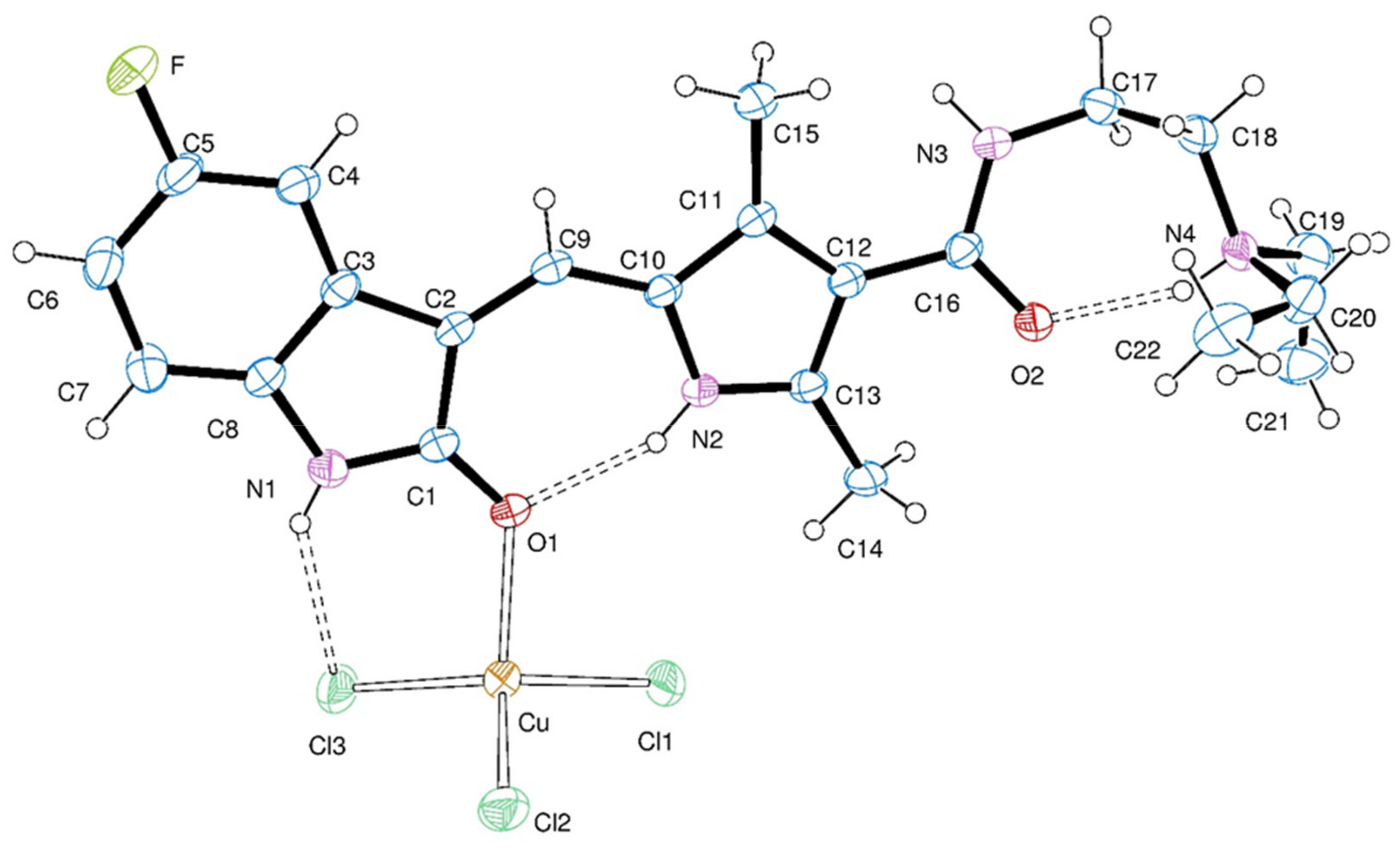
| Empirical formula | C22 H28 Cl3 Cu F N4 O2 | |
| Formula weight | 569.37 | |
| Temperature | 293(2) K | |
| Wavelength | 0.71073 Å | |
| Crystal system | Triclinic | |
| Space group | P-1 | |
| Unit cell dimensions | a = 7.9061(5) Å | α = 105.021(6)° |
| b = 12.412(1) Å | β = 106.744(5)° | |
| c = 13.7005(8) Å | γ = 91.749(5)° | |
| Volume | 1235.4(2) Å | |
| Z, density (calculated) | 2, 1.531 Mg/m3 | |
| Absorption coefficient | 1.243 mm−1 | |
| F(000) | 586 | |
| Crystal size | 0.405 × 0.163 × 0.059 mm3 | |
| ϑ-range for data collection | 3.208 to 29.153° | |
| Index ranges | −10 ≤ h ≤ 10, −16 ≤ k ≤ 16, −18 ≤ l ≤ 15 | |
| Reflections collected | 9998 | |
| Independent reflections | 5335 [R(int) = 0.0310] | |
| Observed reflections [I > 2σ(I)] | 3879 | |
| Completeness to ϑ = 25.242° | 99.8% | |
| Refinement method | Full-matrix least-squares on F2 | |
| Data/restraints/parameters | 5335/3/366 | |
| Goodness-of-fit on F2 | 1.031 | |
| Final R indices a [I > 2σ(I)] | R1 = 0.0474, wR2 = 0.1112 | |
| R indices (all data) | R1 = 0.0726, wR2 = 0.1275 | |
| Largest diff. peak and hole | 0.458 and −0.404 e.Å−3 | |
| Bond Lengths [Å] | Angles [°] | ||
|---|---|---|---|
| O(1)-Cu | 2.082(2) | O(1)-Cu-Cl(1) | 93.48(7) |
| Cu-Cl(1) | 2.179(1) | O(1)-Cu-Cl(2) | 121.29(8) |
| Cu-Cl(2) | 2.193(1) | Cl(1)-Cu-Cl(2) | 104.24(4) |
| Cu-Cl(3) | 2.231(1) | O(1)-Cu-Cl(3) | 100.23(7) |
| Cl(1)-Cu-Cl(3) | 133.95(5) | ||
| Cl(2)-Cu-Cl(3) | 105.39(5) | ||
| Tentative Assignations | Sun/cm−1 | CuSun/cm−1 | |||
|---|---|---|---|---|---|
| FTIR | Calculated | FTIR | Raman | Calculated | |
| ν(N-H)a | 3455 (w), 3300 (m), 3228 (m) | 3516 (w) | 3422 (br,s) 3284 (br,s) 3178 (sh,m) | - | 3387 (w) |
| ν(N-H)o | 3506 (w) | - | 3445 (m) | ||
| ν(N-H)p | 3160 (m) | - | 3436 (w) | ||
| ν(N-H)k | - | - | 3600–2500 * (m, vb) | - | 2790 (s) |
| ν(C-H)o,en,p | 3138–3078 (w) | 3076–3030 (w) | 3048 * (w) | - | 3076–3060 (w) |
| ν(C-H)e,m | 2969–2814 (m) | 3004–2787 (m) | 2983–2853 * (m) | - | 3030–2908 (m) |
| ν(C=O)o + ν(C=C)en + δ(CNH)o | 1677 (s) | 1685 (s) | 1645 (s) | 1649 (w) | 1668 (s) |
| ν(C=O)a | 1656 (s) | 1615 (s) | 1615 (w) | 1596 (s) | |
| ν(C=C)o,en + δ(CNH)a,p + ν(CN)o,p | 1589 (s), 1543 (s), 1480 (m) | 1570 (s), 1541 (m), 1504 (m) | 1564 (s), 1536 (s), 1513 (m) | 1553 (s), 1530 (m), 1512 (sh) | 1577 (s), 1552 (m), 1506 (m) |
| δ(CCH)o + δ(CCC)o,p | 1445 (m) | 1462 (m) | 1478 (m) | 1481 (w), 1460 (w) | 1463 (m) |
| δ(HCH)e,m | 1385 (w) | 1409 (w) | 1427 (m) | 1428 (m) | 1408 (w) |
| ν(CN)o + δ(HCC)en | 1330 (s) | 1304 (m) | 1319 (s) | 1300 (s) | 1315 (m) |
| t(HCCH)o + ω(CH2)e | 1295 (m) | 1276 (s) | 1261 (w) | 1250 (m) | 1260 (w) |
| ν(CN)o + δ(HCC)o | 1190 (m) | 1160 (m) | 1193 (m) | 1190 (m) | 1169 (w) |
| ν(Cu-Cl) | - | - | - | 226 (m) | 338 (w)–251 (w) |
| Ligand | Binding Energy/kcal mol−1 | Ki/μM |
|---|---|---|
| Sun | −4.71 | 355 |
| HSun+ | −4.94 | 238 |
| CuSun | −3.67 | 2030 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarasi, F.; Lanza, P.A.; Ferretti, V.; Echeverría, G.A.; Piro, O.E.; Cacicedo, M.; Gehring, S.; León, I.E.; Islas, M.S. Synthesis and Characterization of Novel Copper(II)-Sunitinib Complex: Molecular Docking, DFT Studies, Hirshfeld Analysis and Cytotoxicity Studies. Inorganics 2022, 10, 3. https://doi.org/10.3390/inorganics10010003
Tarasi F, Lanza PA, Ferretti V, Echeverría GA, Piro OE, Cacicedo M, Gehring S, León IE, Islas MS. Synthesis and Characterization of Novel Copper(II)-Sunitinib Complex: Molecular Docking, DFT Studies, Hirshfeld Analysis and Cytotoxicity Studies. Inorganics. 2022; 10(1):3. https://doi.org/10.3390/inorganics10010003
Chicago/Turabian StyleTarasi, Facundo, Priscila Ailín Lanza, Valeria Ferretti, Gustavo Alberto Echeverría, Oscar Enrique Piro, Maximiliano Cacicedo, Stephan Gehring, Ignacio Esteban León, and María Soledad Islas. 2022. "Synthesis and Characterization of Novel Copper(II)-Sunitinib Complex: Molecular Docking, DFT Studies, Hirshfeld Analysis and Cytotoxicity Studies" Inorganics 10, no. 1: 3. https://doi.org/10.3390/inorganics10010003