Exploring Gut Microenvironment in Colorectal Patient with Dual-Omics Platform: A Comparison with Adenomatous Polyp or Occult Blood
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement of Sample Collection
2.2. Extraction of Bacterial Genomic DNA
2.3. Sequencing of 16S Ribosomal RNA Gene
2.4. Extraction of Fecal Metabolites
2.5. UPLC-MS/MS Analysis
2.6. Processing, Annotation, and Statistical Analysis of Sequencing Results
2.7. Processing and Annotation of UPLC-MS/MS Data
2.8. Statistical Analysis
3. Results
3.1. Metadata of Enrolled Participants in This Study
3.2. Statistical Analysis of Sequencing Throughput in Each Enrolled Group
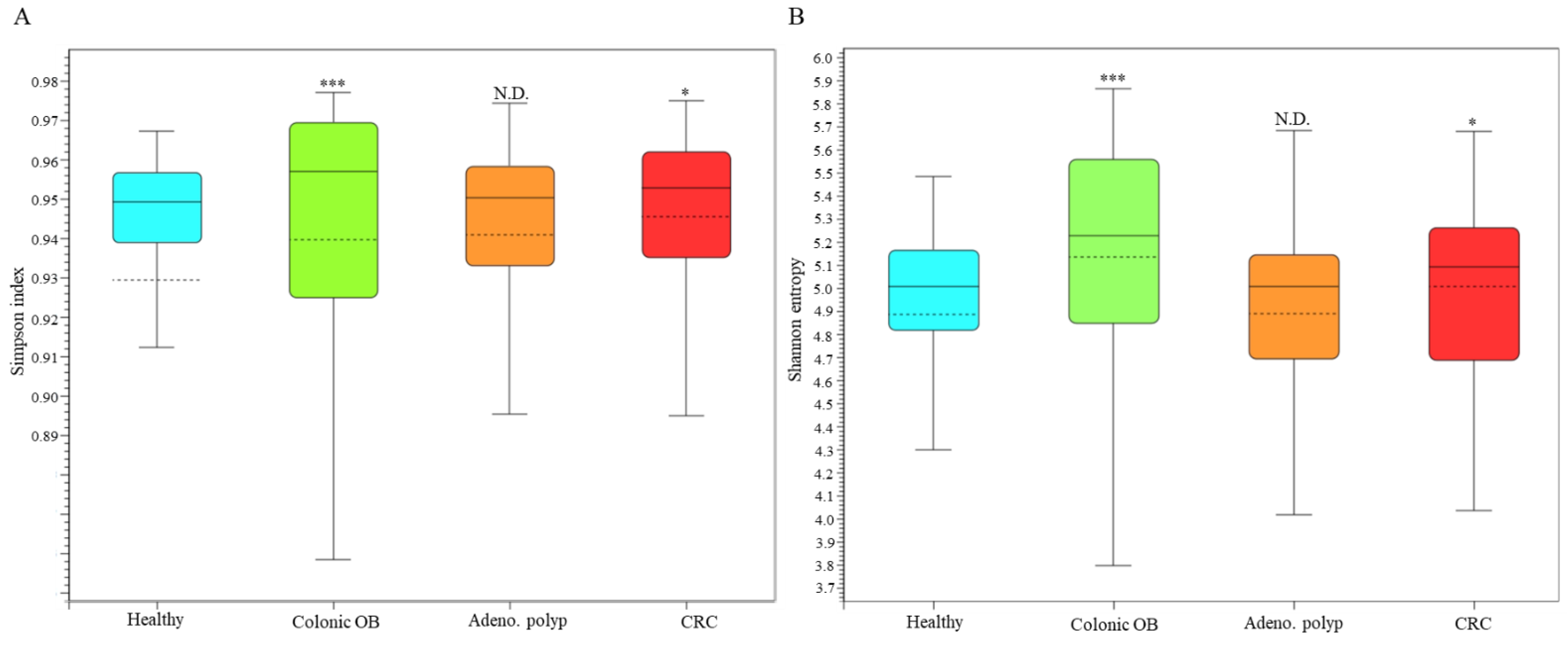
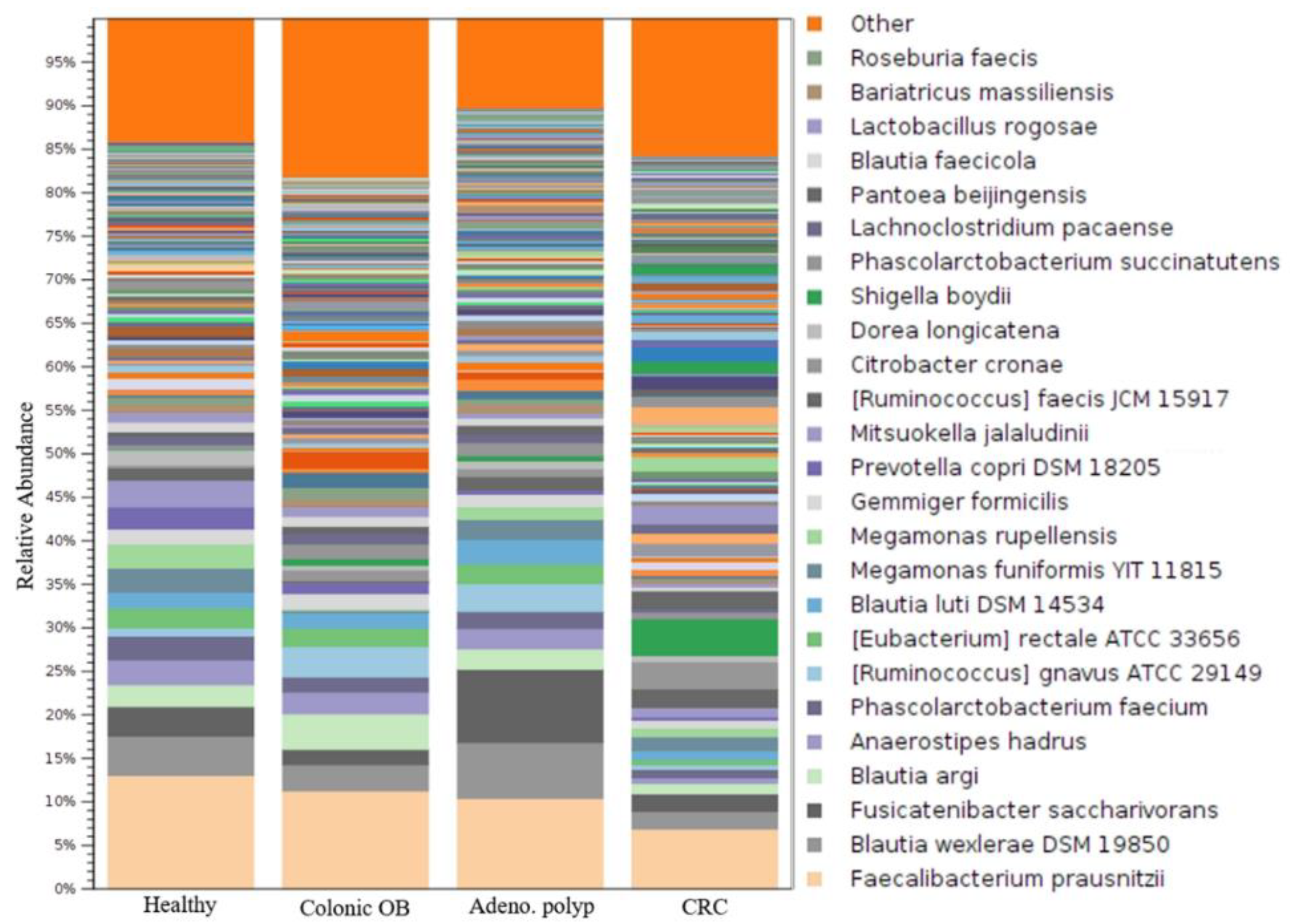
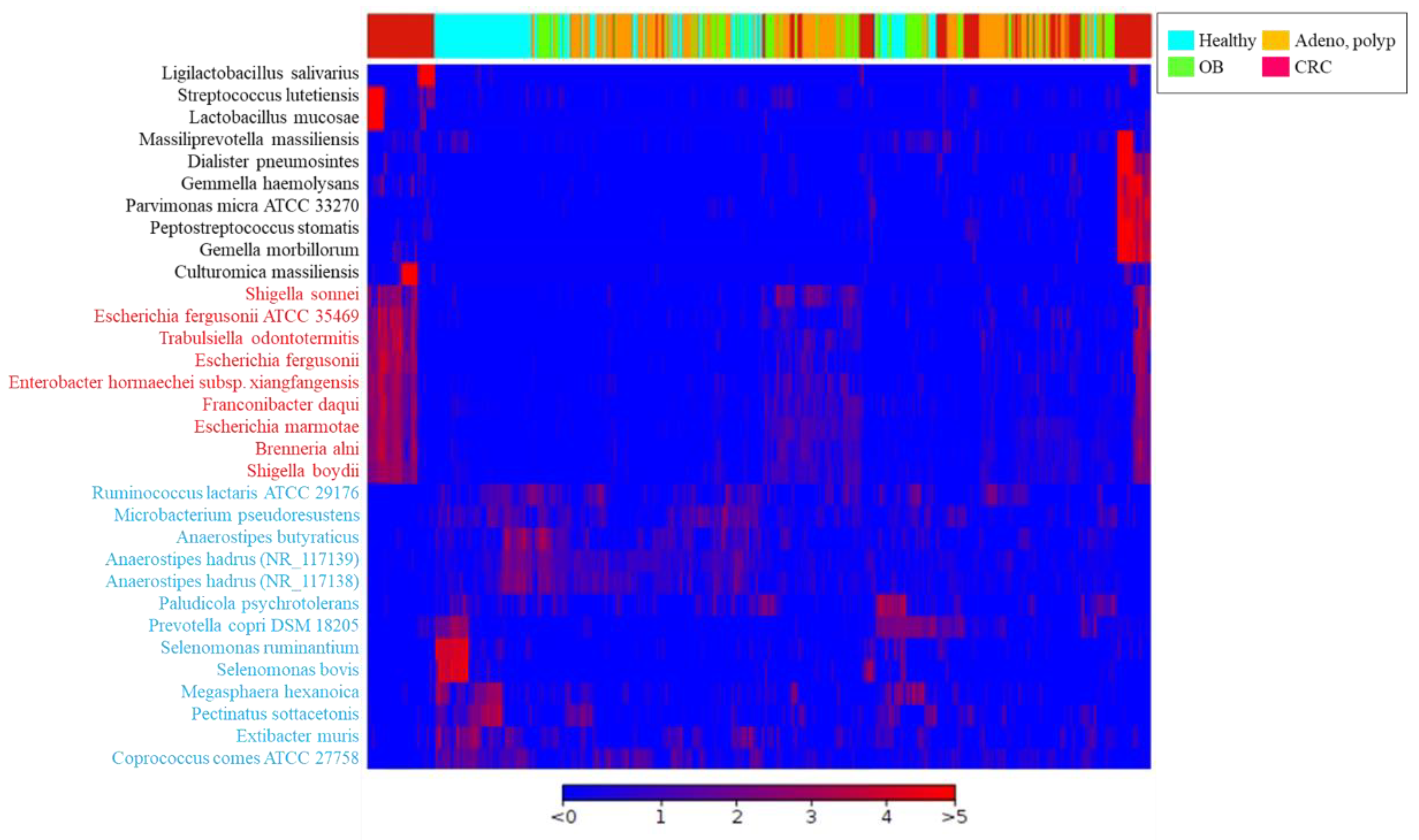
3.3. Identification of CRC-Associated Microbial Community with Result of Long-Read Sequencing
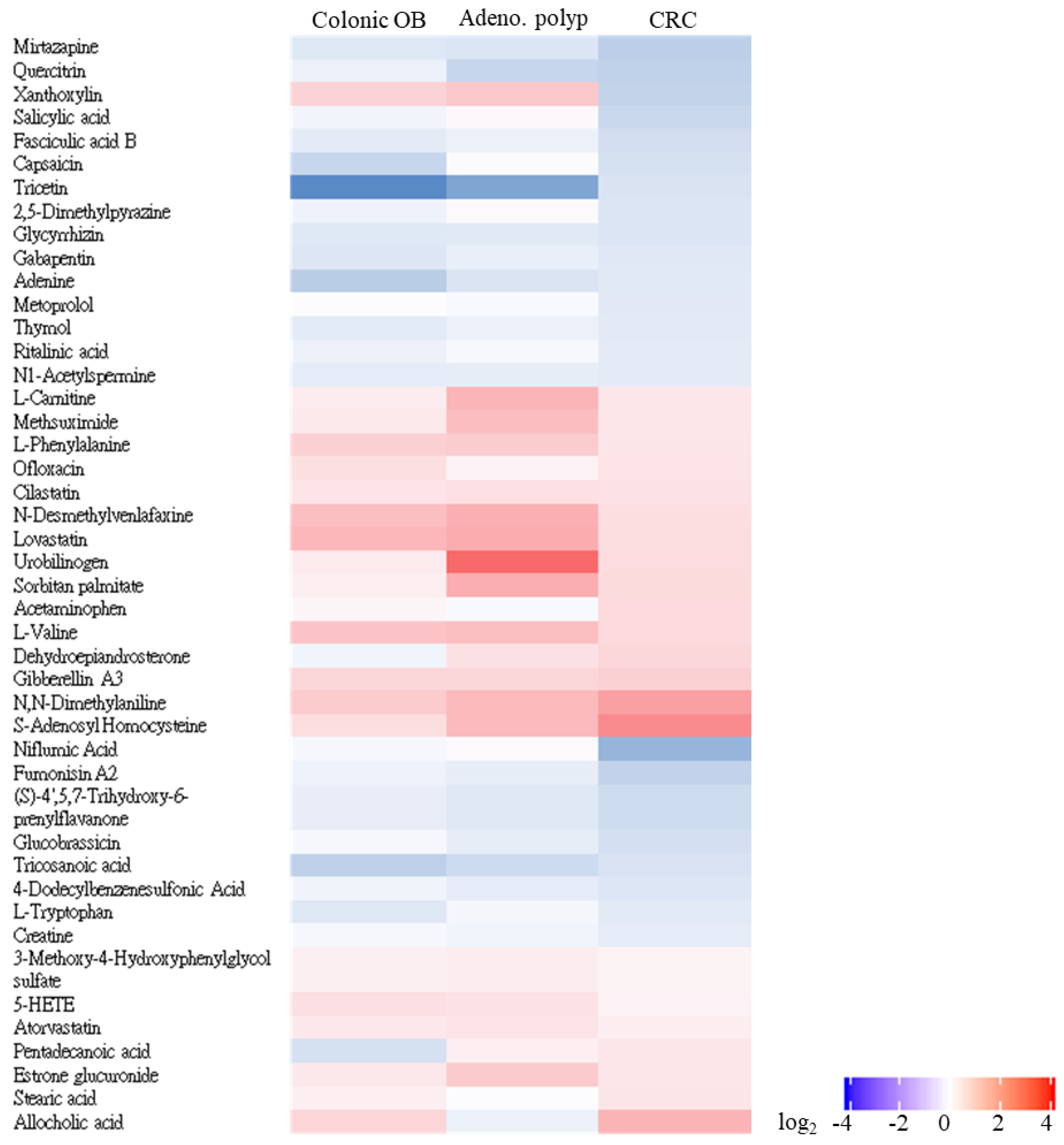
3.4. Untargeted Identification of CRC-Associated Metabolite Profile with LC-MS/MS Analysis
3.5. Predictive Utility of Identified Gut OTUs or Metabolites toward Occurrence of Adenoma or CRC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Sauer, A.G.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Tan, Z.; Hollis-Hansen, K.; Zhang, Y.; Yu, C.; Li, Y. Epidemiological Trends in Colorectal Cancer in China: An Ecological Study. Am. J. Dig. Dis. 2017, 62, 235–243. [Google Scholar] [CrossRef]
- Kuipers, E.J.; Grady, W.M.; Lieberman, D.; Seufferlein, T.; Sung, J.J.; Boelens, P.G.; van de Velde, C.J.; Watanabe, T. Colorectal cancer. Nat. Rev. Dis. Primers. 2015, 1, 15065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.; Jin, Y.; Chen, G.; Ma, X.; Zhang, L. Gut Microbiota Dysbiosis Drives the Development of Colorectal Cancer. Digestion 2021, 102, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Kinugasa, H.; Hirai, M.; Terasawa, H.; Yasutomi, E.; Oka, S.; Ohmori, M.; Yamasaki, Y.; Inokuchi, T.; Harada, K.; et al. Heterogeneous distribution of Fusobacterium nucleatum in the progression of colorectal cancer. J. Gastroenterol. Hepatol. 2021, 36, 1869–1876. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, M.R.; Baik, J.E.; Lagana, S.M.; Han, R.P.; Raab, W.J.; Sahoo, D.; Dalerba, P.; Wang, T.C.; Han, Y.W. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-Catenin modulator Annexin A1. EMBO Rep. 2019, 20, e47638. [Google Scholar] [CrossRef]
- Tsoi, H.; Chu, E.S.H.; Zhang, X.; Sheng, J.; Nakatsu, G.; Ng, S.C.; Chan, A.W.H.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology 2017, 152, 1419–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, X.; Wong, C.C.; Tong, L.; Chu, E.S.H.; Szeto, C.H.; Go, M.Y.Y.; Coker, O.O.; Chan, A.W.H.; Chan, F.K.; Sung, J.J.Y.; et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat. Microbiol. 2019, 4, 2319–2330. [Google Scholar] [CrossRef]
- Ghanavati, R.; Akbari, A.; Mohammadi, F.; Asadollahi, P.; Javadi, A.; Talebi, M.; Rohani, M. Lactobacillus species inhibitory effect on colorectal cancer progression through modulating the Wnt/b-Catenin signaling pathway. Mol. Cell. Biochem. 2020, 470, 1–13. [Google Scholar] [CrossRef]
- Faghfoori, Z.; Faghfoori, M.H.; Saber, A.; Izadi, A.; Khosroushahi, A.Y. Anticancer effects of bifidobacteria on colon cancer cell lines. Cancer Cell Int. 2021, 21, 258. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Jin, D.; Huang, S.; Wu, J.; Xu, M.; Liu, T.; Dong, W.; Liu, X.; Wang, S.; Zhong, W.; et al. Clostridium Butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2020, 469, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Li, J.V.; Swann, J.; Marchesi, J.R. Biology of the Microbiome 2: Metabolic Role. Gastroenterol. Clin. N. Am. 2017, 46, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, H.; Brennan, L. Metabolomics as a tool in the identification of dietary biomarkers. Proc. Nutr. Soc. 2017, 76, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Armitage, E.G.; Ciborowski, M. Applications of Metabolomics in Cancer Studies. Adv. Exp. Med. Biol. 2017, 965, 209–234. [Google Scholar] [CrossRef]
- Gomes, S.D.; Oliveira, C.S.; Azevedo-Silva, J.; Casanova, M.R.; Barreto, J.; Pereira, H.; Chaves, S.R.; Rodrigues, L.R.; Casal, M.; Côrte-Real, M.; et al. The role of diet related short-chain fatty acids in colorectal cancer metabolism and survival: Prevention and therapeutic implications. Curr. Med. Chem. 2020, 27, 4087–4108. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Li, R.; An, J.; Xie, T.; Han, Z.; Xu, R.; Fang, Y.; Zhang, X. Prebiotics-Encapsulated Probiotic Spores Regulate Gut Microbiota and Suppress Colon Cancer. Adv. Mater. 2020, 32, e2004529. [Google Scholar] [CrossRef]
- Beale, M.N.; Leach, H.J.; Baxter, B.A.; Smith, H.V.; Lyden, K.; Ryan, E.P. Physical Activity and Stool Metabolite Relationships Among Adults at High Risk for Colorectal Cancer. J. Phys. Act. Health 2021, 18, 1404–1411. [Google Scholar] [CrossRef]
- Zou, Y.; Xue, W.; Luo, G.; Deng, Z.; Qin, P.; Guo, R.; Sun, H.; Xia, Y.; Liang, S.; Dai, Y.; et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat. Biotechnol. 2019, 37, 179–185. [Google Scholar] [CrossRef]
- Song, X.; An, Y.; Chen, D.; Zhang, W.; Wu, X.; Li, C.; Wang, S.; Dong, W.; Wang, B.; Liu, T.; et al. Microbial metabolite deoxycholic acid promotes vasculogenic mimicry formation in intestinal carcinogenesis. Cancer Sci. 2022, 113, 459–477. [Google Scholar] [CrossRef]
- Fu, T.; Coulter, S.; Yoshihara, E.; Oh, T.G.; Fang, S.; Cayabyab, F.; Zhu, Q.; Zhang, T.; Leblanc, M.; Liu, S.; et al. FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell 2019, 176, 1098–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mo, Z.; Huang, P.; Yang, C.; Xiao, S.; Zhang, G.; Ling, F.; Li, L. Meta-analysis of 16S rRNA Microbial Data Identified Distinctive and Predictive Microbiota Dysbiosis in Colorectal Carcinoma Adjacent Tissue. mSystems 2020, 5, e00138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuo, Y.; Komiya, S.; Yasumizu, Y.; Yasuoka, Y.; Mizushima, K.; Takagi, T.; Kryukov, K.; Fukuda, A.; Morimoto, Y.; Naito, Y.; et al. Full-length 16S rRNA gene amplicon analysis of human gut microbiota using MinION™ nanopore sequencing confers species-level resolution. BMC Microbiol. 2021, 21, 35. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Wagner, B.D.; Harris, J.K.; Fillon, S.A. Zero-inflated negative binomial mixed model: An application to two microbial organisms important in oesophagitis. Epidemiol. Infect. 2016, 144, 2447–2455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Zhang, C.; Gui, Q.; Chen, Y.; Yang, Y. Ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry-based metabolic profiling of human serum prior to and following radical resection of colorectal carcinoma. Mol. Med. Rep. 2015, 12, 6879–6886. [Google Scholar] [CrossRef] [Green Version]
- Robertson, D.J.; Lee, J.K.; Boland, C.R.; Dominitz, J.A.; Giardiello, F.M.; Johnson, D.A.; Kaltenbach, T.; Lieberman, D.; Levin, T.R.; Rex, D.K. Recommendations on Fecal Immunochemical Testing to Screen for Colorectal Neoplasia: A Consensus Statement by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2017, 152, 1217–1237. [Google Scholar] [CrossRef] [Green Version]
- Toes-Zoutendijk, E.; van Leerdam, M.E.; Dekker, E.; van Hees, F.; Penning, C.; Nagtegaal, I.; van der Meulen, M.P.; van Vuuren, A.J.; Kuipers, E.J.; Bonfrer, J.M.; et al. Real-Time Monitoring of Results During First Year of Dutch Colorectal Cancer Screening Program and Optimization by Altering Fecal Immunochemical Test Cut-off Levels. Gastroenterology 2017, 152, 767–775. [Google Scholar] [CrossRef] [Green Version]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012, 22, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Gao, R.; Kong, C.; Li, H.; Huang, L.; Qu, X.; Qin, N.; Qin, H. Dysbiosis signature of mycobiota in colon polyp and colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2457–2468. [Google Scholar] [CrossRef]
- Duvallet, C.; Gibbons, S.M.; Gurry, T.; Irizarry, R.A.; Alm, E.J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017, 8, 1784. [Google Scholar] [CrossRef] [Green Version]
- Laudadio, I.; Fulci, V.; Stronati, L.; Carissimi, C. Next-generation metagenomics: Methodological challenges and opportunities. Omics 2019, 23, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Jiao, N.; Zhu, R.; Zhang, Y.; Wu, D.; Wang, A.-J.; Fang, S.; Tao, L.; Li, Y.; Cheng, S.; et al. Identification of microbial markers across populations in early detection of colorectal cancer. Nat. Commun. 2021, 12, 3063. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.-Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yachida, S.; Mizutani, S.; Shiroma, H.; Shiba, S.; Nakajima, T.; Sakamoto, T.; Watanabe, H.; Masuda, K.; Nishimoto, Y.; Kubo, M.; et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019, 25, 968–976. [Google Scholar] [CrossRef] [PubMed]
- Vettore, L.; Westbrook, R.; Tennant, D.A. New aspects of amino acid metabolism in cancer. Br. J. Cancer 2020, 122, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino acids in cancer. Exp. Mol. Med. 2021, 52, 15–30. [Google Scholar] [CrossRef]
- Ocvirk, S.; O’keefe, S.J.D. Dietary fat, bile acid metabolism and colorectal cancer. Semin. Cancer Biol. 2021, 73, 347–355. [Google Scholar] [CrossRef]
- Winston, J.A.; Theriot, C.M. Diversification of host bile acids by members of the gut microbiota. Gut Microbes 2020, 11, 158–171. [Google Scholar] [CrossRef]
- Cipriani, S.; Mencarelli, A.; Chini, M.G.; Distrutti, E.; Renga, B.; Bifulco, G.; Baldelli, F.; Donini, A.; Fiorucci, S. The Bile Acid Receptor GPBAR-1 (TGR5) Modulates Integrity of Intestinal Barrier and Immune Response to Experimental Colitis. PLoS ONE 2011, 6, e25637. [Google Scholar] [CrossRef]
- Zagato, E.; Pozzi, C.; Bertocchi, A.; Schioppa, T.; Saccheri, F.; Guglietta, S.; Fosso, B.; Melocchi, L.; Nizzoli, G.; Troisi, J.; et al. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat. Microbiol. 2020, 5, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Stoeva, M.K.; Garcia-So, J.; Justice, N.; Myers, J.; Tyagi, S.; Nemchek, M.; McMurdie, P.J.; Kolterman, O.; Eid, J. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes 2021, 13, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Xu, Q.; Sun, L.; Ye, Y.; Ji, G. Short-chain fatty acids administration is protective in colitis-associated colorectal cancer development. J. Nutr. Biochem. 2018, 57, 103–109. [Google Scholar] [CrossRef]
- Chen, X.; Ma, L.; Liu, X.; Wang, J.; Li, Y.; Xie, Q.; Liang, J. Clostridium butyricum alleviates dextran sulfate sodium-induced experimental colitis and promotes intestinal lymphatic vessel regeneration in mice. Ann. Transl. Med. 2022, 10, 341. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, Y.; Gao, L.; Jiang, W.; Lin, W.; Niu, C.; Yuan, K.; Ma, R.; Huang, Z. The Impairment of Methyl Metabolism From luxS Mutation of Streptococcus mutans. Front. Microbiol. 2018, 9, 404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loke, M.F.; Chua, E.G.; Gan, H.M.; Thulasi, K.; Wanyiri, J.W.; Thevambiga, I. Metabolomics and 16S rRNA se-quencing of human colorectal cancers and adjacent mucosa. PLoS ONE 2018, 13, e0208584. [Google Scholar] [CrossRef]
- Dumitrescu, R.G. Alcohol-Induced Epigenetic Changes in Cancer. Methods Mol. Biol. 2018, 1856, 157–172. [Google Scholar] [CrossRef]





| Group | Healthy (n = 60) | Colonic OB (n = 40) | Adenomatous Polyp (n = 67) | CRC (n = 20) | p |
|---|---|---|---|---|---|
| Age (Median(IQR)) | 61 (31–72) | 52 (35–63) | 48 (39–60) | 64 (43–88) | >0.05 |
| Sex (n,%) | >0.05 | ||||
| Female | 35 (58.33) | 22 (55) | 39 (58.2) | 13 (65) | |
| Male | 26 (41.67) | 18 (45) | 28 (41.8) | 7 (35) | |
| History of cancer (n,%) | 6 (11.32) | 3 (8.33) | 5 (11.63) | 4 (20) (p < 0.01) | >0.05 |
| Family history of cancer (n,%) | 10 (16.67) | 8 (20) | 13 (19.4) | 8 (40) (p < 0.01) | >0.05 |
| History of smoking (n,%) | 15 (25) | 8 (20) | 13 (19.4) | 5 (25) | >0.05 |
| History of drinking (n,%) | 6 (10) | 8 (20) | 12 (17.91) | 5 (20) | >0.05 |
| History of regular exercise (n,%) | 27 (45) | 21 (52.5) | 24 (35.82) | 8 (40) | >0.05 |
| Group | Healthy (n = 60) | Colonic OB (n = 40) | Adenomatous Polyp (n = 67) | CRC (n = 20) | p |
|---|---|---|---|---|---|
| Number of Raw reads per sample | 84,534 (±5079) | 87,817 (±4121) | 81,775 (±2719) | 83,756 (±3217) | >0.05 |
| Number of qualified reads per sample | 62,749 (±3226) | 65,292 (±2884) | 51,944 (±2431) | 54,645 (±2005) | >0.05 |
| Reads in identified taxa | 58,505 (±2845) | 60,297 (±2355) | 45,403 (±1977) | 49,446 (±2105) | >0.05 |
| Correctly classified (% (SD)) | 93.24 (±3.64) | 92.35 (±4.97) | 91.59 (±3.55) | 90.49 (±2.69) | >0.05 |
| Number of identified taxa per sample | 1114 | 1075 | 931 | 948 | >0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, P.-L.; Wu, M.-S.; Huang, C.-K.; Ho, Y.-H.; Hung, C.-S.; Lin, Y.-C.; Tsao, M.-F.; Lin, J.-C. Exploring Gut Microenvironment in Colorectal Patient with Dual-Omics Platform: A Comparison with Adenomatous Polyp or Occult Blood. Biomedicines 2022, 10, 1741. https://doi.org/10.3390/biomedicines10071741
Wei P-L, Wu M-S, Huang C-K, Ho Y-H, Hung C-S, Lin Y-C, Tsao M-F, Lin J-C. Exploring Gut Microenvironment in Colorectal Patient with Dual-Omics Platform: A Comparison with Adenomatous Polyp or Occult Blood. Biomedicines. 2022; 10(7):1741. https://doi.org/10.3390/biomedicines10071741
Chicago/Turabian StyleWei, Po-Li, Ming-Shun Wu, Chun-Kai Huang, Yi-Hsien Ho, Ching-Sheng Hung, Ying-Chin Lin, Mei-Fen Tsao, and Jung-Chun Lin. 2022. "Exploring Gut Microenvironment in Colorectal Patient with Dual-Omics Platform: A Comparison with Adenomatous Polyp or Occult Blood" Biomedicines 10, no. 7: 1741. https://doi.org/10.3390/biomedicines10071741







