Using Introspection to Collect Provenance in R
Abstract
:1. Introduction
2. Related Work
3. Provenance Collected by RDataTracker
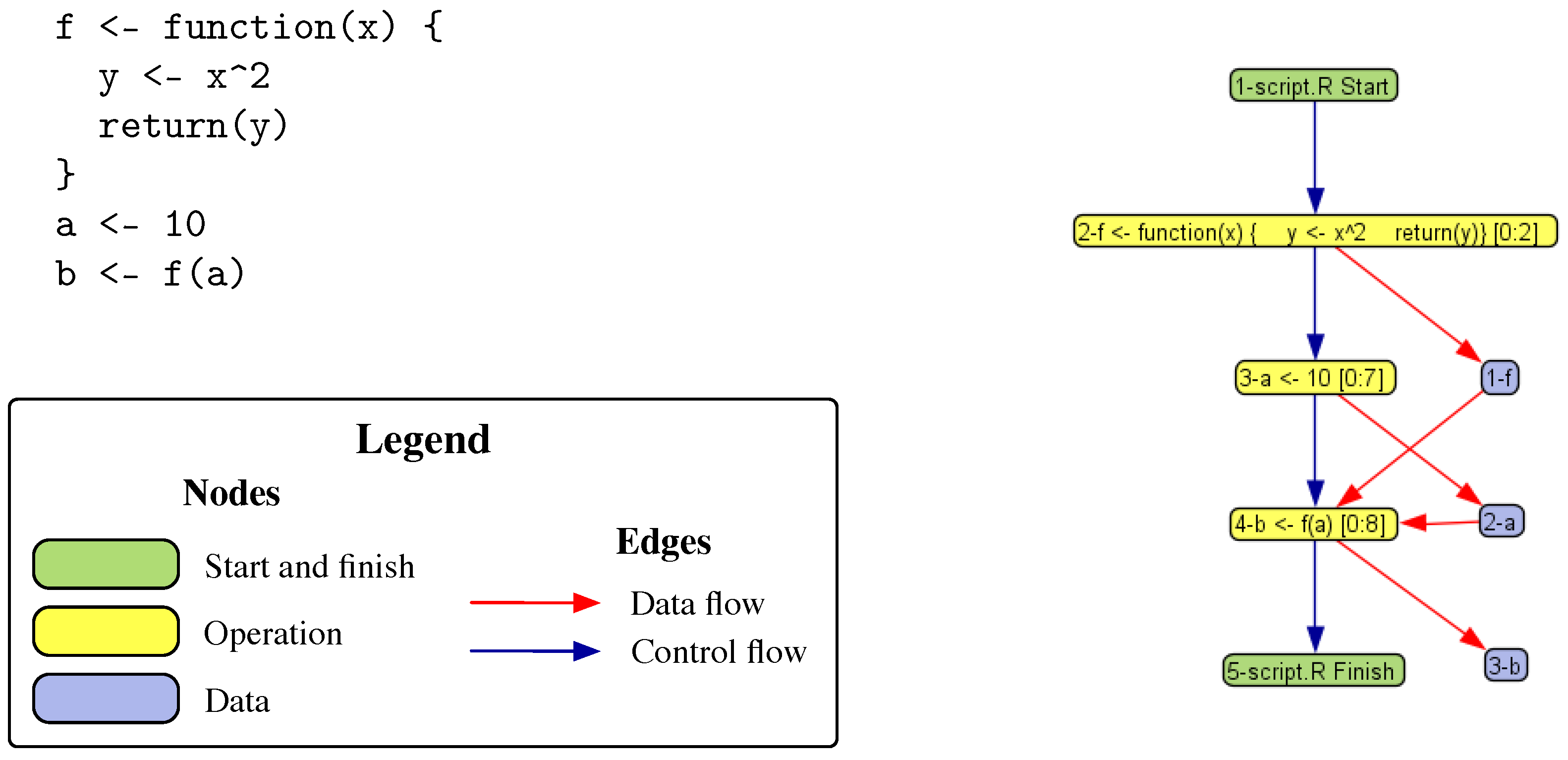
3.1. Provenance Model
3.2. DDG Examples
4. Implementation of RDataTracker
4.1. Background on R
4.2. Techniques Used to Collect Provenance
4.2.1. Gathering Static Information About Statements
4.2.2. Inserting Annotations
f <- function(x) {
y <- x^2
return(y)
}
f <- function(x) {
if (ddg.should.run.annotated("f")) {
ddg.function()
ddg.eval("y <- x^2")
return(ddg.return.value(ddg.eval("y")))
}
else {
y <- x^2
return(y)
}
}
4.2.3. Recording Provenance During Execution
Collecting the Provenance
Collecting Provenance Internal to Functions
read.csv (file, header = TRUE, sep = ",", quote = "\"", dec = ".", fill = TRUE, comment.char = "", ...)
read.csv("mydata.csv")
read.csv("mydata.csv", FALSE)
read.csv(header = FALSE, file = "mydata.csv")
read.csv("mydata.csv", s=".", d=",", h=F)
read.csv("mydata.csv", s=".", d=",", h=F, nrows=100)
Return Values
4.2.4. Granularity
- Limit the size of intermediate values that are saved.
- Control whether nodes are created during the execution of a function or whether a function call is represented with a single procedural node. This can be done globally, so that all functions are treated the same, or this feature can be enabled or disabled for individual functions.
- Control whether fine-grained provenance is collected internal to control constructs, and if it is, limit the number of iterations of a loop for which provenance is collected.
5. Evaluation
- No DDG: The original script with no provenance collected.
- Detail level 0: Collect provenance only for top-level statements.
- Detail level 1: Collect provenance internal to functions and internal to loops but only for a single loop iteration.
- Detail level 2: Collect provenance internal to functions and internal to loops for up to 10 iterations of each loop.
- Detail level 3: Collect provenance internal to functions and internal to loops for all loop iterations.
- No DDG: The original script with no provenance collected.
- No snapshots: Collect provenance but save no snapshots.
- 10K snapshots: Save snapshots up to 10K bytes per snapshot.
- 100K snapshots: Save snapshots up to 100K bytes per snapshot.
- Max snapshots: Save snapshots of any size.
6. Future Work
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A. Using RDataTracker
> ddg.run("script.R")
> ddg.display()
References
- Pasquier, T.; Lau, M.K.; Trisovic, A.; Boose, E.R.; Couturier, B.; Crosas, M.; Ellison, A.M.; Gibson, V.; Jones, C.R.; Seltzer, M. If these data could talk. Sci. Data 2017, 4, 170114. [Google Scholar] [CrossRef] [PubMed]
- Miles, S.; Groth, P.; Branco, M.; Moreau, L. The requirements of recording and using provenance in e-Science experiments. J. Grid Comput. 2005, 5, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Lerner, B.S.; Boose, E.R. RDataTracker: Collecting provenance in an interactive scripting environment. In Proceedings of the 6th USENIX Workshop on the Theory and Practice of Provenance, Cologne, Germany, 12–13 June 2014; USENIX Association: Cologne, Germany, 2014. [Google Scholar]
- Lerner, B.S.; Boose, E.R. RDataTracker and DDG Explorer—Capture, visualization and querying of provenance from R scripts. In Proceedings of the 5th International Provenance and Annotation Workshop, Cologne, Germany, 9–13 June 2014; IPAW: Cologne, Germany, 2014. [Google Scholar]
- Altintas, I.; Barney, O.; Jaeger-Frank, E. Provenance Collection Support in the Kepler Scientific Workflow System. In Proceedings of the 1st International Provenance and Annotation Workshop, Chicago, IL, USA, 3–5 May 2006; Springer: Berlin/Heidelberg, Germany, 2006; pp. 118–132. [Google Scholar]
- Zhao, J.; Goble, C.; Stevens, R.; Turi, D. Mining Taverna’s semantic web of provenance. Concurr. Comput. Pract. Exp. 2008, 20, 463–472. [Google Scholar] [CrossRef]
- Silva, C.T.; Anderson, E.; Santos, E.; Freire, J. Using VisTrails and Provenance for Teaching Scientific Visualization. Comput. Graph. Forum 2011, 30, 75–84. [Google Scholar] [CrossRef]
- Baumer, B.; Cetinkaya-Rundel, M.; Bray, A.; Loi, L.; Horton, N.J. R Markdown: Integrating a Reproducible Analysis Tool into Introductory Statistics. Technol. Innov. Stat. Educ. 2014, 8. Available online: https://escholarship.org/uc/item/90b2f5xh (accessed on 28 February 2018). [CrossRef]
- Acuña, R.; Lacroix, Z.; Bazzi, R.A. Instrumentation and Trace Analysis for Ad-hoc Python Workflows in Cloud Environments. In Proceedings of the 2015 IEEE 8th International Conference on Cloud Computing, New York, NY, USA, 27 June–2 July 2015; pp. 114–121. [Google Scholar]
- Guo, P.J.; Seltzer, M. BURRITO: Wrapping Your Lab Notebook in Computational Infrastructure. In Proceedings of the 4th USENIX Workshop on the Theory and Practice of Provenance, Boston, MA, USA, 14–15 June 2012; USENIX Association: Boston, MA, USA, 2012. [Google Scholar]
- Miao, H.; Chavan, A.; Deshpande, A. ProvDB: Lifecycle Management of Collaborative Analysis Workflows. In Proceedings of the 2nd Workshop on Human-In-the-Loop Data Analytics, Chicago, IL, USA, 14 May 2017; ACM: New York, NY, USA, 2017; pp. 7:1–7:6. [Google Scholar]
- Hellerstein, J.M.; Sreekanti, V.; Gonzalez, J.E.; Dalton, J.; Dey, A.; Nag, S.; Ramachandran, K.; Arora, S.; Bhattacharyya, A.; Das, S.; et al. Ground: A Data Context Service. In Proceedings of the Conference on Innovative Data Systems Research ’17, Chaminade, CA, USA, 8–11 January 2017; CIDR: Chaminade, CA, USA, 2017. [Google Scholar]
- McPhillips, T.; Aulenbach, S.; Song, T.; Belhajjame, K.; Kolisnik, T.; Bocinsky, K.; Dey, S.; Jones, C.; Cao, Y.; Freire, J.; et al. YesWorkflow: A User-Oriented, Language-Independent Tool for Recovering Workflow Information from Scripts. Int. J. Digit. Curation 2015, 10, 298–313. [Google Scholar] [CrossRef]
- Becker, R.; Chambers, J. Auditing of data analyses. SIAM J. Sci. Stat. Comput. 1988, 9, 747–760. [Google Scholar] [CrossRef]
- Slaughter, P.; Jones, M.B.; Jones, C.; Palmer, L. Recordr. Available online: https://github.com/NCEAS/recordr (accessed on 27 February 2018).
- Liu, Z.; Pounds, S. An R package that automatically collects and archives details for reproducible computing. BMC Bioinform. 2014, 15, 138. [Google Scholar] [CrossRef] [PubMed]
- Michaelides, D.T.; Parker, R.; Charlton, C.; Browne, W.J.; Moreau, L. Intermediate Notation for Provenance and Workflow Reproducibility. In International Provenance and Annotation Workshop; number 9672 in Lecture Notes in Computer Science; Mattoso, M., Glavic, B., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 83–94. [Google Scholar]
- Tariq, D.; Ali, M.; Gehani, A. Towards Automated Collection of Application-level Data Provenance. In Proceedings of the 4th USENIX Conference on Theory and Practice of Provenance, Boston, MA, USA, 14–15 June 2012; USENIX Association: Berkeley, CA, USA, 2012. [Google Scholar]
- Guo, P.J.; Engler, D. Towards Practical Incremental Recomputation for Scientists: An Implementation for the Python Language. In Proceedings of the 2nd USENIX Workshop on the Theory and Practice of Provenance, San Jose, CA, USA, 22 February 2010; USENIX Association: Berkeley, CA, USA, 2010. [Google Scholar]
- Guo, P.J.; Engler, D. Using Automatic Persistent Memoization to Facilitate Data Analysis Scripting. In Proceedings of the 2011 International Symposium on Software Testing and Analysis, Toronto, ON, Canada, 17–21 July 2011; ACM: New York, NY, USA, 2011; pp. 287–297. [Google Scholar]
- Silles, C.A.; Runnalls, A.R. Provenance-Awareness in R. In Proceedings of the 3rd International Provenance and Annotation Workshop, Troy, NY, USA, 15–16 June 2010; IPAW: Troy, NY, 2010; pp. 64–72. [Google Scholar]
- Pasquier, T. CamFlow/cytoscape.js-prov: Initial release. Zenodo 2017. [Google Scholar] [CrossRef]
- Murta, L.; Braganholo, V.; Chirigati, F.; Koop, D.; Freire, J. NoWorkflow: Capturing and Analyzing Provenance of Scripts. In Proceedings of the 5th International Provenance and Annotation Workshop, Cologne, Germany, 9–13 June 2014; IPAW: Cologne, Germany, 2014. [Google Scholar]
- Pimentel, J.F.N.; Braganholo, V.; Murta, L.; Freire, J. Collecting and Analyzing Provenance on Interactive Notebooks: When IPython meets noWorkflow. In Proceedings of the 7th USENIX Workshop on the Theory and Practice of Provenance, Edinburgh, Scotland, 8–9 July 2015; USENIX Association: Edinburgh, Scotland, 2015. [Google Scholar]
- Pimentel, J.F.; Freire, J.; Murta, L.; Braganholo, V. Fine-Grained Provenance Collection over Scripts Through Program Slicing. In Proceedings of the 6th International Provenance and Annotation Workshop, McLean, VA, USA, 7–8 June 2016; Mattoso, M., Glavic, B., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2016; Volume 9672. [Google Scholar]
- Wickham, H. Advanced R; The R Series; Chapman and Hall/CRC: Boca Raton, FL, USA, 2014. [Google Scholar]
- Grolemund, G.; Wickham, H. R for Data Science; Chapter 18: Pipes; O’Reilly: Sebastopol, CA, USA, 2017. [Google Scholar]
- Pasquier, T.; Lau, M.K.; Han, X.; Fong, E.; Lerner, B.S.; Boose, E.; Crosas, M.; Ellison, A.; Seltzer, M. Sharing and Preserving Computational Analyses for Posterity with encapsulator. IEEE Comput. Sci. Eng. 2018. under review. [Google Scholar]




| Node Type | Primary Attribute | Use |
|---|---|---|
| Data | Value | Scalar values and short strings |
| File | Location of a saved copy of the file | Files that are read or written |
| Snapshot | Location of the snapshot file | Complex data structures and long strings |
| URL | URL | URLs that are read from |
| Exception | Error message | Warnings or errors generated during execution |
| Node Type | Primary Attribute | Use |
|---|---|---|
| Operation | Location of the statement | Execute a statement |
| Binding | Parameter being bound and argument | Capture parameter passing |
| Start | None | Precedes execution of a control construct or function |
| Finish | None | Follows execution of a control construct or function |
| Incomplete | None | Indicates that detailed provenance is not collected on all executions of a loop |
| DDG Representation | PROV Representation |
|---|---|
| Procedure node | activity |
| Data node | entity |
| Control flow edge | wasInformedBy |
| Input edge | used |
| Output edge | wasGeneratedBy |
| Script | Input Files | Data Values | Output Files | Top-Level Statements | Functions | Loops |
|---|---|---|---|---|---|---|
| Met | 2 | 27,320 | 6 | 149 | 0 | 7 |
| Hydro | 6 | 82,294 | 3 | 141 | 3 | 8 |
| Hydro | Hydro | Hydro | Met | Met | Met | Met | |
|---|---|---|---|---|---|---|---|
| Detail 0 | Detail 1 | Detail 2 | Detail 0 | Detail 1 | Detail 2 | Detail 3 | |
| Procedure Nodes | 141 | 296 | 1282 | 149 | 199 | 564 | 18270 |
| Data Nodes | 162 | 224 | 738 | 168 | 186 | 367 | 6306 |
| Edges | 493 | 913 | 4045 | 557 | 663 | 1653 | 25928 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lerner, B.; Boose, E.; Perez, L. Using Introspection to Collect Provenance in R. Informatics 2018, 5, 12. https://doi.org/10.3390/informatics5010012
Lerner B, Boose E, Perez L. Using Introspection to Collect Provenance in R. Informatics. 2018; 5(1):12. https://doi.org/10.3390/informatics5010012
Chicago/Turabian StyleLerner, Barbara, Emery Boose, and Luis Perez. 2018. "Using Introspection to Collect Provenance in R" Informatics 5, no. 1: 12. https://doi.org/10.3390/informatics5010012




