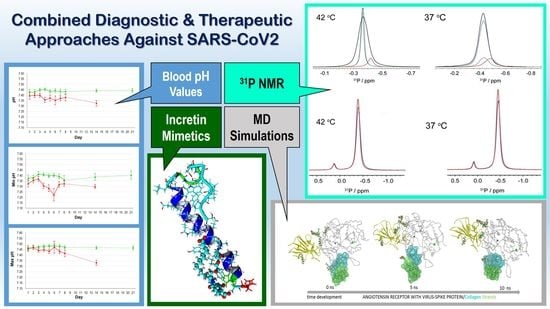
Blood pH Analysis in Combination with Molecular Medical Tools in Relation to COVID-19 Symptoms †
Abstract
:1. Introduction
2. Results
2.1. Clinical Studies
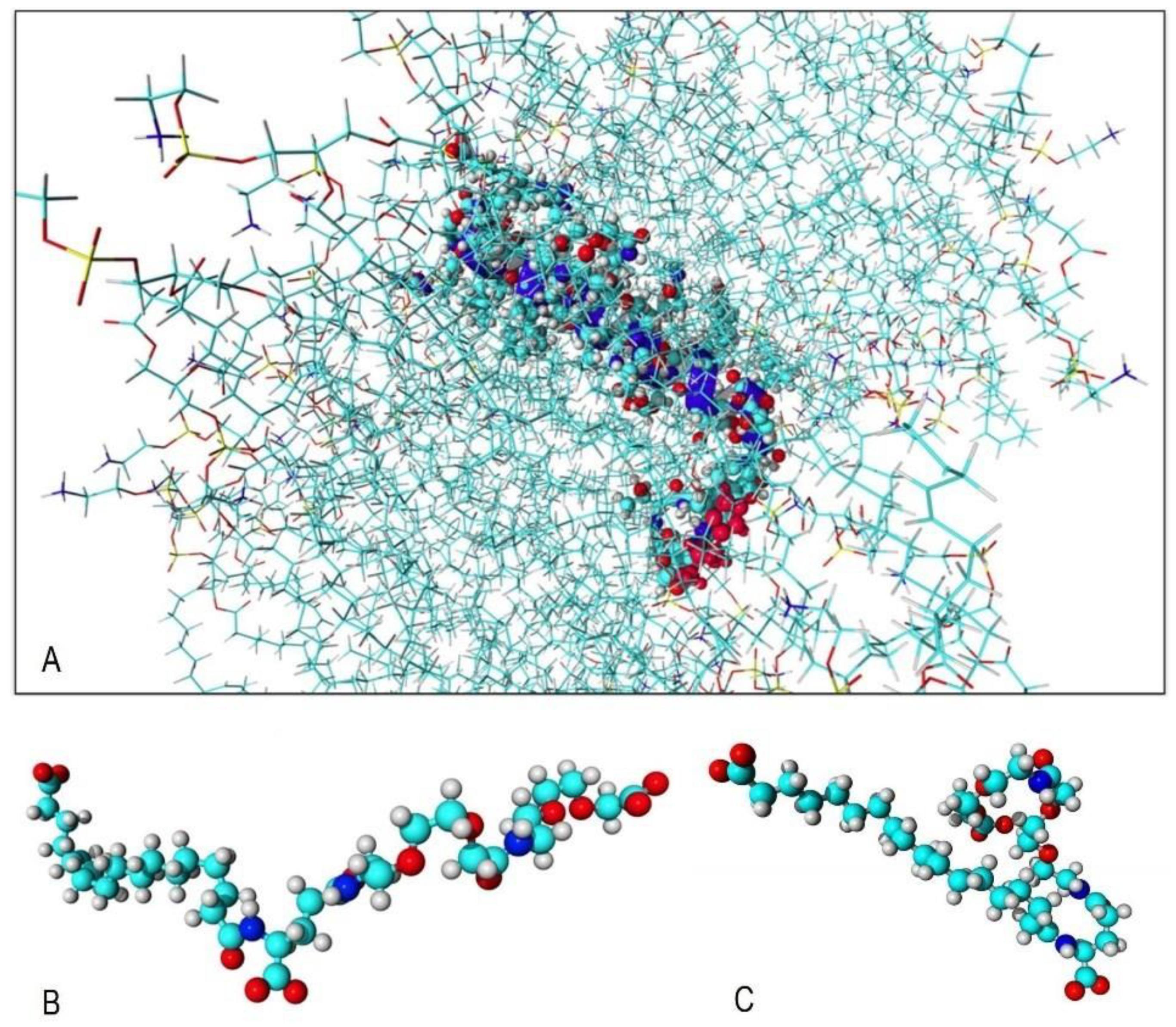
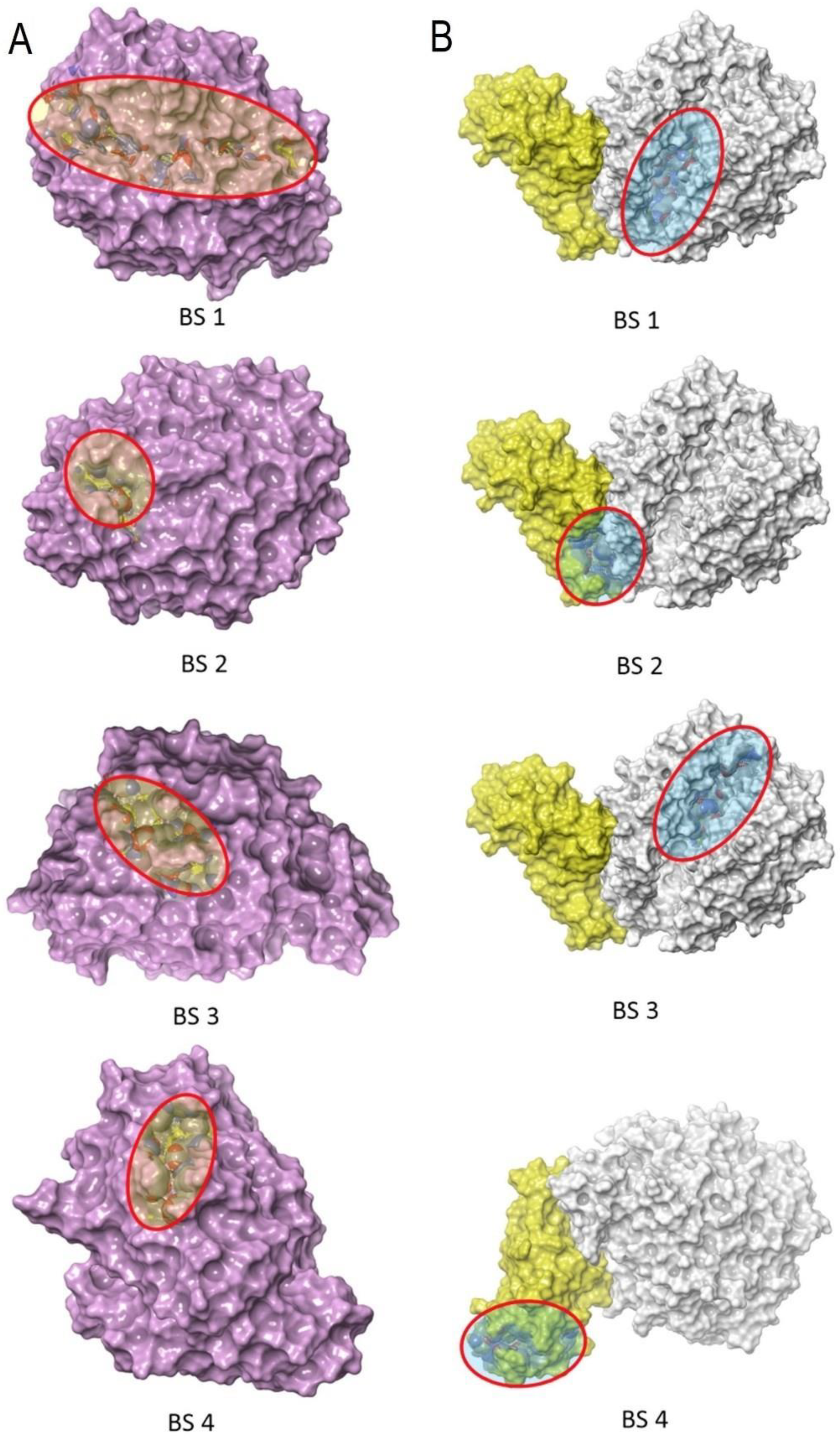
2.2. CAMM: ACE2 Receptor and Incretin Mimetics
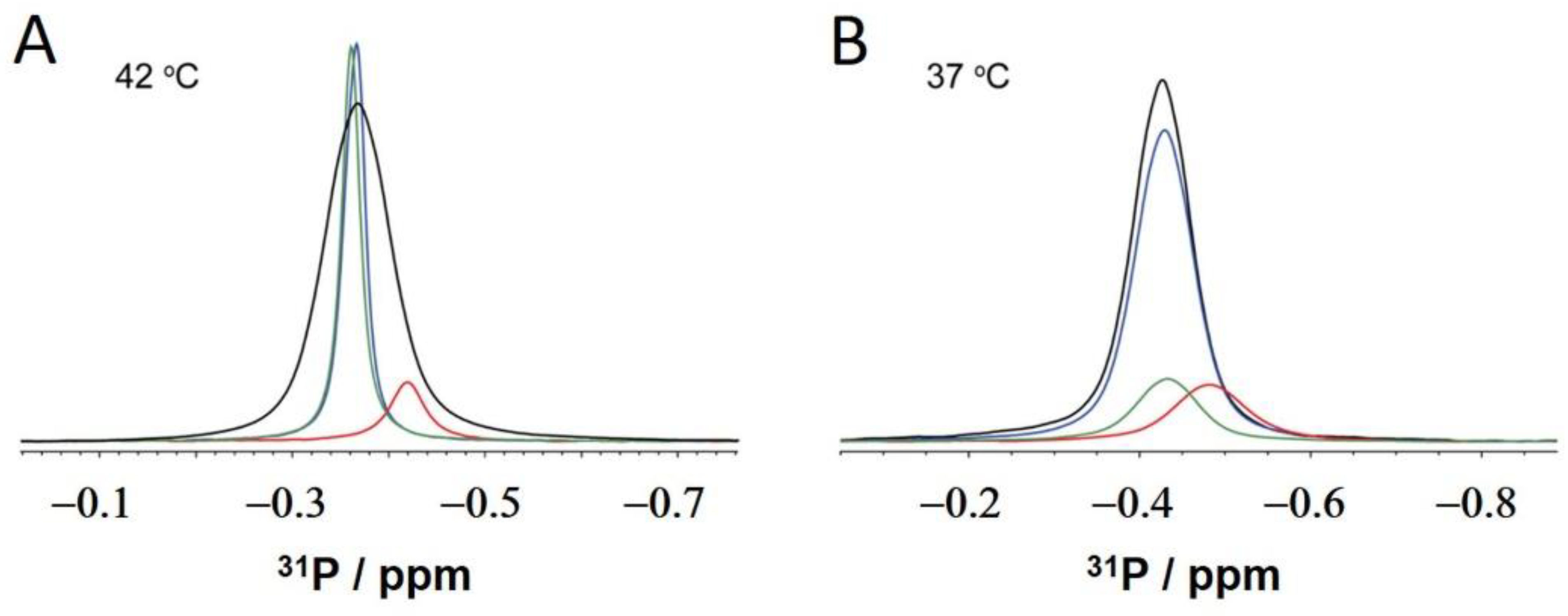
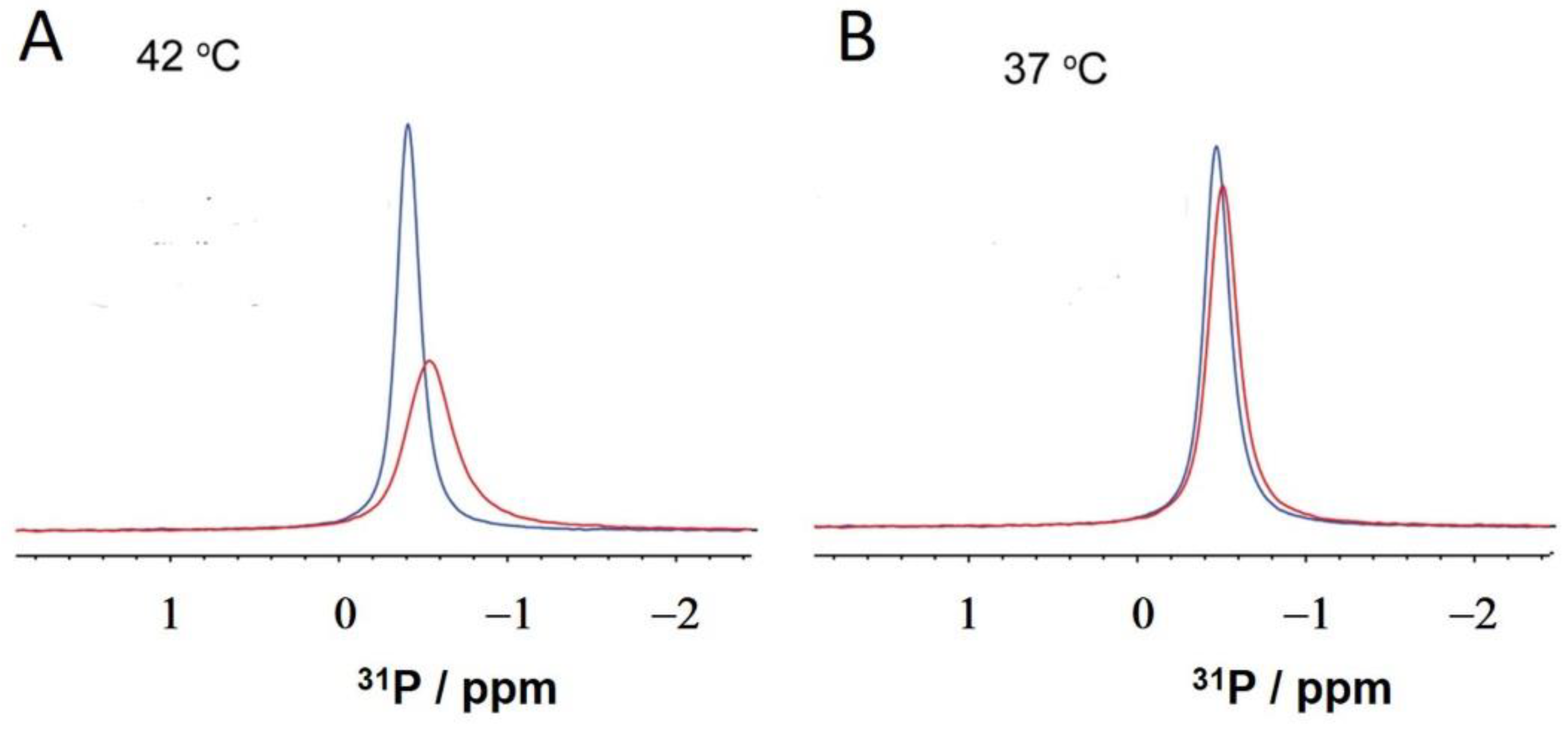
2.3. NMR: Semaglutide and Pt2His
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. NMR Spectroscopy
4.3. Computer-Aided Molecular Modeling (CAMM)
4.3.1. ACE2-Receptor/Spike Protein and Collagen Complexes
4.3.2. De Novo Protein Structure Prediction and Homology Modeling of Pt2His and Semaglutide
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siebert, H.C.; Reuter, G.; Schauer, R.; von der Lieth, C.W.; Dabrowski, J. Solution conformations of GM3 gangliosides containing different sialic acid residues as revealed by NOE-based distance mapping, molecular mechanics, and molecular dynamics calculations. Biochemistry 1992, 31, 6962–6971. [Google Scholar] [CrossRef]
- Mahajan, M.; Bhattacharjya, S. NMR structures and localization of the potential fusion peptides and the pre-transmembrane region of SARS-CoV: Implications in membrane fusion. Biochim. Biophys. Acta 2015, 1848, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, H.; Bhattacharjya, S. Mechanistic insights of host cell fusion of SARS-CoV-1 and SARS-CoV-2 from atomic resolution structure and membrane dynamics. Biophys. Chem. 2020, 265, 106438. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Liu, M.; Wang, C.; Xu, W.; Lan, Q.; Feng, S.; Qi, F.; Bao, L.; Du, L.; Liu, S.; et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020, 30, 343–355. [Google Scholar] [CrossRef]
- Tang, T.; Bidon, M.; Jaimes, J.A.; Whittaker, G.R.; Daniel, S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020, 178, 104792. [Google Scholar] [CrossRef] [PubMed]
- Monda, V.M.; Porcellati, F.; Strollo, F.; Gentile, S. ACE2 and SARS-CoV-2 Infection: Might GLP-1 Receptor Agonists Play a Role? Diabetes Ther. Res. Treat. Educ. Diabetes Relat. Disord. 2020, 11, 1909–1914. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wu, L.; Eckert, T.; Burg-Roderfeld, M.; Rojas-Macias, M.A.; Lütteke, T.; Krylov, V.B.; Argunov, D.A.; Datta, A.; Markart, P.; et al. Lysozyme’s lectin-like characteristics facilitates its immune defense function. Q. Rev. Biophys. 2017, 50, e9. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, N.; Mohri, M.; Wu, L.; Eckert, T.; Krylov, V.B.; Antosova, A.; Ponikova, S.; Bednarikova, Z.; Markart, P.; et al. Nanomedical Relevance of the Intermolecular Interaction Dynamics-Examples from Lysozymes and Insulins. ACS Omega 2019, 4, 4206–4220. [Google Scholar] [CrossRef]
- Jiménez-Barbero, J.; Javier Cañada, F.; Asensio, J.L.; Aboitiz, N.; Vidal, P.; Canales, A.; Groves, P.; Gabius, H.-J.; Siebert, H.-C. Hevein Domains: An Attractive Model to Study Carbohydrate–Protein Interactions at Atomic Resolution. Adv. Carbohydr. Chem. Biochem. 2006, 60, 303–354. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Jung, G.; Ruchala, P.; Andre, S.; Gabius, H.J.; Lu, W. Multivalent binding of carbohydrates by the human alpha-defensin, HD5. J. Immunol. 2009, 183, 480–490. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Lu, W. alpha-Defensins in human innate immunity. Immunol. Rev. 2012, 245, 84–112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Eckert, T.; Lütteke, T.; Hanstein, S.; Scheidig, A.; Bonvin, A.M.; Nifantiev, N.E.; Kozar, T.; Schauer, R.; Enani, M.A.; et al. Structure-Function Relationships of Antimicrobial Peptides and Proteins with Respect to Contact Molecules on Pathogen Surfaces. Curr. Top. Med. Chem. 2016, 16, 89–98. [Google Scholar] [CrossRef]
- Li, H.; Liu, L.; Zhang, D.; Xu, J.; Dai, H.; Tang, N.; Su, X.; Cao, B. SARS-CoV-2 and viral sepsis: Observations and hypotheses. Lancet 2020, 395, 1517–1520. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Yu, M.M.; Chai, Y.F.; Shou, S.T. Sialic Acids in the Immune Response during Sepsis. Front. Immunol. 2017, 8, 1601. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, L.; Bao, L.; Liu, J.; Zhu, H.; Lv, Q.; Liu, R.; Chen, W.; Tong, W.; Wei, Q.; et al. SARS-CoV-2 crosses the blood-brain barrier accompanied with basement membrane disruption without tight junctions alteration. Sign. Transduct. Target 2021, 6, 337. [Google Scholar] [CrossRef]
- Zougbede, S.; Miller, F.; Ravassard, P.; Rebollo, A.; Ciceron, L.; Couraud, P.O.; Mazier, D.; Moreno, A. Metabolic acidosis induced by Plasmodium falciparum intraerythrocytic stages alters blood-brain barrier integrity. J. Cereb. Blood Flow Metab. 2011, 31, 514–526. [Google Scholar] [CrossRef] [PubMed]
- Soares, C.O.; Grosso, A.S.; Ereno-Orbea, J.; Coelho, H.; Marcelo, F. Molecular Recognition Insights of Sialic Acid Glycans by Distinct Receptors Unveiled by NMR and Molecular Modeling. Front. Mol. Biosci. 2021, 8, 727847. [Google Scholar] [CrossRef]
- Siebert, H.C.; Born, K.; Andre, S.; Frank, M.; Kaltner, H.; von der Lieth, C.W.; Heck, A.J.; Jiménez-Barbero, J.; Kopitz, J.; Gabius, H.J. Carbohydrate chain of ganglioside GM1 as a ligand: Identification of the binding strategies of three 15 mer peptides and their divergence from the binding modes of growth-regulatory galectin-1 and cholera toxin. Chemistry 2005, 12, 388–402. [Google Scholar] [CrossRef]
- Siebert, H.C.; Lu, S.Y.; Wechselberger, R.; Born, K.; Eckert, T.; Liang, S.; von der Lieth, C.W.; Jiménez-Barbero, J.; Schauer, R.; Vliegenthart, J.F.; et al. A lectin from the Chinese bird-hunting spider binds sialic acids. Carbohydr. Res. 2009, 344, 1515–1525. [Google Scholar] [CrossRef]
- Zhang, R.; Loers, G.; Schachner, M.; Boelens, R.; Wienk, H.; Siebert, S.; Eckert, T.; Kraan, S.; Rojas-Macias, M.A.; Lütteke, T.; et al. Molecular Basis of the Receptor Interactions of Polysialic Acid (polySia), polySia Mimetics, and Sulfated Polysaccharides. Chem. Med. Chem. 2016, 11, 990–1002. [Google Scholar] [CrossRef]
- Siebert, H.C.; von der Lieth, C.W.; Dong, X.; Reuter, G.; Schauer, R.; Gabius, H.J.; Vliegenthart, J.F. Molecular dynamics-derived conformation and intramolecular interaction analysis of the N-acetyl-9-O-acetylneuraminic acid-containing ganglioside GD1a and NMR-based analysis of its binding to a human polyclonal immunoglobulin G fraction with selectivity for O-acetylated sialic acids. Glycobiology 1996, 6, 561–572. [Google Scholar] [CrossRef]
- Siebert, H.C.; Rosen, J.; Seyrek, K.; Kaltner, H.; Andre, S.; Bovin, N.V.; Nyholm, P.G.; Sinowatz, F.; Gabius, H.J. alpha2,3/alpha2,6-Sialylation of N-glycans: Non-synonymous signals with marked developmental regulation in bovine reproductive tracts. Biochimie 2006, 88, 399–410. [Google Scholar] [CrossRef]
- Zhang, R.; Jin, L.; Zhang, N.; Petridis, A.K.; Eckert, T.; Scheiner-Bobis, G.; Bergmann, M.; Scheidig, A.; Schauer, R.; Yan, M.; et al. The Sialic Acid-Dependent Nematocyst Discharge Process in Relation to Its Physical-Chemical Properties Is A Role Model for Nanomedical Diagnostic and Therapeutic Tools. Mar. Drugs 2019, 17, 469. [Google Scholar] [CrossRef] [PubMed]
- Sato, C.; Kitajima, K. Polysialylation and disease. Mol. Asp. Med. 2021, 79, 100892. [Google Scholar] [CrossRef]
- van Eijk, M.; Rynkiewicz, M.J.; Khatri, K.; Leymarie, N.; Zaia, J.; White, M.R.; Hartshorn, K.L.; Cafarella, T.R.; van Die, I.; Hessing, M.; et al. Lectin-mediated binding and sialoglycans of porcine surfactant protein D synergistically neutralize influenza A virus. J. Biol. Chem. 2018, 293, 10646–10662. [Google Scholar] [CrossRef]
- Gorgun, D.; Lihan, M.; Kapoor, K.; Tajkhorshid, E. Binding mode of SARS-CoV-2 fusion peptide to human cellular membrane. Biophys. J. 2021, 120, 2914–2926. [Google Scholar] [CrossRef]
- Lenza, M.P.; Oyenarte, I.; Diercks, T.; Quintana, J.I.; Gimeno, A.; Coelho, H.; Diniz, A.; Peccati, F.; Delgado, S.; Bosch, A.; et al. Structural Characterization of N-Linked Glycans in the Receptor Binding Domain of the SARS-CoV-2 Spike Protein and their Interactions with Human Lectins. Angew. Chem. Int. Ed. Eng. 2020, 59, 23763–23771. [Google Scholar] [CrossRef] [PubMed]
- Unione, L.; Arda, A.; Jiménez-Barbero, J.; Millet, O. NMR of glycoproteins: Profiling, structure, conformation and interactions. Curr. Opin. Struct. Biol. 2021, 68, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Harikishore, A.; Bhunia, A. Targeting C-terminal Helical bundle of NCOVID19 Envelope (E) protein. Int. J. Biol. Macromol. 2021, 175, 131–139. [Google Scholar] [CrossRef]
- Siebert, H.C.; Lu, S.Y.; Frank, M.; Kramer, J.; Wechselberger, R.; Joosten, J.; Andre, S.; Rittenhouse-Olson, K.; Roy, R.; von der Lieth, C.W.; et al. Analysis of protein-carbohydrate interaction at the lower size limit of the protein part (15-mer peptide) by NMR spectroscopy, electrospray ionization mass spectrometry, and molecular modeling. Biochemstry 2002, 41, 9707–9717. [Google Scholar] [CrossRef]
- Zhang, N.; Jin, L.; Liu, C.; Zhang, R.; Siebert, H.-C.; Li, Y.; Loers, G.; Petridis, A.K.; Xia, Z.; Dong, H.; et al. An antarctic krill oil-based diet elicits neuroprotective effects by inhibiting oxidative stress and rebalancing the M1/M2 microglia phenotype in a cuprizone model for demyelination. J. Funct. Foods 2021, 76, 104309. [Google Scholar] [CrossRef]
- Gu, R.X.; Ingolfsson, H.I.; de Vries, A.H.; Marrink, S.J.; Tieleman, D.P. Ganglioside-Lipid and Ganglioside-Protein Interactions Revealed by Coarse-Grained and Atomistic Molecular Dynamics Simulations. J. Phys. Chem. B 2017, 121, 3262–3275. [Google Scholar] [CrossRef] [PubMed]
- Siebert, H.C.; Andre, S.; Lu, S.Y.; Frank, M.; Kaltner, H.; van Kuik, J.A.; Korchagina, E.Y.; Bovin, N.; Tajkhorshid, E.; Kaptein, R.; et al. Unique conformer selection of human growth-regulatory lectin galectin-1 for ganglioside GM1 versus bacterial toxins. Biochemistry 2003, 42, 14762–14773. [Google Scholar] [CrossRef]
- Siebert, H.C.; Burg-Roderfeld, M.; Eckert, T.; Stötzel, S.; Kirch, U.; Diercks, T.; Humphries, M.J.; Frank, M.; Wechselberger, R.; Tajkhorshid, E.; et al. Interaction of the alpha2A domain of integrin with small collagen fragments. Protein Cell 2010, 1, 393–405. [Google Scholar] [CrossRef]
- Stötzel, S.; Schurink, M.; Wienk, H.; Siebler, U.; Burg-Roderfeld, M.; Eckert, T.; Kulik, B.; Wechselberger, R.; Sewing, J.; Steinmeyer, J.; et al. Molecular organization of various collagen fragments as revealed by atomic force microscopy and diffusion-ordered NMR spectroscopy. Chemphyschem 2012, 13, 3117–3125. [Google Scholar] [CrossRef] [PubMed]
- Schadow, S.; Simons, V.S.; Lochnit, G.; Kordelle, J.; Gazova, Z.; Siebert, H.C.; Steinmeyer, J. Metabolic Response of Human Osteoarthritic Cartilage to Biochemically Characterized Collagen Hydrolysates. Int. J. Mol. Sci. 2017, 18, 207. [Google Scholar] [CrossRef]
- Schadow, S.; Siebert, H.C.; Lochnit, G.; Kordelle, J.; Rickert, M.; Steinmeyer, J. Collagen metabolism of human osteoarthritic articular cartilage as modulated by bovine collagen hydrolysates. PLoS ONE 2013, 8, e53955. [Google Scholar] [CrossRef]
- Eckert, T.; Jährling-Butkus, M.; Louton, H.; Burg-Roderfeld, M.; Zhang, R.; Zhang, N.; Hesse, K.; Petridis, A.; Kožár, T.; Steinmeyer, J.; et al. Efficacy of Chondroprotective Food Supplements Based on Collagen Hydrolysate and Compounds Isolated from Marine Organisms. Mar. Drugs 2021, 19, 542. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Li, Z. Angiotensin-converting enzyme 2 (ACE2): SARS-CoV-2 receptor and RAS modulator. Acta Pharm. Sin. B 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Ng, A.W.; Bidani, A.; Heming, T.A. Innate host defense of the lung: Effects of lung-lining fluid pH. Lung 2004, 182, 297–317. [Google Scholar] [CrossRef]
- Choudhury, D.; Tanner, M.G.; McAughtrie, S.; Yu, F.; Mills, B.; Choudhary, T.R.; Seth, S.; Craven, T.H.; Stone, J.M.; Mati, I.K.; et al. Endoscopic sensing of alveolar pH. Biomed. Opt. Express 2017, 8, 243–259. [Google Scholar] [CrossRef]
- Alsaadi, E.A.J.; Neuman, B.W.; Jones, I.M. A Fusion Peptide in the Spike Protein of MERS Coronavirus. Viruses 2019, 11, 825. [Google Scholar] [CrossRef]
- Baker, A.N.; Richards, S.-J.; Guy, C.S.; Congdon, T.R.; Hasan, M.; Zwetsloot, A.J.; Straube, A.; Walker, M.; Chessa, S.; Pergolizzi, G.; et al. The SARS-CoV-2 Spike Protein Binds Sialic Acids, and Enables Rapid Detection in a Lateral Flow Point of Care Diagnostic Device; InvivoGen: San Diego, CA, USA, 2020. [Google Scholar] [CrossRef]
- Morniroli, D.; Gianni, M.L.; Consales, A.; Pietrasanta, C.; Mosca, F. Human Sialome and Coronavirus Disease-2019 (COVID-19) Pandemic: An Understated Correlation? Front. Immunol. 2020, 11, 1480. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Mahendran, A.S.K.; Lim, Y.S.; Fang, C.M.; Loh, H.S.; Le, C.F. The Potential of Antiviral Peptides as COVID-19 Therapeutics. Front. Pharm. 2020, 11, 575444. [Google Scholar] [CrossRef]
- Vilas Boas, L.C.P.; Campos, M.L.; Berlanda, R.L.A.; de Carvalho Neves, N.; Franco, O.L. Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. CMLS 2019, 76, 3525–3542. [Google Scholar] [CrossRef]
- Schutz, D.; Ruiz-Blanco, Y.B.; Munch, J.; Kirchhoff, F.; Sanchez-Garcia, E.; Muller, J.A. Peptide and peptide-based inhibitors of SARS-CoV-2 entry. Adv. Drug Deliv. Rev. 2020, 167, 47–65. [Google Scholar] [CrossRef]
- Ricchetti, M. Replication of mitochondrial DNA: The art of staying paired to avoid dangerous changes (comment on DOI 10.1002/bies.201400052). Bioessays 2014, 36, 1016. [Google Scholar] [CrossRef] [PubMed]
- Schauer, R.; Kamerling, J.P. Exploration of the Sialic Acid World. Adv. Carbohydr. Chem. BioChem. 2018, 75, 1–213. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Bhattacharyya, D.; Bhunia, A. Structural insights of a self-assembling 9-residue peptide from the C-terminal tail of the SARS corona virus E-protein in DPC and SDS micelles: A combined high and low resolution spectroscopic study. Biochim. Biophys. Acta Biomembr. 2018, 1860, 335–346. [Google Scholar] [CrossRef]
- Mukherjee, S.; Bhattacharyya, D.; Bhunia, A. Host-membrane interacting interface of the SARS coronavirus envelope protein: Immense functional potential of C-terminal domain. Biophys. Chem. 2020, 266, 106452. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.; Holscher, C. Receptors for the incretin glucagon-like peptide-1 are expressed on neurons in the central nervous system. Neuroreport 2009, 20, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, C.P.; Antunes, V.L.; Moreira, E.L.; de Mello, N.; Medeiros, R.; Di Giunta, G.; Lobao-Soares, B.; Linhares, M.; Lin, K.; Mazzuco, T.L.; et al. Glucose-dependent insulinotropic peptide receptor expression in the hippocampus and neocortex of mesial temporal lobe epilepsy patients and rats undergoing pilocarpine induced status epilepticus. Peptides 2011, 32, 781–789. [Google Scholar] [CrossRef]
- Tabuchi, A.; Sakaya, H.; Kisukeda, T.; Fushiki, H.; Tsuda, M. Involvement of an upstream stimulatory factor as well as cAMP-responsive element-binding protein in the activation of brain-derived neurotrophic factor gene promoter I. J. Biol. Chem. 2002, 277, 35920–35931. [Google Scholar] [CrossRef] [PubMed]
- Perry, T.; Greig, N.H. The glucagon-like peptides: A double-edged therapeutic sword? Trends Pharm. Sci. 2003, 24, 377–383. [Google Scholar] [CrossRef]
- Cui, Q.; So, K.F. Involvement of cAMP in neuronal survival and axonal regeneration. Anat Sci. Int. 2004, 79, 209–212. [Google Scholar] [CrossRef]
- Stoian, A.P.; Papanas, N.; Prazny, M.; Rizvi, A.A.; Rizzo, M. Incretin-Based Therapies Role in COVID-19 Era: Evolving Insights. J. Cardiovasc. Pharm. 2020, 25, 494–496. [Google Scholar] [CrossRef] [PubMed]
- Pantanetti, P.; Cangelosi, G.; Ambrosio, G. Potential role of incretins in diabetes and COVID-19 infection: A hypothesis worth exploring. Int. Emerg. Med. 2020, 15, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, A.A.; Kathuria, A.; Al Mahmeed, W.; Al-Rasadi, K.; Al-Alawi, K.; Banach, M.; Banerjee, Y.; Ceriello, A.; Cesur, M.; Cosentino, F.; et al. Post-COVID syndrome, inflammation, and diabetes. J. Diabetes Complicat. 2022, 36, 108336. [Google Scholar] [CrossRef] [PubMed]
- Drucker, D.J. Diabetes, obesity, metabolism, and SARS-CoV-2 infection: The end of the beginning. Cell Metab. 2021, 33, 479–498. [Google Scholar] [CrossRef]
- Tan, Q.; Akindehin, S.E.; Orsso, C.E.; Waldner, R.C.; DiMarchi, R.D.; Muller, T.D.; Haqq, A.M. Recent Advances in Incretin-Based Pharmacotherapies for the Treatment of Obesity and Diabetes. Front. Endocrinol. 2022, 13, 838410. [Google Scholar] [CrossRef] [PubMed]
- Niemann, H.; Klenk, H.D. Coronavirus glycoprotein E1, a new type of viral glycoprotein. J. Mol. Biol. 1981, 153, 993–1010. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.; Herrler, G.; Klenk, H.D. Sialic Acid Receptors of Viruses. Top. Curr. Chem. 2015, 367, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.L.; Freed, J.H. SARS-CoV-2 Fusion Peptide has a Greater Membrane Perturbating Effect than SARS-CoV with Highly Specific Dependence on Ca(2). J. Mol. Biol. 2021, 433, 166946. [Google Scholar] [CrossRef]
- Min, T.; Bain, S.C. The Role of Tirzepatide, Dual GIP and GLP-1 Receptor Agonist, in the Management of Type 2 Diabetes: The SURPASS Clinical Trials. Diabetes Ther. Res. Treat. Educ. Diabetes Relat. Disord. 2021, 12, 143–157. [Google Scholar] [CrossRef]
- Johnson, R.M.; Zhang, X.; Piper, S.J.; Nettleton, T.J.; Vandekolk, T.H.; Langmead, C.J.; Danev, R.; Sexton, P.M.; Wootten, D. Cryo-EM structure of the dual incretin receptor agonist, peptide-19, in complex with the glucagon-like peptide-1 receptor. Biochem. Biophys. Res. Commun. 2021, 578, 84–90. [Google Scholar] [CrossRef]
- Underwood, C.R.; Garibay, P.; Knudsen, L.B.; Hastrup, S.; Peters, G.H.; Rudolph, R.; Reedtz-Runge, S. Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J. Biol. Chem. 2010, 285, 723–730. [Google Scholar] [CrossRef]
- Kim, Y.H.; Min, K.H.; Wang, Z.; Kim, J.; Jacobson, O.; Huang, P.; Zhu, G.; Liu, Y.; Yung, B.; Niu, G.; et al. Development of Sialic Acid-coated Nanoparticles for Targeting Cancer and Efficient Evasion of the Immune System. Theranostics 2017, 7, 962–973. [Google Scholar] [CrossRef]
- de Matos, A.M.; Blazquez-Sanchez, M.T.; Sousa, C.; Oliveira, M.C.; de Almeida, R.F.M.; Rauter, A.P. C-Glucosylation as a tool for the prevention of PAINS-induced membrane dipole potential alterations. Sci. Rep. 2021, 11, 4443. [Google Scholar] [CrossRef]
- Dondorp, A.M.; Lee, S.J.; Faiz, M.A.; Mishra, S.; Price, R.; Tjitra, E.; Than, M.; Htut, Y.; Mohanty, S.; Yunus, E.B.; et al. The Relationship between Age and the Manifestations of and Mortality Associated with Severe Malaria. Clin. Infect. Dis. 2008, 47, 151–157. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, N.; Zhang, R.; Jin, L.; Petridis, A.K.; Loers, G.; Zheng, X.; Wang, Z.; Siebert, H.C. Cuprizone-Induced Demyelination in Mouse Hippocampus Is Alleviated by Ketogenic Diet. J. Agric. Food Chem. 2020, 68, 11215–11228. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, C.; Zhang, R.; Jin, L.; Yin, X.; Zheng, X.; Siebert, H.C.; Li, Y.; Wang, Z.; Loers, G.; et al. Amelioration of clinical course and demyelination in the cuprizone mouse model in relation to ketogenic diet. Food Funct. 2020, 11, 5647–5663. [Google Scholar] [CrossRef] [PubMed]
- Andre, S.; Kaltner, H.; Lensch, M.; Russwurm, R.; Siebert, H.C.; Fallsehr, C.; Tajkhorshid, E.; Heck, A.J.; von Knebel Doeberitz, M.; Gabius, H.J.; et al. Determination of structural and functional overlap/divergence of five proto-type galectins by analysis of the growth-regulatory interaction with ganglioside GM1 in silico and in vitro on human neuroblastoma cells. Int. J. Cancer 2005, 114, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Raghav, P.K.; Kalyanaraman, K.; Kumar, D. Human cell receptors: Potential drug targets to combat COVID-19. Amino Acids 2021, 53, 813–842. [Google Scholar] [CrossRef]
- Fung, T.S.; Liu, D.X. Post-translational modifications of coronavirus proteins: Roles and function. Future Virol. 2018, 13, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Buzhdygan, T.P.; DeOre, B.J.; Baldwin-Leclair, A.; Bullock, T.A.; McGary, H.M.; Khan, J.A.; Razmpour, R.; Hale, J.F.; Galie, P.A.; Potula, R.; et al. The SARS-CoV-2 spike protein alters barrier function in 2D static and 3D microfluidic in-vitro models of the human blood–brain barrier. NeuroBiol. Dis. 2020, 146, 105131. [Google Scholar] [CrossRef]
- Pellegrini, L.; Albecka, A.; Mallery, D.L.; Kellner, M.J.; Paul, D.; Carter, A.P.; James, L.C.; Lancaster, M.A. SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids. Cell Stem. Cell 2020, 27, 951–961.e955. [Google Scholar] [CrossRef]
- Prieto-Fernández, E.; Egia-Mendikute, L.; Vila-Vecilla, L.; Bosch, A.; Barreira-Manrique, A.; Lee, S.Y.; García-Del Río, A.; Antoñana-Vildosola, A.; Jiménez-Lasheras, B.; Moreno-Cugnon, L.; et al. Hypoxia reduces cell attachment of SARS-CoV-2 spike protein by modulating the expression of ACE2, neuropilin-1, syndecan-1 and cellular heparan sulfate. Emerg. Microbes Infect. 2021, 10, 1065–1076. [Google Scholar] [CrossRef]
- Whitaker, G.M.; Lynn, F.C.; McIntosh, C.H.; Accili, E.A. Regulation of GIP and GLP1 receptor cell surface expression by N-glycosylation and receptor heteromerization. PLoS ONE 2012, 7, e32675. [Google Scholar] [CrossRef]
- Schroeder, J.T.; Bieneman, A.P. The S1 Subunit of the SARS-CoV-2 Spike Protein Activates Human Monocytes to Produce Cytokines Linked to COVID-19: Relevance to Galectin-3. Front. Immunol. 2022, 13, 831763. [Google Scholar] [CrossRef]
- Klein, M.L.; Romero, A.; Kaltner, H.; Percec, V.; Gabius, H.J. From examining the relationship between (corona)viral adhesins and galectins to glyco-perspectives. Biophys. J. 2021, 120, 1031–1039. [Google Scholar] [CrossRef]
- von Hundelshausen, P.; Wichapong, K.; Gabius, H.J.; Mayo, K.H. The marriage of chemokines and galectins as functional heterodimers. Cell. Mol. Life Sci. CMLS 2021, 78, 8073–8095. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, N.; Liu, B.; Yang, J.; Loers, G.; Siebert, H.C.; Wen, M.; Zheng, X.; Wang, Z.; Han, J.; et al. HDAC3 Inhibitor RGFP966 Ameliorated Neuroinflammation in the Cuprizone-Induced Demyelinating Mouse Model and LPS-Stimulated BV2 Cells by Downregulating the P2X7R/STAT3/NF-kappaB65/NLRP3 Activation. ACS Chem. Neurosci. 2022, 13, 2579–2598. [Google Scholar] [CrossRef] [PubMed]
- Haak, T.; Golz, S.; Fritsche, A.; Fuchtenbusch, M.; Siegmund, T.; Schnellbacher, E.; Klein, H.H.; Uebel, T.; Drossel, D. Therapy of Type 1 Diabetes. Exp. Clin. Endocrinol. Diabetes 2019, 127, S27–S38. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shao, Z.; Xia, Y.; Qin, J.; Xiao, Y.; Zhou, Z.; Mei, Z. Incretin-based therapies for patients with type 1 diabetes: A meta-analysis. Endocr. Connect 2019, 8, 277–288. [Google Scholar] [CrossRef]
- Bader, M.; Li, Y.; Tweedie, D.; Shlobin, N.A.; Bernstein, A.; Rubovitch, V.; Tovar, Y.R.L.B.; DiMarchi, R.D.; Hoffer, B.J.; Greig, N.H.; et al. Neuroprotective Effects and Treatment Potential of Incretin Mimetics in a Murine Model of Mild Traumatic Brain Injury. Front. Cell Dev. Biol. 2019, 7, 356. [Google Scholar] [CrossRef]
- Glotfelty, E.J.; Delgado, T.; Tovar, Y.R.L.B.; Luo, Y.; Hoffer, B.; Olson, L.; Karlsson, T.; Mattson, M.P.; Harvey, B.; Tweedie, D.; et al. Incretin Mimetics as Rational Candidates for the Treatment of Traumatic Brain Injury. ACS Pharm. Trans. Sci. 2019, 2, 66–91. [Google Scholar] [CrossRef] [PubMed]
- Eckert, T.; Cosel, J.v.; Kamps, B.; Siebert, H.-C.; Zhang, R.; Gousias, N.Z.K.; Falahati, A.P.K. Evidence for quantum chemical effects in receptor-ligand binding between integrin and collagen fragments—A computational investigation with an impact on tissue repair, neuro-oncolgy and glycobiology. Front. Mol. Biosci. 2021; in print. [Google Scholar] [CrossRef]
- Israelsen, S.B.; Pottegard, A.; Sandholdt, H.; Madsbad, S.; Thomsen, R.W.; Benfield, T. Comparable COVID-19 outcomes with current use of GLP-1 receptor agonists, DPP-4 inhibitors or SGLT-2 inhibitors among patients with diabetes who tested positive for SARS-CoV-2. Diabetes Obes Metab. 2021, 23, 1397–1401. [Google Scholar] [CrossRef]
- Bhunia, A.; Vivekanandan, S.; Eckert, T.; Burg-Roderfeld, M.; Wechselberger, R.; Romanuka, J.; Bachle, D.; Kornilov, A.V.; von der Lieth, C.W.; Jiménez-Barbero, J.; et al. Why structurally different cyclic peptides can be glycomimetics of the HNK-1 carbohydrate antigen. J. Am. Chem. Soc. 2010, 132, 96–105. [Google Scholar] [CrossRef]
- Eckert, T.; Stötzel, S.; Burg-Roderfeld, M.; Sewing, J.; Lütteke, T.; Nifantiev, N.E.; Vliegenthart, J.F.G.; Siebert, H.-C. In silico Study on Sulfated and Non-Sulfated Carbohydrate Chains from Proteoglycans in Cnidaria and Interaction with Collagen. Open J. Phys. Chem. 2012, 2, 123–133. [Google Scholar] [CrossRef]
- Tsvetkov, Y.E.; Burg-Roderfeld, M.; Loers, G.; Arda, A.; Sukhova, E.V.; Khatuntseva, E.A.; Grachev, A.A.; Chizhov, A.O.; Siebert, H.C.; Schachner, M.; et al. Synthesis and molecular recognition studies of the HNK-1 trisaccharide and related oligosaccharides. The specificity of monoclonal anti-HNK-1 antibodies as assessed by surface plasmon resonance and STD NMR. J. Am. Chem. Soc. 2012, 134, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Siebert, H.C.; Tajkhorshid, E.; Dabrowski, J. Barrier to Rotation around the Csp2-Csp2 Bond of the Ketoaldehyde Enol Ether MeC(O)CHCH−OEt As Determined by 13C NMR and ab Initio Calculations. J. Phys. Chem. A 2001, 105, 8488–8494. [Google Scholar] [CrossRef]
- van Lenthe, J.H.; den Boer, D.H.W.; Havenith, R.W.A.; Schauer, R.; Siebert, H.-C. Ab initio calculations on various sialic acids provide valuable information about sialic acid-specific enzymes. J. Mol. Struct. Theochem. 2004, 677, 29–37. [Google Scholar] [CrossRef]
- Gao, W.; Jin, L.; Liu, C.; Zhang, N.; Zhang, R.; Bednarikova, Z.; Gazova, Z.; Bhunia, A.; Siebert, H.C.; Dong, H. Inhibition behavior of Sennoside A and Sennoside C on amyloid fibrillation of human lysozyme and its possible mechanism. Int. J. Biol. Macromol. 2021, 178, 424–433. [Google Scholar] [CrossRef]
- Jin, L.; Gao, W.; Liu, C.; Zhang, N.; Mukherjee, S.; Zhang, R.; Dong, H.; Bhunia, A.; Bednarikova, Z.; Gazova, Z.; et al. Investigating the inhibitory effects of entacapone on amyloid fibril formation of human lysozyme. Int. J. Biol. Macromol. 2020, 161, 1393–1404. [Google Scholar] [CrossRef]
- Bestle, D.; Heindl, M.R.; Limburg, H.; Van Lam, T.; Pilgram, O.; Moulton, H.; Stein, D.A.; Hardes, K.; Eickmann, M.; Dolnik, O.; et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 2020, 3, e202000786. [Google Scholar] [CrossRef]
- Limburg, H.; Harbig, A.; Bestle, D.; Stein, D.A.; Moulton, H.M.; Jaeger, J.; Janga, H.; Hardes, K.; Koepke, J.; Schulte, L.; et al. TMPRSS2 Is the Major Activating Protease of Influenza A Virus in Primary Human Airway Cells and Influenza B Virus in Human Type II Pneumocytes. J. Virol. 2019, 93, e00649-19. [Google Scholar] [CrossRef]
- Naeem, A.; Hamed, M.E.; Alghoribi, M.F.; Aljabr, W.; Alsaran, H.; Enani, M.A.; Alosaimi, B. Molecular Evolution and Structural Mapping of N-Terminal Domain in Spike Gene of Middle East Respiratory Syndrome Coronavirus (MERS-CoV). Viruses 2020, 12, 502. [Google Scholar] [CrossRef]
- Dennis, E.A.; Plückthun, A. Phosphorus-31 NMR of Phospholipids in Micelles. Phosphorous-31 NMR 1984, 14, 423–446. [Google Scholar] [CrossRef]
- Kallick, D.A.; Tessmer, M.R.; Watts, C.R.; Li, C.Y. The use of dodecylphosphocholine micelles in solution NMR. J. Magn. Reson. Ser. B 1995, 109, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Siebert, H.C.; Adar, R.; Arango, R.; Burchert, M.; Kaltner, H.; Kayser, G.; Tajkhorshid, E.; von der Lieth, C.W.; Kaptein, R.; Sharon, N.; et al. Involvement of laser photo-CIDNP (chemically induced dynamic nuclear polarization)-reactive amino acid side chains in ligand binding by galactoside-specific lectins in solution. Eur. J. Biochem. 1997, 249, 27–38. [Google Scholar] [CrossRef]
- Warschawski, D.E.; Arnold, A.A.; Beaugrand, M.; Gravel, A.; Chartrand, E.; Marcotte, I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim. Biophys. Acta 2011, 1808, 1957–1974. [Google Scholar] [CrossRef]
- de Jesus, A.J.; White, O.R.; Flynn, A.D.; Yin, H. Determinants of Curvature-Sensing Behavior for MARCKS-Fragment Peptides. Biophys. J. 2016, 110, 1980–1992. [Google Scholar] [CrossRef] [PubMed]
- Mineev, K.S.; Nadezhdin, K.D. Membrane mimetics for solution NMR studies of membrane proteins. Nanotechnol. Rev. 2017, 6, 15–32. [Google Scholar] [CrossRef]
- Jayawardena, H.S.; Jayawardana, K.W.; Chen, X.; Yan, M. Maltoheptaose promotes nanoparticle internalization by Escherichia coli. Chem. Commun. 2013, 49, 3034–3036. [Google Scholar] [CrossRef]
- Halgren, T.A. New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 2007, 69, 146–148. [Google Scholar] [CrossRef]
- Halgren, T.A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009, 49, 377–389. [Google Scholar] [CrossRef]
- SiteMap, Schrödinger Release 2021-4; Schrödinger, LLC: New York, NY, USA, 2021.
- MAESTRO 2021-2; Schrödinger, LLC,: New York, NY, USA, 2021.
- Desmond Molecular Dynamics System; D. E. Shaw Research: New York, NY, USA, 2020; Available online: https://www.deshawresearch.com/downloads/download_desmond.cgi/ (accessed on 5 March 2021).
- Lamiable, A.; Thevenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tuffery, P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucl. Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Kallberg, M.; Wang, H.; Wang, S.; Peng, J.; Wang, Z.; Lu, H.; Xu, J. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012, 7, 1511–1522. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucl. Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Dunbrack, R.L.; Hooft, R.W.W.; Krieger, B. Assignment of Protonation States in Proteins and Ligands: Combining pKa Prediction with Hydrogen Bonding Network Optimization. Methods Mol. Biol. 2011, 819, 405–421. [Google Scholar] [CrossRef]
- Krieger, E.; Nielsen, J.E.; Spronk, C.A.; Vriend, G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model 2006, 25, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef]
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Anal Chem. 2020, 92, 9895–9900. [Google Scholar] [CrossRef]
- Düzgüneş, N.; Fernandez-Fuentes, N.; Konopka, K. Inhibition of Viral Membrane Fusion by Peptides and Approaches to Peptide Design. Pathogens 2021, 10, 1599. [Google Scholar] [CrossRef]
- Yang, K.; Wang, C.; Kreutzberger, A.J.B.; Ojha, R.; Kuivanen, S.; Couoh-Cardel, S.; Muratcioglu, S.; Eisen, T.J.; White, K.I.; Held, R.G.; et al. Nanomolar inhibition of SARS-CoV-2 infection by an unmodified peptide targeting the prehairpin intermediate of the spike protein. Proc. Natl. Acad. Sci. USA 2022, 119, e2210990119. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, M.; Yamamoto, M.; Tani, H.; Kobayashi, A.; Gohda, J.; Kawaguchi, Y.; Park, B.K.; Kwon, H.J.; Inoue, J.I.; Alkattan, A. Discovery of New Fusion Inhibitor Peptides against SARS-CoV-2 by Targeting the Spike S2 Subunit. Biomolecules 2021, 29, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Wen, M.; Liu, M.; Wang, Q.; Liu, Q.; Li, L.; Siebert, H.C.; Loers, G.; Zhang, R.; Zhang, N. Effect of beta-hydroxybutyrate on behavioral alterations, molecular and morphological changes in CNS of multiple sclerosis mouse model. Front. Aging Neurosci. 2022, 14, 1075161. [Google Scholar] [CrossRef] [PubMed]
- Sango, K.; Takaku, S.; Tsukamoto, M.; Niimi, N.; Yako, H. Glucagon-Like Peptide-1 Receptor Agonists as Potential Myelination-Inducible and Anti-Demyelinating Remedies. Front. Cell Dev. Biol. 2022, 10, 950623. [Google Scholar] [CrossRef] [PubMed]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siebert, H.-C.; Eckert, T.; Bhunia, A.; Klatte, N.; Mohri, M.; Siebert, S.; Kozarova, A.; Hudson, J.W.; Zhang, R.; Zhang, N.; et al. Blood pH Analysis in Combination with Molecular Medical Tools in Relation to COVID-19 Symptoms. Biomedicines 2023, 11, 1421. https://doi.org/10.3390/biomedicines11051421
Siebert H-C, Eckert T, Bhunia A, Klatte N, Mohri M, Siebert S, Kozarova A, Hudson JW, Zhang R, Zhang N, et al. Blood pH Analysis in Combination with Molecular Medical Tools in Relation to COVID-19 Symptoms. Biomedicines. 2023; 11(5):1421. https://doi.org/10.3390/biomedicines11051421
Chicago/Turabian StyleSiebert, Hans-Christian, Thomas Eckert, Anirban Bhunia, Nele Klatte, Marzieh Mohri, Simone Siebert, Anna Kozarova, John W. Hudson, Ruiyan Zhang, Ning Zhang, and et al. 2023. "Blood pH Analysis in Combination with Molecular Medical Tools in Relation to COVID-19 Symptoms" Biomedicines 11, no. 5: 1421. https://doi.org/10.3390/biomedicines11051421