Anticancer Drug Discovery Based on Natural Products: From Computational Approaches to Clinical Studies
Abstract
:1. Introduction
2. Historical Perspective
3. The Importance and Potential of Natural Products in Drug Discovery
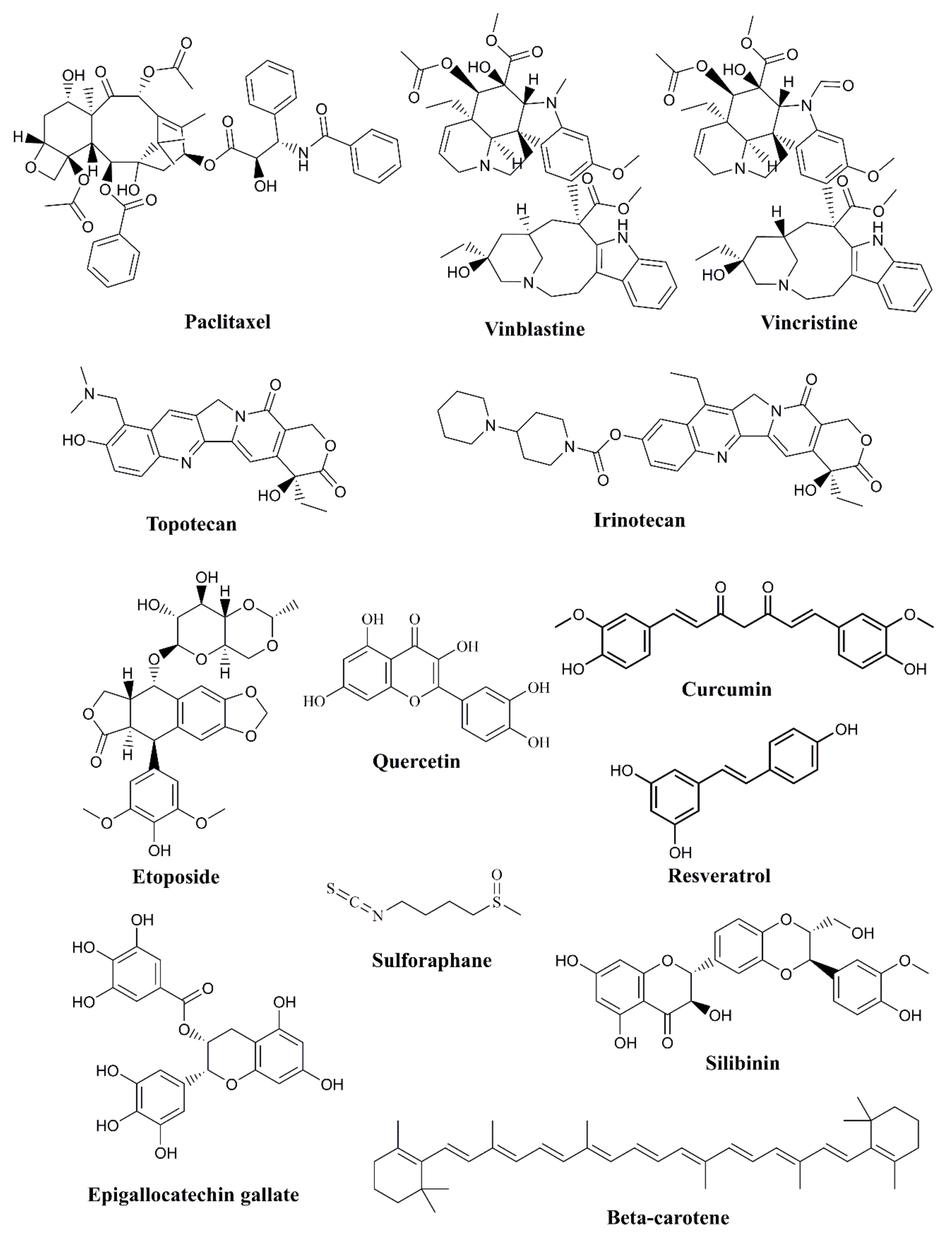
- Paclitaxel: derived from the Pacific yew tree, and is used to treat various cancers, including breast, ovarian, and lung cancers [59].
- Vinblastine and Vincristine: these alkaloids are derived from the Madagascar periwinkle plant and are used in the treatment of leukemia and lymphoma [60].
- Etoposide: derived from the mayapple plant, etoposide is used to treat lung and testicular cancer [63].
| Natural Compound | Source | Mechanism of Action | Target Genes | Cancer | Reference |
|---|---|---|---|---|---|
| Curcumin | Turmeric (Curcuma longa) | Inhibits cell proliferation, induces apoptosis. | TNF, IL-1, VEGF, EGF, FGF, EGFR, HER-2, AR, NF-κB, AP-1, STAT | Breast, lung, skin, gastrointestinal, colorectal, prostate, head and neck. | [81] |
| Resveratrol | Grapes, berries, peanuts | Antioxidant, affects cell cycle regulation. | APE1/Ref-1, NF-κB, LSD1, MCP-1 | Breast, cervical, uterine, blood, kidney, liver, eye, bladder, thyroid, esophageal, prostate, brain, lung, skin, gastric, colon, head and neck, bone, ovarian, and cervical. | [82] |
| Paclitaxel (Taxol) | Pacific yew tree (Taxus brevifolia) | Disrupts microtubule function. | AP-1, JNK1, p38, ERK1, IL-1α, IL-1β, TNF-α | Breast, ovarian, lung cancers. | [83] |
| Epigallocatechin gallate (EGCG) | Green tea | Antioxidant, induces apoptosis, inhibits proliferation. | retinoic acid receptor β (RARβ), CDH1 (e-cadherine gene), DAPK1, DNMT1, DNMT3B, HDAC1 | Breast, lung, bladder, head and neck, prostate, colorectal. | [84] |
| Sulforaphane | Cruciferous vegetables | Induces detoxification enzymes, pro-apoptotic. | TIMP1, AURKA, CEP55, CRYAB, PLCE1, and MMP28, CRC | Colorectal. | [85] |
| Genistein | Soybeans | Inhibits angiogenesis, modulates hormone activity. | p21-WAF1, p16-INK4a, p21-WAF1 and p16-INK4a | Breast, colorectal, lung, pancreatic. | [86] |
| Quercetin | Apples, onions, tea, red wine | Antioxidant, anti-inflammatory, inhibits proliferation. | bcl-2-associated X protein (BAX), Cytochrome c release, Cysteine-aspartic proteases (caspase)-3, Caspase-9, Transforming growth factor β (TGF-β), Anti-apoptotic Bcl-2 | Breast, prostate, colorectal, lung. | [87] |
| Capsaicin | Chili peppers | Induces apoptosis, inhibits cell growth. | c-myc, c-Ha-ras, p53 | Breast, lung, bladder, colon and pancreatic, colorectal. | [88] |
| Silymarin (Silibinin) | Milk thistle | Antioxidant, anti-inflammatory, cell regeneration. | NF-кB, TGF-β, TNF-α, interferon-gamma, IL-2, IL-4, and COX-2 | Breast, lung, colorectal, skin, pancreatic, prostate, gastrointestinal. | [89] |
| Berberine | Berberis plants | Inhibits cell progression, promotes apoptosis. | IL-1, TNF, IL-6, cyclooxygenase 2 and prostaglandin E2 | Colon. | [90] |
| Ellagic acid | Pomegranates, berries, nuts | Antioxidant, anti-proliferative. | p53-dependent genes, NF-kB p50, p65, and the PPAR family | Colorectal, prostate, lung, bladder, ovarian, breast. | [91] |
| Lycopene | Tomatoes, watermelon, pink grapefruit | Antioxidant, anti-proliferative. | IGFBP-3, c-fos, and uPAR | Breast, colorectal, lung, pancreatic, ovarian, cervical. | [92] |
| Indole-3-carbinol | Cruciferous vegetables | Modulates estrogen metabolism, apoptosis. | CYP1A1, CYP1B1 and AhR | Lung, head and neck, bladder, breast. | [93] |
| Beta-glucans | Oats, barley, mushrooms | Stimulates immune response. | TLR-2/6, CR3 | Breast, colorectal, prostate, ovarian. | [94] |
| Allicin | Garlic | Antioxidant, anti-proliferative, pro-apoptotic. | E2F1, E2F2, and E2F3 | Breast, bladder, lung, colorectal, prostate. | [95] |
| Catechins | Tea, cocoa, fruits | Antioxidant, anti-inflammatory, anti-proliferative. | JNK, MAP kinase, JAKs, BCL-2, and Nrf2 | Colorectal, pancreatic, lung, breast. | [96] |
| Ursolic acid | Apples, basil, cranberries | Inhibits metastasis, induces apoptosis. | MMP-9, CT45A2, Bcl-2, Bcl-xL, and BAX | Breast. | [97] |
| Limonene | Citrus peels | Induces detoxification enzymes, anti-proliferative. | Bcl-2-associated X protein (BAX), Cytochrome c release, Cysteine-aspartic proteases (caspase)-3, Caspase-9, Transforming growth factor β (TGF-β), Anti-apoptotic Bcl-2 | These are not directly associated with causing specific cancers, but rather are involved in cellular pathways related to apoptosis (programmed cell death) and regulation of cell survival. | [98] |
| Vinblastine | Periwinkle plant (Catharanthus roseus) | Inhibits microtubule assembly. | CCNB1 and AURKA | Breast, colorectal, lung, ovarian, prostate. | [99] |
| Vincristine | Periwinkle plant (Catharanthus roseus) | Binds to tubulin, inhibits microtubule formation. | CYP3A4, CYP3A5 | Liver. | [60] |
| Topotecan | Happy tree (Camptotheca acuminata) | Inhibits DNA topoisomerase I. | ABCB1, ABCG2, ALDH1A1, IFIH1, SAMD4 and EPHA3 | Breast, ovarian, colon. | [100] |
| Irinotecan | Happy tree (Camptotheca acuminata) | Inhibits DNA topoisomerase I. | UGT1A1 | It is not directly responsible for causing cance; variations in this gene can influence how the body processes certain chemotherapy drugs used in cancer treatment. | [101] |
| Etoposide | Mayapple plant (Podophyllum peltatum) | Inhibits DNA topoisomerase II. | SEMA5A, SLC7A6 and PRMT7 | For these genes, ongoing research might reveal their specific associations with certain cancers or their roles in cancer biology. The understanding of their involvement in cancer development and progression might evolve as more studies uncover their molecular mechanisms and connections to different cancer types. | [102] |
| Beta-carotene | Carrots, sweet potatoes, spinach | Antioxidant, modulates immune response. | CD38, NCF1B, and ITGAL | These genes are involved in various biological processes, including immune response and cell signalling. Their associations with specific cancers are not as prominent as some other genes, but they have been implicated in certain contexts. | [103] |
4. Present Status of Natural Compounds
5. Computational Approaches in Drug Discovery
5.1. Molecular Modelling and Drug Design
5.1.1. Molecular Dynamics Simulations
5.1.2. Virtual Screening and Docking Studies
5.1.3. Pharmacophore Mapping and QSAR Analysis
5.1.4. From Computational Studies to the Bench: Experimental Validation
6. In Vitro Assays
7. In Vivo Studies
8. Clinical Trials: The Journey to Therapeutics
8.1. Preclinical Studies
8.2. Clinical Trials
Integration of Biomarkers and Personalized Medicine in Clinical Trials
9. Challenges and Limitations
Strategies to Overcome the Challenges
10. Conclusions and Future Direction
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Magalhaes, L.G.; Ferreira, L.L.G.; Andricopulo, A.D. Recent Advances and Perspectives in Cancer Drug Design. An. Acad. Bras. Ciências 2018, 90, 1233–1250. [Google Scholar] [CrossRef]
- Kaleem, M.; Perwaiz, M.; Nur, S.M.; Abdulrahman, A.O.; Ahmad, W.; Al-Abbasi, F.A.; Kumar, V.; Kamal, M.A.; Anwar, F. Epigenetics of Triple-Negative Breast Cancer via Natural Compounds. Curr. Med. Chem. 2021, 29, 1436–1458. [Google Scholar] [CrossRef] [PubMed]
- Karn, V.; Sandhya, S.; Hsu, W.; Parashar, D.; Singh, H.N.; Jha, N.K.; Gupta, S.; Dubey, N.K.; Kumar, S. CRISPR/Cas9 System in Breast Cancer Therapy: Advancement, Limitations and Future Scope. Cancer Cell Int. 2022, 22, 234. [Google Scholar] [CrossRef] [PubMed]
- Alhosin, M.; Sharif, T.; Mousli, M.; Etienne-Selloum, N.; Fuhrmann, G.; Schini-Kerth, V.B.; Bronner, C. Down-Regulation of UHRF1, Associated with Re-Expression of Tumor Suppressor Genes, Is a Common Feature of Natural Compounds Exhibiting Anti-Cancer Properties. J. Exp. Clin. Cancer Res. 2011, 30, 41. [Google Scholar] [CrossRef] [PubMed]
- Parvez, A.; Choudhary, F.; Mudgal, P.; Khan, R.; Qureshi, K.A.; Aspatwar, A.; Farooqi, H. PD-1 and PD-L1: Architects of Immune Symphony and Immunotherapy Breakthroughs in Cancer Treatment. Front. Immunol. 2023, 14, 1296341. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Mambo, E.; Sidransky, D. Mitochondrial DNA Mutations in Human Cancer. Oncogene 2006, 25, 4663–4674. [Google Scholar] [CrossRef]
- Watson, I.R.; Takahashi, K.; Futreal, P.A.; Chin, L. Emerging Patterns of Somatic Mutations in Cancer. Nat. Rev. Genet. 2013, 14, 703–718. [Google Scholar] [CrossRef]
- Piñeros, M.; Parkin, D.M.; Ward, K.; Chokunonga, E.; Ervik, M.; Farrugia, H.; Gospodarowicz, M.; O’Sullivan, B.; Soerjomataram, I.; Swaminathan, R.; et al. Essential TNM: A Registry Tool to Reduce Gaps in Cancer Staging Information. Lancet Oncol. 2019, 20, e103–e111. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Loud, J.T.; Murphy, J. Cancer Screening and Early Detection in the 21 St Century. Semin. Oncol. Nurs. 2017, 33, 121–128. [Google Scholar] [CrossRef]
- Zugazagoitia, J.; Guedes, C.; Ponce, S.; Ferrer, I.; Molina-Pinelo, S.; Paz-Ares, L. Current Challenges in Cancer Treatment. Clin. Ther. 2016, 38, 1551–1566. [Google Scholar] [CrossRef]
- Wang, D.R.; Wu, X.L.; Sun, Y.L. Therapeutic Targets and Biomarkers of Tumor Immunotherapy: Response versus Non-Response. Signal Transduct. Target. Ther. 2022, 7, 331. [Google Scholar] [CrossRef] [PubMed]
- Debela, D.T.; Muzazu, S.G.Y.; Heraro, K.D.; Ndalama, M.T.; Mesele, B.W.; Haile, D.C.; Kitui, S.K.; Manyazewal, T. New Approaches and Procedures for Cancer Treatment: Current Perspectives. SAGE Open Med. 2021, 9, 20503121211034366. [Google Scholar] [CrossRef] [PubMed]
- Tohme, S.; Simmons, R.L.; Tsung, A. Surgery for Cancer: A Trigger for Metastases. Cancer Res. 2017, 77, 1548–1552. [Google Scholar] [CrossRef]
- Tiwari, P.K.; Ko, T.-H.; Dubey, R.; Chouhan, M.; Tsai, L.-W.; Singh, H.N.; Chaubey, K.K.; Dayal, D.; Chiang, C.-W.; Kumar, S. CRISPR/Cas9 as a Therapeutic Tool for Triple Negative Breast Cancer: From Bench to Clinics. Front. Mol. Biosci. 2023, 10, 1214489. [Google Scholar] [CrossRef]
- Kumar, M.; Dubey, R.; Kumar Shukla, P.; Dayal, D.; Kumar Chaubey, K.; Tsai, L.-W.; Kumar, S. Identification of Small Molecule Inhibitors of RAD52 for Breast Cancer Therapy: In Silico Approach. J. Biomol. Struct. Dyn. 2023, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Baskar, R.; Lee, K.A.; Yeo, R.; Yeoh, K.W. Cancer and Radiation Therapy: Current Advances and Future Directions. Int. J. Med. Sci. 2012, 9, 193. [Google Scholar] [CrossRef] [PubMed]
- Salas-Benito, D.; Pérez-Gracia, J.L.; Ponz-Sarvisé, M.; Rodriguez-Ruiz, M.E.; Martínez-Forero, I.; Castañón, E.; López-Picazo, J.M.; Sanmamed, M.F.; Melero, I. Paradigms on Immunotherapy Combinations with Chemotherapy. Cancer Discov. 2021, 11, 1353–1367. [Google Scholar] [CrossRef]
- Esfahani, K.; Roudaia, L.; Buhlaiga, N.; Del Rincon, S.V.; Papneja, N.; Miller, W.H. A Review of Cancer Immunotherapy: From the Past, to the Present, to the Future. Curr. Oncol. 2020, 27, 87–97. [Google Scholar] [CrossRef]
- Choi, H.Y.; Chang, J.-E. Targeted Therapy for Cancers: From Ongoing Clinical Trials to FDA-Approved Drugs. Int. J. Mol. Sci. 2023, 24, 13618. [Google Scholar] [CrossRef]
- Deli, T.; Orosz, M.; Jakab, A. Hormone Replacement Therapy in Cancer Survivors—Review of the Literature. Pathol. Oncol. Res. 2020, 26, 63–78. [Google Scholar] [CrossRef]
- Chu, D.-T.; Nguyen, T.T.; Tien, N.L.B.; Tran, D.-K.; Jeong, J.-H.; Anh, P.G.; Van Thanh, V.; Truong, D.T.; Dinh, T.C. Recent Progress of Stem Cell Therapy in Cancer Treatment: Molecular Mechanisms and Potential Applications. Cells 2020, 9, 563. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Fountzilas, E.; Nikanjam, M.; Kurzrock, R. Review of Precision Cancer Medicine: Evolution of the Treatment Paradigm. Cancer Treat. Rev. 2020, 86, 102019. [Google Scholar] [CrossRef] [PubMed]
- Dumanovsky, T.; Augustin, R.; Rogers, M.; Lettang, K.; Meier, D.E.; Morrison, R.S. The Growth of Palliative Care in U.S. Hospitals: A Status Report. J. Palliat. Med. 2016, 19, 8–15. [Google Scholar] [CrossRef]
- Ma, D.-L.; Chan, D.S.-H.; Leung, C.-H. Molecular Docking for Virtual Screening of Natural Product Databases. Chem. Sci. 2011, 2, 1656–1665. [Google Scholar] [CrossRef]
- Schuster, D.; Wolber, G. Identification of Bioactive Natural Products by Pharmacophore-Based Virtual Screening. Curr. Pharm. Des. 2010, 16, 1666–1681. [Google Scholar] [CrossRef]
- Cavalcanti, É.B.V.S.; Félix, M.B.; Scotti, L.; Scotti, M.T. Virtual Screening of Natural Products to Select Compounds with Potential Anticancer Activity. Anti-Cancer Agents Med. Chem. 2019, 19, 154–171. [Google Scholar] [CrossRef] [PubMed]
- Anjum, F.; Ali, F.; Mohammad, T.; Shafie, A.; Akhtar, O.; Abdullaev, B.; Hassan, I. Discovery of Natural Compounds as Potential Inhibitors of Human Carbonic Anhydrase II: An Integrated Virtual Screening, Docking, and Molecular Dynamics Simulation Study. OMICS J. Integr. Biol. 2021, 25, 513–524. [Google Scholar] [CrossRef]
- Yu, E.; Xu, Y.; Shi, Y.; Yu, Q.; Liu, J.; Xu, L. Discovery of Novel Natural Compound Inhibitors Targeting Estrogen Receptor α by an Integrated Virtual Screening Strategy. J. Mol. Model. 2019, 25, 278. [Google Scholar] [CrossRef] [PubMed]
- Poornima, P.; Kumar, J.D.; Zhao, Q.; Blunder, M.; Efferth, T. Network Pharmacology of Cancer: From Understanding of Complex Interactomes to the Design of Multi-Target Specific Therapeutics from Nature. Pharmacol. Res. 2016, 111, 290–302. [Google Scholar] [CrossRef]
- Iqbal, M.J.; Javed, Z.; Sadia, H.; Qureshi, I.A.; Irshad, A.; Ahmed, R.; Malik, K.; Raza, S.; Abbas, A.; Pezzani, R.; et al. Clinical Applications of Artificial Intelligence and Machine Learning in Cancer Diagnosis: Looking into the Future. Cancer Cell Int. 2021, 21, 270. [Google Scholar] [CrossRef]
- Ravishankar, B.; Shukla, V. Indian Systems of Medicine: A Brief Profile. Afr. J. Tradit. Complement. Altern. Med. 2008, 4, 319. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Lo, S.-H.; Dubey, R.; Kumar, S.; Chaubey, K.K.; Kumar, S. Plant-Derived Natural Compounds as an Emerging Antiviral in Combating COVID-19. Indian J. Microbiol. 2023, 63, 429–446. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, A.D.B.; Devasagayam, T.P.A. Current Status of Herbal Drugs in India: An Overview. J. Clin. Biochem. Nutr. 2007, 41, 1–11. [Google Scholar] [CrossRef]
- Pirintsos, S.; Panagiotopoulos, A.; Bariotakis, M.; Daskalakis, V.; Lionis, C.; Sourvinos, G.; Karakasiliotis, I.; Kampa, M.; Castanas, E. From Traditional Ethnopharmacology to Modern Natural Drug Discovery: A Methodology Discussion and Specific Examples. Molecules 2022, 27, 4060. [Google Scholar] [CrossRef] [PubMed]
- Dhyani, P.; Quispe, C.; Sharma, E.; Bahukhandi, A.; Sati, P.; Attri, D.C.; Szopa, A.; Sharifi-Rad, J.; Docea, A.O.; Mardare, I.; et al. Anticancer Potential of Alkaloids: A Key Emphasis to Colchicine, Vinblastine, Vincristine, Vindesine, Vinorelbine and Vincamine. Cancer Cell Int. 2022, 22, 206. [Google Scholar] [CrossRef] [PubMed]
- Zulkipli, I.N.; David, S.R.; Rajabalaya, R.; Idris, A. Medicinal Plants: A Potential Source of Compounds for Targeting Cell Division. Drug Target Insights 2015, 9, DTI.S24946. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Kumar, S.; Sandeep, K.; Patel, S.K.S. Therapeutic Approaches in Pancreatic Cancer: Recent Updates. Biomedicines 2023, 11, 1611. [Google Scholar] [CrossRef]
- Dias, D.A.; Urban, S.; Roessner, U. A Historical Overview of Natural Products in Drug Discovery. Metabolites 2012, 2, 303–336. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Lou, H.-X. Strategies to Diversify Natural Products for Drug Discovery. Med. Res. Rev. 2018, 38, 1255–1294. [Google Scholar] [CrossRef]
- Cragg, G.M.; Pezzuto, J.M. Natural Products as a Vital Source for the Discovery of Cancer Chemotherapeutic and Chemopreventive Agents. Med. Princ. Pract. 2016, 25, 41–59. [Google Scholar] [CrossRef]
- Ponphaiboon, J.; Krongrawa, W.; Aung, W.W.; Chinatangkul, N.; Limmatvapirat, S.; Limmatvapirat, C. Advances in Natural Product Extraction Techniques, Electrospun Fiber Fabrication, and the Integration of Experimental Design: A Comprehensive Review. Molecules 2023, 28, 5163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-W.; Lin, L.-G.; Ye, W.-C. Techniques for Extraction and Isolation of Natural Products: A Comprehensive Review. Chin. Med. 2018, 13, 20. [Google Scholar] [CrossRef]
- Mushtaq, S.; Abbasi, B.H.; Uzair, B.; Abbasi, R. Natural Products as Reservoirs of Novel Therapeutic Agents. EXCLI J. 2018, 17, 420–451. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Sayed, A.M.; Elmaidomy, A.H. Natural Products’ Extraction and Isolation-Between Conventional and Modern Techniques. Front. Nat. Prod. 2022, 1, 873808. [Google Scholar] [CrossRef]
- Cech, N.B.; Oberlies, N.H. From Plant to Cancer Drug: Lessons Learned from the Discovery of Taxol. Nat. Prod. Rep. 2023, 40, 1153–1157. [Google Scholar] [CrossRef] [PubMed]
- Shaik, B.B.; Katari, N.K.; Jonnalagadda, S.B. Role of Natural Products in Developing Novel Anticancer Agents: A Perspective. Chem. Biodivers. 2022, 19, e202200535. [Google Scholar] [CrossRef] [PubMed]
- Wani, M.C.; Horwitz, S.B. Nature as a Remarkable Chemist. Anti-Cancer Drugs 2014, 25, 482–487. [Google Scholar] [CrossRef]
- Gallego-Jara, J.; Lozano-Terol, G.; Sola-Martínez, R.A.; Cánovas-Díaz, M.; de Diego Puente, T. A Compressive Review about Taxol®: History and Future Challenges. Molecules 2020, 25, 5986. [Google Scholar] [CrossRef]
- Weaver, B.A. How Taxol/Paclitaxel Kills Cancer Cells. Mol. Biol. Cell 2014, 25, 2677–2681. [Google Scholar] [CrossRef]
- Donehower, R.C. The Clinical Development of Paclitaxel: A Successful Collaboration of Academia, Industry and the National Cancer Institute. Oncologist 1996, 1, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Ma, Q.; Ye, L.; Piao, G. The Traditional Medicine and Modern Medicine from Natural Products. Molecules 2016, 21, 559. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Ongnok, B.; Chattipakorn, N.; Chattipakorn, S.C. Doxorubicin and Cisplatin Induced Cognitive Impairment: The Possible Mechanisms and Interventions. Exp. Neurol. 2020, 324, 113118. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Su, H.; Zhou, B.; Liu, S. The Function of Natural Compounds in Important Anticancer Mechanisms. Front. Oncol. 2022, 12, 1049888. [Google Scholar] [CrossRef] [PubMed]
- Desilets, A.; Adam, J.-P.; Soulières, D. Management of Cisplatin-Associated Toxicities in Bladder Cancer Patients. Curr. Opin. Support. Palliat. Care 2020, 14, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Tait, S.W.G. Apoptosis and Cancer: Force Awakens, Phantom Menace, or Both? Int. Rev. Cell Mol. Biol. 2018, 337, 135–152. [Google Scholar] [CrossRef]
- Naeem, A.; Hu, P.; Yang, M.; Zhang, J.; Liu, Y.; Zhu, W.; Zheng, Q. Natural Products as Anticancer Agents: Current Status and Future Perspectives. Molecules 2022, 27, 8367. [Google Scholar] [CrossRef]
- Zhu, L.; Chen, L. Progress in Research on Paclitaxel and Tumor Immunotherapy. Cell. Mol. Biol. Lett. 2019, 24, 40. [Google Scholar] [CrossRef]
- Paul, A.; Acharya, K.; Chakraborty, N. Biosynthesis, Extraction, Detection and Pharmacological Attributes of Vinblastine and Vincristine, Two Important Chemotherapeutic Alkaloids of Catharanthus Roseus (L.) G. Don: A Review. S. Afr. J. Bot. 2023, 161, 365–376. [Google Scholar] [CrossRef]
- Avemann, K.; Knippers, R.; Koller, T.; Sogo, J.M. Camptothecin, a Specific Inhibitor of Type I DNA Topoisomerase, Induces DNA Breakage at Replication Forks. Mol. Cell. Biol. 1988, 8, 3026–3034. [Google Scholar] [CrossRef] [PubMed]
- Behera, A.; Padhi, S. Passive and Active Targeting Strategies for the Delivery of the Camptothecin Anticancer Drug: A Review. Environ. Chem. Lett. 2020, 18, 1557–1567. [Google Scholar] [CrossRef]
- Wu, C.-C.; Li, T.-K.; Farh, L.; Lin, L.-Y.; Lin, T.-S.; Yu, Y.-J.; Yen, T.-J.; Chiang, C.-W.; Chan, N.-L. Structural Basis of Type II Topoisomerase Inhibition by the Anticancer Drug Etoposide. Science 2011, 333, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Shahar, N.; Larisch, S. Inhibiting the Inhibitors: Targeting Anti-Apoptotic Proteins in Cancer and Therapy Resistance. Drug Resist. Updates 2020, 52, 100712. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, L.; Hei, R.; Li, X.; Cai, H.; Wu, X.; Zheng, Q.; Cai, C. CDK Inhibitors in Cancer Therapy, an Overview of Recent Development. Am. J. Cancer Res. 2021, 11, 1913–1935. [Google Scholar] [PubMed]
- Yang, Y.; Cao, Y. The Impact of VEGF on Cancer Metastasis and Systemic Disease. Semin. Cancer Biol. 2022, 86, 251–261. [Google Scholar] [CrossRef] [PubMed]
- DiDonato, J.A.; Mercurio, F.; Karin, M. NF-κB and the Link between Inflammation and Cancer. Immunol. Rev. 2012, 246, 379–400. [Google Scholar] [CrossRef]
- Liou, G.-Y.; Storz, P. Reactive Oxygen Species in Cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef]
- Halder, A.K.; Saha, A.; Jha, T. Exploring QSAR and Pharmacophore Mapping of Structurally Diverse Selective Matrix Metalloproteinase-2 Inhibitors. J. Pharm. Pharmacol. 2013, 65, 1541–1554. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Lu, F.; Liu, L.; Ji, Q.; Song, K.; Yin, Q.; Lerner, R.A.; Yang, G.; Xu, H.; et al. A DNA-Encoded Library for the Identification of Natural Product Binders That Modulate Poly (ADP-Ribose) Polymerase 1, a Validated Anti-Cancer Target. Biochem. Biophys. Res. Commun. 2020, 533, 241–248. [Google Scholar] [CrossRef]
- Akone, S.H.; Ntie-Kang, F.; Stuhldreier, F.; Ewonkem, M.B.; Noah, A.M.; Mouelle, S.E.M.; Müller, R. Natural Products Impacting DNA Methyltransferases and Histone Deacetylases. Front. Pharmacol. 2020, 11, 992. [Google Scholar] [CrossRef]
- Malhotra, M.; Suman, S.K. Laccase-Mediated Delignification and Detoxification of Lignocellulosic Biomass: Removing Obstacles in Energy Generation. Environ. Sci. Pollut. Res. 2021, 28, 58929–58944. [Google Scholar] [CrossRef] [PubMed]
- Polivka, J.; Janku, F. Molecular Targets for Cancer Therapy in the PI3K/AKT/MTOR Pathway. Pharmacol. Ther. 2014, 142, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Long, H.; Zhou, Z.; Fu, Y.; Jiang, B. PI3K–AKT-Targeting Breast Cancer Treatments: Natural Products and Synthetic Compounds. Biomolecules 2023, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Nandi, S.; Dey, R.; Samadder, A.; Saxena, A.; Saxena, A.K. Natural Sourced Inhibitors of EGFR, PDGFR, FGFR and VEGFRMediated Signaling Pathways as Potential Anticancer Agents. Curr. Med. Chem. 2022, 29, 212–234. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, K.; Xu, B. Telomerase Inhibitors from Natural Products and Their Anticancer Potential. Int. J. Mol. Sci. 2017, 19, 13. [Google Scholar] [CrossRef]
- Cerella, C.; Radogna, F.; Dicato, M.; Diederich, M. Natural Compounds as Regulators of the Cancer Cell Metabolism. Int. J. Cell Biol. 2013, 2013, 639401. [Google Scholar] [CrossRef]
- Martinez-Outschoorn, U.E.; Peiris-Pagés, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer Metabolism: A Therapeutic Perspective. Nat. Rev. Clin. Oncol. 2017, 14, 11–31. [Google Scholar] [CrossRef]
- Jain, C.; Majumder, H.; Roychoudhury, S. Natural Compounds as Anticancer Agents Targeting DNA Topoisomerases. Curr. Genom. 2016, 18, 75–92. [Google Scholar] [CrossRef]
- Ottaviani, A.; Iacovelli, F.; Fiorani, P.; Desideri, A. Natural Compounds as Therapeutic Agents: The Case of Human Topoisomerase IB. Int. J. Mol. Sci. 2021, 22, 4138. [Google Scholar] [CrossRef]
- Prakobwong, S.; Gupta, S.C.; Kim, J.H.; Sung, B.; Pinlaor, P.; Hiraku, Y.; Wongkham, S.; Sripa, B.; Pinlaor, S.; Aggarwal, B.B. Curcumin Suppresses Proliferation and Induces Apoptosis in Human Biliary Cancer Cells through Modulation of Multiple Cell Signaling Pathways. Carcinogenesis 2011, 32, 1372–1380. [Google Scholar] [CrossRef]
- Rauf, A.; Imran, M.; Butt, M.S.; Nadeem, M.; Peters, D.G.; Mubarak, M.S. Resveratrol as an Anti-Cancer Agent: A Review. Crit. Rev. Food Sci. Nutr. 2018, 58, 1428–1447. [Google Scholar] [CrossRef] [PubMed]
- Swamy, M.K.; Vasamsetti, B.M.K. 2-Taxol: Occurrence, Chemistry, and Understanding Its Molecular Mechanisms. In Paclitaxel; Swamy, M.K., Pullaiah, T., Chen, Z.-S., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 29–45. ISBN 978-0-323-90951-8. [Google Scholar]
- Fang, C.-Y.; Wu, C.-C.; Hsu, H.-Y.; Chuang, H.-Y.; Huang, S.-Y.; Tsai, C.-H.; Chang, Y.; Tsao, G.S.-W.; Chen, C.-L.; Chen, J.-Y. EGCG Inhibits Proliferation, Invasiveness and Tumor Growth by up-Regulation of Adhesion Molecules, Suppression of Gelatinases Activity, and Induction of Apoptosis in Nasopharyngeal Carcinoma Cells. Int. J. Mol. Sci. 2015, 16, 2530–2558. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.-T.; Yen, G.-C. Chemopreventive Functions of Sulforaphane: A Potent Inducer of Antioxidant Enzymes and Apoptosis. J. Funct. Foods 2009, 1, 23–32. [Google Scholar] [CrossRef]
- Varinska, L.; Gal, P.; Mojzisova, G.; Mirossay, L.; Mojzis, J. Soy and Breast Cancer: Focus on Angiogenesis. Int. J. Mol. Sci. 2015, 16, 11728–11749. [Google Scholar] [CrossRef] [PubMed]
- Lotfi, N.; Yousefi, Z.; Golabi, M.; Khalilian, P.; Ghezelbash, B.; Montazeri, M.; Shams, M.H.; Baghbadorani, P.Z.; Eskandari, N. The Potential Anti-Cancer Effects of Quercetin on Blood, Prostate and Lung Cancers: An Update. Front. Immunol. 2023, 14, 1077531. [Google Scholar] [CrossRef] [PubMed]
- Lavorgna, M.; Orlo, E.; Nugnes, R.; Piscitelli, C.; Russo, C.; Isidori, M. Capsaicin in Hot Chili Peppers: In Vitro Evaluation of Its Antiradical, Antiproliferative and Apoptotic Activities. Plant Foods Hum. Nutr. 2019, 74, 164–170. [Google Scholar] [CrossRef]
- Olajide, P.; Adetuyi, B.; Oloke, J.; Omolabi, F. Pharmacological, Biochemical and Therapeutic Potential of Milk Thistle (Silymarin): A Review. World News Nat. Sci. 2022, 37, 75–91. [Google Scholar]
- Chidambara Murthy, K.N.; Jayaprakasha, G.K.; Patil, B.S. The Natural Alkaloid Berberine Targets Multiple Pathways to Induce Cell Death in Cultured Human Colon Cancer Cells. Eur. J. Pharmacol. 2012, 688, 14–21. [Google Scholar] [CrossRef]
- Ceci, C.; Lacal, P.M.; Tentori, L.; De Martino, M.G.; Miano, R.; Graziani, G. Experimental Evidence of the Antitumor, Antimetastatic and Antiangiogenic Activity of Ellagic Acid. Nutrients 2018, 10, 1756. [Google Scholar] [CrossRef]
- Kelkel, M.; Schumacher, M.; Dicato, M.; Diederich, M. Antioxidant and Anti-Proliferative Properties of Lycopene. Free Radic Res. 2011, 45, 925–940. [Google Scholar] [CrossRef] [PubMed]
- Katz, E.; Nisani, S.; Chamovitz, D.A. Indole-3-Carbinol: A Plant Hormone Combatting Cancer. F1000Research 2018, 7, 689. [Google Scholar] [CrossRef] [PubMed]
- Cerletti, C.; Esposito, S.; Iacoviello, L. Edible Mushrooms and Beta-Glucans: Impact on Human Health. Nutrients 2021, 13, 2195. [Google Scholar] [CrossRef] [PubMed]
- Özkan, İ.; Koçak, P.; Yıldırım, M.; Ünsal, N.; Yılmaz, H.; Telci, D.; Şahin, F. Garlic (Allium Sativum)-Derived SEVs Inhibit Cancer Cell Proliferation and Induce Caspase Mediated Apoptosis. Sci. Rep. 2021, 11, 14773. [Google Scholar] [CrossRef] [PubMed]
- Zanwar, A.A.; Badole, S.L.; Shende, P.S.; Hegde, M.V.; Bodhankar, S.L. Chapter 21—Antioxidant Role of Catechin in Health and Disease. In Polyphenols in Human Health and Disease; Watson, R.R., Preedy, V.R., Zibadi, S., Eds.; Academic Press: San Diego, CA, USA, 2014; pp. 267–271. ISBN 978-0-12-398456-2. [Google Scholar]
- Iqbal, J.; Abbasi, B.A.; Ahmad, R.; Mahmood, T.; Kanwal, S.; Ali, B.; Khalil, A.T.; Shah, S.A.; Alam, M.M.; Badshah, H. Ursolic Acid a Promising Candidate in the Therapeutics of Breast Cancer: Current Status and Future Implications. Biomed. Pharmacother. 2018, 108, 752–756. [Google Scholar] [CrossRef] [PubMed]
- ALaqeel, N.K. Antioxidants from Different Citrus Peels Provide Protection against Cancer. Braz. J. Biol. 2023, 84, e271619. [Google Scholar] [CrossRef] [PubMed]
- Moudi, M.; Go, R.; Yien, C.Y.S.; Nazre, M. Vinca Alkaloids. Int. J. Prev. Med. 2013, 4, 1231–1235. [Google Scholar]
- Martino, E.; Della Volpe, S.; Terribile, E.; Benetti, E.; Sakaj, M.; Centamore, A.; Sala, A.; Collina, S. The Long Story of Camptothecin: From Traditional Medicine to Drugs. Bioorg. Med. Chem. Lett. 2017, 27, 701–707. [Google Scholar] [CrossRef]
- Basili, S.; Moro, S. Novel Camptothecin Derivatives as Topoisomerase I Inhibitors. Expert Opin. Ther. Pat. 2009, 19, 555–574. [Google Scholar] [CrossRef]
- Zhang, W.; Gou, P.; Dupret, J.-M.; Chomienne, C.; Rodrigues-Lima, F. Etoposide, an Anticancer Drug Involved in Therapy-Related Secondary Leukemia: Enzymes at Play. Transl. Oncol. 2021, 14, 101169. [Google Scholar] [CrossRef]
- Toti, E.; Chen, C.-Y.O.; Palmery, M.; Villaño Valencia, D.; Peluso, I. Non-Provitamin A and Provitamin A Carotenoids as Immunomodulators: Recommended Dietary Allowance, Therapeutic Index, or Personalized Nutrition? Oxid. Med. Cell Longev. 2018, 2018, 4637861. [Google Scholar] [CrossRef]
- Salehi, B.; Stojanović-Radić, Z.; Matejić, J.; Sharifi-Rad, M.; Anil Kumar, N.V.; Martins, N.; Sharifi-Rad, J. The Therapeutic Potential of Curcumin: A Review of Clinical Trials. Eur. J. Med. Chem. 2019, 163, 527–545. [Google Scholar] [CrossRef] [PubMed]
- Berman, A.Y.; Motechin, R.A.; Wiesenfeld, M.Y.; Holz, M.K. The Therapeutic Potential of Resveratrol: A Review of Clinical Trials. NPJ Precis. Oncol. 2017, 1, 35. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, V.; Tuli, H.S.; Tania, M.; Srivastava, S.; Ritzer, E.E.; Pandey, A.; Aggarwal, D.; Barwal, T.S.; Jain, A.; Kaur, G.; et al. Molecular Mechanisms of Action of Epigallocatechin Gallate in Cancer: Recent Trends and Advancement. Semin. Cancer Biol. 2022, 80, 256–275. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-H.; Mao, J.-W.; Tan, X.-L. Research Progress on the Source, Production, and Anti-Cancer Mechanisms of Paclitaxel. Chin. J. Nat. Med. 2020, 18, 890–897. [Google Scholar] [CrossRef]
- Tang, S.-M.; Deng, X.-T.; Zhou, J.; Li, Q.-P.; Ge, X.-X.; Miao, L. Pharmacological Basis and New Insights of Quercetin Action in Respect to Its Anti-Cancer Effects. Biomed. Pharmacother. 2020, 121, 109604. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Ohe, Y.; Shibata, T.; Seto, T.; Takahashi, T.; Nakagawa, K.; Tanaka, H.; Takeda, K.; Nishio, M.; Mori, K.; et al. Combined Chemotherapy with Cisplatin, Etoposide, and Irinotecan versus Topotecan Alone as Second-Line Treatment for Patients with Sensitive Relapsed Small-Cell Lung Cancer (JCOG0605): A Multicentre, Open-Label, Randomised Phase 3 Trial. Lancet Oncol. 2016, 17, 1147–1157. [Google Scholar] [CrossRef]
- Chen, Q.-H.; Wu, B.-K.; Pan, D.; Sang, L.-X.; Chang, B. Beta-Carotene and Its Protective Effect on Gastric Cancer. World J. Clin. Cases 2021, 9, 6591–6607. [Google Scholar] [CrossRef]
- Mangla, B.; Javed, S.; Sultan, M.H.; Kumar, P.; Kohli, K.; Najmi, A.; Alhazmi, H.A.; Al Bratty, M.; Ahsan, W. Sulforaphane: A Review of Its Therapeutic Potentials, Advances in Its Nanodelivery, Recent Patents, and Clinical Trials. Phytother. Res. 2021, 35, 5440–5458. [Google Scholar] [CrossRef]
- Mashhadi Akbar Boojar, M.; Mashhadi Akbar Boojar, M.; Golmohammad, S. Overview of Silibinin Anti-Tumor Effects. J. Herb. Med. 2020, 23, 100375. [Google Scholar] [CrossRef]
- Thomford, N.; Senthebane, D.; Rowe, A.; Munro, D.; Seele, P.; Maroyi, A.; Dzobo, K. Natural Products for Drug Discovery in the 21st Century: Innovations for Novel Drug Discovery. Int. J. Mol. Sci. 2018, 19, 1578. [Google Scholar] [CrossRef]
- Singh, I.P.; Ahmad, F.; Chatterjee, D.; Bajpai, R.; Sengar, N. Natural Products: Drug Discovery and Development. In Drug Discovery and Development: From Targets and Molecules to Medicines; Poduri, R., Ed.; Springer: Singapore, 2021; pp. 11–65. ISBN 9789811555343. [Google Scholar]
- Calixto, J.B. The Role of Natural Products in Modern Drug Discovery. An. Acad. Bras. Ciências 2019, 91, 1–7. [Google Scholar] [CrossRef]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W. Computational Methods in Drug Discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Yao, X.J.; Gu, R.-X. Editorial: Computational Approaches in Drug Discovery and Precision Medicine. Front. Chem. 2021, 8, 639449. [Google Scholar] [CrossRef] [PubMed]
- Ou-Yang, S.; Lu, J.; Kong, X.; Liang, Z.; Luo, C.; Jiang, H. Computational Drug Discovery. Acta Pharmacol. Sin. 2012, 33, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Tutone, M.; Almerico, A.M. Computational Approaches: Drug Discovery and Design in Medicinal Chemistry and Bioinformatics. Molecules 2021, 26, 7500. [Google Scholar] [CrossRef] [PubMed]
- Iwaloye, O.; Ottu, P.O.; Olawale, F.; Babalola, O.O.; Elekofehinti, O.O.; Kikiowo, B.; Adegboyega, A.E.; Ogbonna, H.N.; Adeboboye, C.F.; Folorunso, I.M.; et al. Computer-Aided Drug Design in Anti-Cancer Drug Discovery: What Have We Learnt and What Is the Way Forward? Inform. Med. Unlocked 2023, 41, 101332. [Google Scholar] [CrossRef]
- Issa, N.T.; Stathias, V.; Schürer, S.; Dakshanamurthy, S. Machine and Deep Learning Approaches for Cancer Drug Repurposing. Semin. Cancer Biol. 2021, 68, 132–142. [Google Scholar] [CrossRef]
- Prada-Gracia, D.; Huerta-Yépez, S.; Moreno-Vargas, L.M. Application of Computational Methods for Anticancer Drug Discovery, Design, and Optimization. Boletín Médico Hosp. Infant. México 2016, 73, 411–423. [Google Scholar] [CrossRef]
- Li, K.; Du, Y.; Li, L.; Wei, D.-Q. Bioinformatics Approaches for Anti-Cancer Drug Discovery. Curr. Drug Targets 2019, 21, 3–17. [Google Scholar] [CrossRef]
- Goetz, L.H.; Schork, N.J. Personalized Medicine: Motivation, Challenges, and Progress. Fertil. Steril. 2018, 109, 952–963. [Google Scholar] [CrossRef]
- Qureshi, R.; Irfan, M.; Gondal, T.M.; Khan, S.; Wu, J.; Hadi, M.U.; Heymach, J.; Le, X.; Yan, H.; Alam, T. AI in Drug Discovery and Its Clinical Relevance. Heliyon 2023, 9, e17575. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, R.; Suvarna, K.; Yamada, M.; Kobayashi, K.; Shinkai, N.; Miyake, M.; Takahashi, M.; Jinnai, S.; Shimoyama, R.; Sakai, A.; et al. Application of Artificial Intelligence Technology in Oncology: Towards the Establishment of Precision Medicine. Cancers 2020, 12, 3532. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Yu, P.; Yang, S.; Li, Z.; Zhang, Q.; Liu, Z.; Li, H. Discovery of Novel Mono-Carbonyl Curcumin Derivatives as Potential Anti-Hepatoma Agents. Molecules 2023, 28, 6796. [Google Scholar] [CrossRef] [PubMed]
- Nadendla, R.R. Molecular Modeling: A Powerful Tool for Drug Design and Molecular Docking. Resonance 2004, 9, 51–60. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput. Aided-Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Adelusi, T.I.; Oyedele, A.-Q.K.; Boyenle, I.D.; Ogunlana, A.T.; Adeyemi, R.O.; Ukachi, C.D.; Idris, M.O.; Olaoba, O.T.; Adedotun, I.O.; Kolawole, O.E.; et al. Molecular Modeling in Drug Discovery. Inform. Med. Unlocked 2022, 29, 100880. [Google Scholar] [CrossRef]
- Salo-Ahen, O.M.H.; Alanko, I.; Bhadane, R.; Bonvin, A.M.J.J.; Honorato, R.V.; Hossain, S.; Juffer, A.H.; Kabedev, A.; Lahtela-Kakkonen, M.; Larsen, A.S.; et al. Molecular Dynamics Simulations in Drug Discovery and Pharmaceutical Development. Processes 2020, 9, 71. [Google Scholar] [CrossRef]
- Behl, T.; Kaur, I.; Sehgal, A.; Singh, S.; Bhatia, S.; Al-Harrasi, A.; Zengin, G.; Babes, E.E.; Brisc, C.; Stoicescu, M.; et al. Bioinformatics Accelerates the Major Tetrad: A Real Boost for the Pharmaceutical Industry. Int. J. Mol. Sci. 2021, 22, 6184. [Google Scholar] [CrossRef]
- Panda, S.S.; Tran, Q.L.; Rajpurohit, P.; Pillai, G.G.; Thomas, S.J.; Bridges, A.E.; Capito, J.E.; Thangaraju, M.; Lokeshwar, B.L. Design, Synthesis, and Molecular Docking Studies of Curcumin Hybrid Conjugates as Potential Therapeutics for Breast Cancer. Pharmaceuticals 2022, 15, 451. [Google Scholar] [CrossRef]
- Xu, X.; Liu, A.; Liu, S.; Ma, Y.; Zhang, X.; Zhang, M.; Zhao, J.; Sun, S.; Sun, X. Application of Molecular Dynamics Simulation in Self-Assembled Cancer Nanomedicine. Biomater. Res. 2023, 27, 39. [Google Scholar] [CrossRef]
- De Vivo, M.; Masetti, M.; Bottegoni, G.; Cavalli, A. Role of Molecular Dynamics and Related Methods in Drug Discovery. J. Med. Chem. 2016, 59, 4035–4061. [Google Scholar] [CrossRef]
- Durrant, J.D.; McCammon, J.A. Molecular Dynamics Simulations and Drug Discovery. BMC Biol. 2011, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Ikemura, S.; Yasuda, H.; Matsumoto, S.; Kamada, M.; Hamamoto, J.; Masuzawa, K.; Kobayashi, K.; Manabe, T.; Arai, D.; Nakachi, I.; et al. Molecular Dynamics Simulation-Guided Drug Sensitivity Prediction for Lung Cancer with Rare EGFR Mutations. Proc. Natl. Acad. Sci. USA 2019, 116, 10025–10030. [Google Scholar] [CrossRef] [PubMed]
- Bunker, A.; Róg, T. Mechanistic Understanding From Molecular Dynamics Simulation in Pharmaceutical Research 1: Drug Delivery. Front. Mol. Biosci. 2020, 7, 604770. [Google Scholar] [CrossRef] [PubMed]
- Bag, S.; Burman, M.D.; Bhowmik, S. Structural Insights and Shedding Light on Preferential Interactions of Dietary Flavonoids with G-Quadruplex DNA Structures: A New Horizon. Heliyon 2023, 9, e13959. [Google Scholar] [CrossRef] [PubMed]
- Tawani, A.; Kumar, A. Structural Insight into the Interaction of Flavonoids with Human Telomeric Sequence. Sci. Rep. 2015, 5, 17574. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, H.; Lin, H.; Wen, R.; Yang, F. High-Throughput Screening and Molecular Dynamics Simulation of Natural Products for the Identification of Anticancer Agents against MCM7 Protein. Appl. Bionics Biomech. 2022, 2022, 8308192. [Google Scholar] [CrossRef] [PubMed]
- Kontoyianni, M. Docking and Virtual Screening in Drug Discovery. In Proteomics for Drug Discovery: Methods and Protocols; Lazar, I.M., Kontoyianni, M., Lazar, A.C., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 255–266. ISBN 978-1-4939-7201-2. [Google Scholar] [CrossRef]
- Berry, M.; Fielding, B.; Gamieldien, J. Practical Considerations in Virtual Screening and Molecular Docking. In Emerging Trends in Computational Biology, Bioinformatics, and Systems Biology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 487–502. [Google Scholar] [CrossRef]
- Maia, E.H.B.; Assis, L.C.; de Oliveira, T.A.; da Silva, A.M.; Taranto, A.G. Structure-Based Virtual Screening: From Classical to Artificial Intelligence. Front. Chem. 2020, 8, 343. [Google Scholar] [CrossRef]
- De Oliveira, T.A.; da Silva, M.P.; Maia, E.H.B.; da Silva, A.M.; Taranto, A.G. Virtual Screening Algorithms in Drug Discovery: A Review Focused on Machine and Deep Learning Methods. Drugs Drug Candidates 2023, 2, 71. [Google Scholar] [CrossRef]
- Cheng, T.; Li, Q.; Zhou, Z.; Wang, Y.; Bryant, S.H. Structure-Based Virtual Screening for Drug Discovery: A Problem-Centric Review. AAPS J. 2012, 14, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-J.; Leung, K.-H.; Chan, D.S.-H.; Wang, Y.-T.; Ma, D.-L.; Leung, C.-H. Identification of a Natural Product-like STAT3 Dimerization Inhibitor by Structure-Based Virtual Screening. Cell Death Dis. 2014, 5, e1293. [Google Scholar] [CrossRef] [PubMed]
- Cherkasov, A.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.I.; Cronin, M.; Dearden, J.; Gramatica, P.; Martin, Y.C.; Todeschini, R.; et al. QSAR Modeling: Where Have You Been? Where Are You Going To? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef]
- Sen, D.; Chatterjee, T. Pharmacophore Modeling and 3D Quantitative Structure-Activity Relationship Analysis of Febrifugine Analogues as Potent Antimalarial Agent. J. Adv. Pharm. Technol. Res. 2013, 4, 50. [Google Scholar] [CrossRef] [PubMed]
- Giordano, D.; Biancaniello, C.; Argenio, M.A.; Facchiano, A. Drug Design by Pharmacophore and Virtual Screening Approach. Pharmaceuticals 2022, 15, 646. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Chandra, S.; Tiwari, N.; Subbarao, N. 3D QSAR, Pharmacophore and Molecular Docking Studies of Known Inhibitors and Designing of Novel Inhibitors for M18 Aspartyl Aminopeptidase of Plasmodium Falciparum. BMC Struct. Biol. 2016, 16, 12. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; MacKerell, A.D. Computer-Aided Drug Design Methods; Humana Press: New York, NY, USA, 2017; pp. 85–106. [Google Scholar] [CrossRef]
- Neves, B.J.; Braga, R.C.; Melo-Filho, C.C.; Moreira-Filho, J.T.; Muratov, E.N.; Andrade, C.H. QSAR-Based Virtual Screening: Advances and Applications in Drug Discovery. Front. Pharmacol. 2018, 9, 1275. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.; Bae, H.; Jo, J.; Yoon, S. Comprehensive Ensemble in QSAR Prediction for Drug Discovery. BMC Bioinform. 2019, 20, 521. [Google Scholar] [CrossRef]
- Danishuddin; Khan, A.U. Descriptors and Their Selection Methods in QSAR Analysis: Paradigm for Drug Design. Drug Discov. Today 2016, 21, 1291–1302. [Google Scholar] [CrossRef]
- Haji Agha Bozorgi, A.; Zarghi, A. Search for the Pharmacophore of Histone Deacetylase Inhibitors Using Pharmacophore Query and Docking Study. Iran. J. Pharm. Res. 2014, 13, 1165–1172. [Google Scholar]
- Qiu, X.; Zhu, L.; Wang, H.; Tan, Y.; Yang, Z.; Yang, L.; Wan, L. From Natural Products to HDAC Inhibitors: An Overview of Drug Discovery and Design Strategy. Bioorg. Med. Chem. 2021, 52, 116510. [Google Scholar] [CrossRef]
- Shirbhate, E.; Pandey, J.; Patel, V.K.; Veerasamy, R.; Rajak, H. Exploration of Structure-Activity Relationship Using Integrated Structure and Ligand Based Approach: Hydroxamic Acid-Based HDAC Inhibitors and Cytotoxic Agents. Turk. J. Pharm. Sci. 2023, 20, 270–284. [Google Scholar] [CrossRef] [PubMed]
- Pai, P.; Kumar, A.; Shetty, M.G.; Kini, S.G.; Krishna, M.B.; Satyamoorthy, K.; Babitha, K.S. Identification of Potent HDAC 2 Inhibitors Using E-Pharmacophore Modelling, Structure-Based Virtual Screening and Molecular Dynamic Simulation. J. Mol. Model. 2022, 28, 119. [Google Scholar] [CrossRef]
- Xiang, Y.; Hou, Z.; Zhang, Z. Pharmacophore and QSAR Studies to Design Novel Histone Deacetylase 2 Inhibitors. Chem. Biol. Drug Des. 2012, 79, 760–770. [Google Scholar] [CrossRef] [PubMed]
- AbdElmoniem, N.; Abdallah, M.H.; Mukhtar, R.M.; Moutasim, F.; Rafie Ahmed, A.; Edris, A.; Ibraheem, W.; Makki, A.A.; Elshamly, E.M.; Elhag, R.; et al. Identification of Novel Natural Dual HDAC and Hsp90 Inhibitors for Metastatic TNBC Using E-Pharmacophore Modeling, Molecular Docking, and Molecular Dynamics Studies. Molecules 2023, 28, 1771. [Google Scholar] [CrossRef]
- Lin, X.; Li, X.; Lin, X. A Review on Applications of Computational Methods in Drug Screening and Design. Molecules 2020, 25, 1375. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Lou, L.; Xie, Z.-R. A Pilot Study of All-Computational Drug Design Protocol–From Structure Prediction to Interaction Analysis. Front. Chem. 2020, 8, 81. [Google Scholar] [CrossRef]
- Moshari, M.; Wang, Q.; Michalak, M.; Klobukowski, M.; Tuszynski, J.A. Computational Prediction and Experimental Validation of the Unique Molecular Mode of Action of Scoulerine. Molecules 2022, 27, 3991. [Google Scholar] [CrossRef]
- Mohs, R.C.; Greig, N.H. Drug Discovery and Development: Role of Basic Biological Research. Alzheimer’s Dement. 2017, 3, 651–657. [Google Scholar] [CrossRef]
- Rosier, J.A.; Martens, M.A.; Thomas, J.R. Methods and Techniques Used in Drug Development. In Global New Drug Development; Wiley: Hoboken, NJ, USA, 2014; pp. 91–168. [Google Scholar]
- Cummings, J.; Ward, T.H.; Greystoke, A.; Ranson, M.; Dive, C. Biomarker Method Validation in Anticancer Drug Development. Br. J. Pharmacol. 2008, 153, 646–656. [Google Scholar] [CrossRef]
- Levin, V.A.; Abrey, L.E.; Heffron, T.P.; Tonge, P.J.; Dar, A.C.; Weiss, W.A.; Gallo, J.M. CNS Anticancer Drug Discovery and Development: 2016 Conference Insights. CNS Oncol. 2017, 6, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Khadempar, S.; Lotfi, M.; Haghiralsadat, F.; Saidijam, M.; Ghasemi, N.; Afshar, S. Lansoprazole as a Potent HDAC2 Inhibitor for Treatment of Colorectal Cancer: An in-Silico Analysis and Experimental Validation. Comput. Biol. Med. 2023, 166, 107518. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, H.-L.; Chen, X.-B.; Jin, W.-Y.; Zhou, H.; Ma, Y.; Wang, R.-L. 3D QSAR Pharmacophore Based Virtual Screening for Identification of Potential Inhibitors for CDC25B. Comput. Biol. Chem. 2018, 73, 1–12. [Google Scholar] [CrossRef]
- Robert, B.M.; Brindha, G.R.; Santhi, B.; Kanimozhi, G.; Prasad, N.R. Computational Models for Predicting Anticancer Drug Efficacy: A Multi Linear Regression Analysis Based on Molecular, Cellular and Clinical Data of Oral Squamous Cell Carcinoma Cohort. Comput. Methods Programs Biomed. 2019, 178, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Wang, L. In Vitro Human Cell Line Models to Predict Clinical Response to Anticancer Drugs. Pharmacogenomics 2015, 16, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Larsson, P.; Engqvist, H.; Biermann, J.; Werner Rönnerman, E.; Forssell-Aronsson, E.; Kovács, A.; Karlsson, P.; Helou, K.; Parris, T.Z. Optimization of Cell Viability Assays to Improve Replicability and Reproducibility of Cancer Drug Sensitivity Screens. Sci. Rep. 2020, 10, 5798. [Google Scholar] [CrossRef] [PubMed]
- Samarghandian, S.; Boskabady, M.; Davoodi, S. Use of in Vitro Assays to Assess the Potential Antiproliferative and Cytotoxic Effects of Saffron (Crocus sativus L.) in Human Lung Cancer Cell Line. Pharmacogn. Mag. 2010, 6, 309. [Google Scholar] [CrossRef]
- Lovitt, C.; Shelper, T.; Avery, V. Advanced Cell Culture Techniques for Cancer Drug Discovery. Biology 2014, 3, 345–367. [Google Scholar] [CrossRef]
- Hulkower, K.I.; Herber, R.L. Cell Migration and Invasion Assays as Tools for Drug Discovery. Pharmaceutics 2011, 3, 107–124. [Google Scholar] [CrossRef]
- Caicedo, J.C.; Cooper, S.; Heigwer, F.; Warchal, S.; Qiu, P.; Molnar, C.; Vasilevich, A.S.; Barry, J.D.; Bansal, H.S.; Kraus, O.; et al. Data-Analysis Strategies for Image-Based Cell Profiling. Nat. Methods 2017, 14, 849–863. [Google Scholar] [CrossRef]
- Fulda, S. Betulinic Acid for Cancer Treatment and Prevention. Int. J. Mol. Sci. 2008, 9, 1096–1107. [Google Scholar] [CrossRef]
- Jiang, W.; Li, X.; Dong, S.; Zhou, W. Betulinic Acid in the Treatment of Tumour Diseases: Application and Research Progress. Biomed. Pharmacother. 2021, 142, 111990. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Liang, N.; Liu, Y.; Cheng, M.-S. Recent Progress on Betulinic Acid and Its Derivatives as Antitumor Agents: A Mini Review. Chin. J. Nat. Med. 2021, 19, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Hordyjewska, A.; Ostapiuk, A.; Horecka, A.; Kurzepa, J. Betulin and Betulinic Acid: Triterpenoids Derivatives with a Powerful Biological Potential. Phytochem. Rev. 2019, 18, 929–951. [Google Scholar] [CrossRef]
- Chrobak, E.; Kadela-Tomanek, M.; Bębenek, E.; Marciniec, K.; Wietrzyk, J.; Trynda, J.; Pawełczak, B.; Kusz, J.; Kasperczyk, J.; Chodurek, E.; et al. New Phosphate Derivatives of Betulin as Anticancer Agents: Synthesis, Crystal Structure, and Molecular Docking Study. Bioorg. Chem. 2019, 87, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Li, H.; Zhang, S.; Lu, H.; Chen, Q. A Review on Preparation of Betulinic Acid and Its Biological Activities. Molecules 2021, 26, 5583. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lee, S.R.; Kang, K.S.; Ko, Y.; Pang, C.; Yamabe, N.; Kim, K.H. Betulinic Acid Suppresses Ovarian Cancer Cell Proliferation through Induction of Apoptosis. Biomolecules 2019, 9, 257. [Google Scholar] [CrossRef]
- Qi, X.; Gao, C.; Yin, C.; Fan, J.; Wu, X.; Guo, C. Improved Anticancer Activity of Betulinic Acid on Breast Cancer through a Grafted Copolymer-Based Micelles System. Drug Deliv. 2021, 28, 1962–1971. [Google Scholar] [CrossRef]
- Ali-Seyed, M.; Jantan, I.; Vijayaraghavan, K.; Bukhari, S.N.A. Betulinic Acid: Recent Advances in Chemical Modifications, Effective Delivery, and Molecular Mechanisms of a Promising Anticancer Therapy. Chem. Biol. Drug Des. 2016, 87, 517–536. [Google Scholar] [CrossRef]
- Wang, W.; Rayburn, E.R.; Velu, S.E.; Nadkarni, D.H.; Murugesan, S.; Zhang, R. In Vitro and In Vivo Anticancer Activity of Novel Synthetic Makaluvamine Analogues. Clin. Cancer Res. 2009, 15, 3511–3518. [Google Scholar] [CrossRef]
- Seifaddinipour, M.; Farghadani, R.; Namvar, F.; Bin Mohamad, J.; Muhamad, N.A. In Vitro and In Vivo Anticancer Activity of the Most Cytotoxic Fraction of Pistachio Hull Extract in Breast Cancer. Molecules 2020, 25, 1776. [Google Scholar] [CrossRef] [PubMed]
- Lawal, B.; Kuo, Y.-C.; Sumitra, M.R.; Wu, A.T.; Huang, H.-S. In Vivo Pharmacokinetic and Anticancer Studies of HH-N25, a Selective Inhibitor of Topoisomerase I, and Hormonal Signaling for Treating Breast Cancer. J. Inflamm. Res. 2021, 14, 4901–4913. [Google Scholar] [CrossRef] [PubMed]
- Workman, P. Bottlenecks in Anticancer Drug Discovery and Development: In Vivo Pharmacokinetic and Pharmacodynamic Issues and the Potential Role of PET. In PET for Drug Development and Evaluation; Springer: Dordrecht, The Netherlands, 1995; pp. 277–285. [Google Scholar]
- Li, Z.; Zheng, W.; Wang, H.; Cheng, Y.; Fang, Y.; Wu, F.; Sun, G.; Sun, G.; Lv, C.; Hui, B. Application of Animal Models in Cancer Research: Recent Progress and Future Prospects. Cancer Manag. Res. 2021, 13, 2455–2475. [Google Scholar] [CrossRef]
- Zhuo, Z.; Yu, Y.; Wang, M.; Li, J.; Zhang, Z.; Liu, J.; Wu, X.; Lu, A.; Zhang, G.; Zhang, B. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017, 18, 2142. [Google Scholar] [CrossRef]
- Atteeq, M. Evaluating Anticancer Properties of Withaferin A—A Potent Phytochemical. Front. Pharmacol. 2022, 13, 975320. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.-C.; Choi, B. Withaferin-A—A Natural Anticancer Agent with Pleitropic Mechanisms of Action. Int. J. Mol. Sci. 2016, 17, 290. [Google Scholar] [CrossRef]
- Hassannia, B.; Logie, E.; Vandenabeele, P.; Vanden Berghe, T.; Vanden Berghe, W. Withaferin A: From Ayurvedic Folk Medicine to Preclinical Anti-Cancer Drug. Biochem. Pharmacol. 2020, 173, 113602. [Google Scholar] [CrossRef]
- Xing, Z.; Su, A.; Mi, L.; Zhang, Y.; He, T.; Qiu, Y.; Wei, T.; Li, Z.; Zhu, J.; Wu, W. Withaferin A: A Dietary Supplement with Promising Potential as an Anti-Tumor Therapeutic for Cancer Treatment—Pharmacology and Mechanisms. Drug Des. Dev. Ther. 2023, 17, 2909–2929. [Google Scholar] [CrossRef]
- Sultana, T.; Okla, M.K.; Ahmed, M.; Akhtar, N.; Al-Hashimi, A.; Abdelgawad, H.; Haq, I. Withaferin A: From Ancient Remedy to Potential Drug Candidate. Molecules 2021, 26, 7696. [Google Scholar] [CrossRef]
- Minami, H.; Kiyota, N.; Kimbara, S.; Ando, Y.; Shimokata, T.; Ohtsu, A.; Fuse, N.; Kuboki, Y.; Shimizu, T.; Yamamoto, N.; et al. Guidelines for Clinical Evaluation of Anti-cancer Drugs. Cancer Sci. 2021, 112, 2563–2577. [Google Scholar] [CrossRef]
- Olivier, T.; Haslam, A.; Prasad, V. Anticancer Drugs Approved by the US Food and Drug Administration From 2009 to 2020 According to Their Mechanism of Action. JAMA Netw. Open 2021, 4, e2138793. [Google Scholar] [CrossRef] [PubMed]
- Choudhari, A.S.; Mandave, P.C.; Deshpande, M.; Ranjekar, P.; Prakash, O. Phytochemicals in Cancer Treatment: From Preclinical Studies to Clinical Practice. Front. Pharmacol. 2020, 10, 1614. [Google Scholar] [CrossRef]
- Wong, C.C.; Cheng, K.-W.; Rigas, B. Preclinical Predictors of Anticancer Drug Efficacy: Critical Assessment with Emphasis on Whether Nanomolar Potency Should Be Required of Candidate Agents: TABLE 1. J. Pharmacol. Exp. Ther. 2012, 341, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Bajaj, S.; Bodla, R. Preclinical Screening Methods in Cancer. Indian J. Pharmacol. 2016, 48, 481. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Aouidate, A.; Wang, S.; Yu, Q.; Li, Y.; Yuan, S. Discovering Anti-Cancer Drugs via Computational Methods. Front. Pharmacol. 2020, 11, 733. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Hwang, B.Y.; Su, B.-N.; Chai, H.; Mi, Q.; Kinghorn, A.D.; Wild, R.; Swanson, S.M. Silvestrol, a Potential Anticancer Rocaglate Derivative from Aglaia Foveolata, Induces Apoptosis in LNCaP Cells through the Mitochondrial/Apoptosome Pathway without Activation of Executioner Caspase-3 or -7. Anticancer Res. 2007, 27, 2175–2183. [Google Scholar]
- Kinghorn, A.D.; Chin, Y.-W.; Swanson, S.M. Discovery of Natural Product Anticancer Agents from Biodiverse Organisms. Curr. Opin. Drug Discov. Dev. 2009, 12, 189–196. [Google Scholar]
- Kandi, V.; Vadakedath, S. Clinical Trials and Clinical Research: A Comprehensive Review. Cureus 2023, 15, 1–15. [Google Scholar] [CrossRef]
- Types and Phases of Clinical Trials|What Are Clinical Trial Phases?|American Cancer Society. Available online: https://www.cancer.org/cancer/managing-cancer/making-treatment-decisions/clinical-trials/what-you-need-to-know/phases-of-clinical-trials.html (accessed on 20 November 2023).
- Xu, C.; Zhang, H.; Mu, L.; Yang, X. Artemisinins as Anticancer Drugs: Novel Therapeutic Approaches, Molecular Mechanisms, and Clinical Trials. Front. Pharmacol. 2020, 11, 529881. [Google Scholar] [CrossRef]
- Augustin, Y.; Staines, H.M.; Krishna, S. Artemisinins as a Novel Anti-Cancer Therapy: Targeting a Global Cancer Pandemic through Drug Repurposing. Pharmacol. Ther. 2020, 216, 107706. [Google Scholar] [CrossRef]
- Krishna, S.; Ganapathi, S.; Ster, I.C.; Saeed, M.E.M.; Cowan, M.; Finlayson, C.; Kovacsevics, H.; Jansen, H.; Kremsner, P.G.; Efferth, T.; et al. A Randomised, Double Blind, Placebo-Controlled Pilot Study of Oral Artesunate Therapy for Colorectal Cancer. EBioMedicine 2015, 2, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Ong, F.S.; Das, K.; Wang, J.; Vakil, H.; Kuo, J.Z.; Blackwell, W.-L.B.; Lim, S.W.; Goodarzi, M.O.; Bernstein, K.E.; Rotter, J.I.; et al. Personalized Medicine and Pharmacogenetic Biomarkers: Progress in Molecular Oncology Testing. Expert Rev. Mol. Diagn. 2012, 12, 593–602. [Google Scholar] [CrossRef]
- Hu, C.; Dignam, J.J. Biomarker-Driven Oncology Clinical Trials: Key Design Elements, Types, Features, and Practical Considerations. JCO Precis. Oncol. 2019, 1, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Morrison, K.C.; Hergenrother, P.J. Natural Products as Starting Points for the Synthesis of Complex and Diverse Compounds. Nat. Prod. Rep. 2014, 31, 6–14. [Google Scholar] [CrossRef]
- Wang, J. Natural Compounds as Anticancer Agents: Experimental Evidence. World J. Exp. Med. 2012, 2, 45. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, M.A.; Andreassend, S.K.; Keyzers, R.A.; Lein, M. Accurate Prediction of the Optical Rotation and NMR Properties for Highly Flexible Chiral Natural Products. Phys. Chem. Chem. Phys. 2016, 18, 24506–24510. [Google Scholar] [CrossRef]
- Kartal, M. Intellectual Property Protection in the Natural Product Drug Discovery, Traditional Herbal Medicine and Herbal Medicinal Products. Phytother. Res. 2007, 21, 113–119. [Google Scholar] [CrossRef]
- Baxi, S.M.; Beall, R.; Yang, J.; Mackey, T.K. A Multidisciplinary Review of the Policy, Intellectual Property Rights, and International Trade Environment for Access and Affordability to Essential Cancer Medications. Glob. Health 2019, 15, 57. [Google Scholar] [CrossRef]
- Spreafico, A.; Hansen, A.R.; Abdul Razak, A.R.; Bedard, P.L.; Siu, L.L. The Future of Clinical Trial Design in Oncology. Cancer Discov. 2021, 11, 822–837. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Kummar, S. Increasing Patient Participation in Oncology Clinical Trials. Cancer Med. 2023, 12, 2219–2226. [Google Scholar] [CrossRef]
- Najmi, A.; Javed, S.A.; Al Bratty, M.; Alhazmi, H.A. Modern Approaches in the Discovery and Development of Plant-Based Natural Products and Their Analogues as Potential Therapeutic Agents. Molecules 2022, 27, 349. [Google Scholar] [CrossRef]
- Katiyar, C.; Kanjilal, S.; Gupta, A.; Katiyar, S. Drug Discovery from Plant Sources: An Integrated Approach. AYU Int. Q. J. Res. Ayurveda 2012, 33, 10. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Lu, J.-J.; Ding, J. Natural Products in Cancer Therapy: Past, Present and Future. Nat. Prod. Bioprospect. 2021, 11, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural Products in Drug Discovery: Advances and Opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Rayan, A.; Raiyn, J.; Falah, M. Nature Is the Best Source of Anticancer Drugs: Indexing Natural Products for Their Anticancer Bioactivity. PLoS ONE 2017, 12, e0187925. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Sanap, G.; Shenoy, S.; Kalyane, D.; Kalia, K.; Tekade, R.K. Artificial Intelligence in Drug Discovery and Development. Drug Discov. Today 2021, 26, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Dara, S.; Dhamercherla, S.; Jadav, S.S.; Babu, C.M.; Ahsan, M.J. Machine Learning in Drug Discovery: A Review. Artif. Intell. Rev. 2022, 55, 1947–1999. [Google Scholar] [CrossRef]
- Sahayasheela, V.J.; Lankadasari, M.B.; Dan, V.M.; Dastager, S.G.; Pandian, G.N.; Sugiyama, H. Artificial Intelligence in Microbial Natural Product Drug Discovery: Current and Emerging Role. Nat. Prod. Rep. 2022, 39, 2215–2230. [Google Scholar] [CrossRef] [PubMed]
- Hallock, Y.; Cragg, G. National Cooperative Drug Discovery Groups (NCDDGs): A Successful Model for Public Private Partnerships in Cancer Drug Discovery. Pharm. Biol. 2003, 41, 78–91. [Google Scholar] [CrossRef]
- Davis, A.M.; Engkvist, O.; Fairclough, R.J.; Feierberg, I.; Freeman, A.; Iyer, P. Public-Private Partnerships: Compound and Data Sharing in Drug Discovery and Development. SLAS Discov. 2021, 26, 604–619. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chunarkar-Patil, P.; Kaleem, M.; Mishra, R.; Ray, S.; Ahmad, A.; Verma, D.; Bhayye, S.; Dubey, R.; Singh, H.N.; Kumar, S. Anticancer Drug Discovery Based on Natural Products: From Computational Approaches to Clinical Studies. Biomedicines 2024, 12, 201. https://doi.org/10.3390/biomedicines12010201
Chunarkar-Patil P, Kaleem M, Mishra R, Ray S, Ahmad A, Verma D, Bhayye S, Dubey R, Singh HN, Kumar S. Anticancer Drug Discovery Based on Natural Products: From Computational Approaches to Clinical Studies. Biomedicines. 2024; 12(1):201. https://doi.org/10.3390/biomedicines12010201
Chicago/Turabian StyleChunarkar-Patil, Pritee, Mohammed Kaleem, Richa Mishra, Subhasree Ray, Aftab Ahmad, Devvret Verma, Sagar Bhayye, Rajni Dubey, Himanshu Narayan Singh, and Sanjay Kumar. 2024. "Anticancer Drug Discovery Based on Natural Products: From Computational Approaches to Clinical Studies" Biomedicines 12, no. 1: 201. https://doi.org/10.3390/biomedicines12010201







