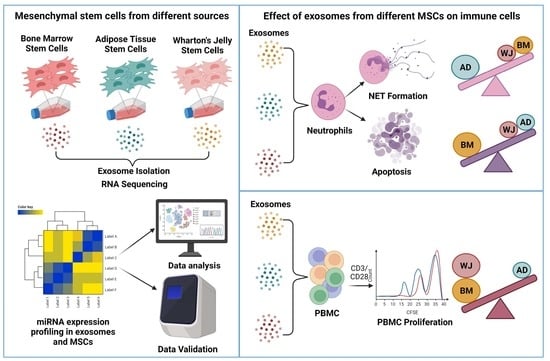
MicroRNA-Enriched Exosomes from Different Sources of Mesenchymal Stem Cells Can Differentially Modulate Functions of Immune Cells and Neurogenesis
Abstract
:1. Introduction
2. Material and Methods
2.1. Ethics Approval and Consent to Participate
2.2. Isolation and Culture of hMSCs from Bone Marrow (BM), Adipose Tissue (AD), and Wharton Jelly (WJ)
2.2.1. Isolation and Culture of Bone Marrow-Derived Stem Cells (BM-hMSCs)
2.2.2. Adipose Tissue-Derived Mesenchymal Stem Cells (AD-hMSCs)
2.2.3. Wharton Jelly (WJ)-Derived Mesenchymal Stem Cells (WJ-hMSCs)
2.3. Trilineage Differentiation Potential of hMSCs
2.4. Isolation of Exosomes from hMSCs
2.5. Nanoparticle Tracking Analysis (NTA)
2.6. Transmission Electron Microscopy (TEM)
2.7. Western Blotting
2.8. Small RNA Sequencing of RNA Isolated from hMSCs and Their Respective Exosomes
2.9. Library Preparation, Sequencing, and Data Analysis
2.10. MicroRNA Profiling
2.11. Neural Stem Cells C17.2 Culture
2.12. The qRT-PCR Assay
2.13. PBMCs’ Isolation and Proliferation
2.14. Neutrophil Isolation
2.15. Neutrophils’ Phagocytosis Assay
2.16. Apoptosis Assay
2.17. Visualization of NETs and Measurement of Extracellular DNA Release
2.18. Angiogenesis Study
2.19. Statistical Analysis
2.20. Data Availability
3. Results
3.1. Characterization of hMSCs and Their Exosomes
3.2. MicroRNAs’ Profiling of hMSCs and Their Exosomes
3.3. Effect of hMSCs-Exosomes on Neurogenesis
3.4. Effect of hMSC-Derived Exosomes on the Expansion of T Cells In Vitro
3.5. The Effects of hMSCs-Exosomes on Neutrophil Phagocytosis
3.6. The Effects of hMSC-Exosomes on Survival and NET Formation of Neutrophils
3.7. Effects of hMSCs-Exosomes on Angiogenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Wei, X.; Yang, X.; Han, Z.-P.; Qu, F.-F.; Shao, L.; Shi, Y.-F. Mesenchymal stem cells: A new trend for cell therapy. Acta Pharmacol. Sin. 2013, 34, 747–754. [Google Scholar] [CrossRef] [Green Version]
- Brown, C.; McKee, C.; Bakshi, S.; Walker, K.; Hakman, E.; Halassy, S.; Svinarich, D.; Dodds, R.; Govind, C.K.; Chaudhry, G.R. Mesenchymal stem cells: Cell therapy and regeneration potential. J. Tissue Eng. Regen. Med. 2019, 13, 1738–1755. [Google Scholar] [CrossRef]
- Kassem, M.; Kristiansen, M.; Abdallah, B.M. Mesenchymal stem cells: Cell biology and potential use in therapy. Basic Clin. Pharmacol. Toxicol. 2004, 95, 209–214. [Google Scholar] [CrossRef]
- Cao, W.; Cao, K.; Cao, J.; Wang, Y.; Shi, Y. Mesenchymal stem cells and adaptive immune responses. Immunol. Lett. 2015, 168, 147–153. [Google Scholar] [CrossRef]
- Fibbe, W.E.; Nauta, A.J.; Roelofs, H. Modulation of immune responses by mesenchymal stem cells. Ann. N. Y. Acad. Sci. 2007, 1106, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ren, G.; Huang, Y.; Su, J.; Han, Y.; Li, J.; Chen, X.; Cao, K.; Chen, Q.; Shou, P.; et al. Mesenchymal stem cells: A double-edged sword in regulating immune responses. Cell Death Differ. 2012, 19, 1505–1513. [Google Scholar] [CrossRef]
- Ns, S.; Na, A. Sources of Mesenchymal Stromal Cells: An Overview. Am. J. Pharmacol. 2018, 1, 1–7. [Google Scholar]
- Camussi, G.; Deregibus, M.C.; Bruno, S.; Cantaluppi, V.; Biancone, L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010, 78, 838–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 1–6. [Google Scholar] [CrossRef]
- Doyle, M.L.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Li, Y.; Cui, Y.; Yang, J.J.; Zhang, Z.G.; Chopp, M. Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. J. Cereb. Blood Flow Metab. 2013, 33, 1711–1715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, M.; Kosaka, N.; Tominaga, N.; Yoshioka, Y.; Takeshita, F.; Takahashi, R.U.; Yoshida, M.; Tsuda, H.; Tamura, K.; Ochiya, T. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 2014, 7, ra63. [Google Scholar] [CrossRef]
- Nakamura, Y.; Miyaki, S.; Ishitobi, H.; Matsuyama, S.; Nakasa, T.; Kamei, N.; Akimoto, T.; Higashi, Y.; Ochi, M. Mesenchymal-stem-cell-derived exosomes accelerate skeletal muscle regeneration. FEBS Lett. 2015, 589, 1257–1265. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.I.; Wang, J.; Zhou, X.; Xiong, Z.; Zhao, J.; Yu, R.; Huang, F.; Zhang, H.; Chen, L. Exosomes derived from human adipose mensenchymal stem cells accelerates cutaneous wound healing via optimizing the characteristics of fibroblasts. Sci. Rep. 2016, 6, 1–11. [Google Scholar]
- He, L.; He, T.; Xing, J.; Zhou, Q.; Fan, L.; Liu, C.; Chen, Y.; Wu, D.; Tian, Z.; Liu, B.; et al. Bone marrow mesenchymal stem cell-derived exosomes protect cartilage damage and relieve knee osteoarthritis pain in a rat model of osteoarthritis. Stem Cell Res. Ther. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Tieu, A.; Lalu, M.M.; Slobodian, M.; Gnyra, C.; Fergusson, D.A.; Montroy, J.; Burger, D.; Stewart, D.J.; Allan, D.S. An Analysis of Mesenchymal Stem Cell-Derived Extracellular Vesicles for Preclinical Use. ACS Nano 2020, 14, 9728–9743. [Google Scholar] [CrossRef] [PubMed]
- Shirazi, S.; Huang, C.-C.; Kang, M.; Lu, Y.; Ravindran, S.; Cooper, L.F. The importance of cellular and exosomal miRNAs in mesenchymal stem cell osteoblastic differentiation. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Li, C.; Ni, Y.-Q.; Xu, H.; Xiang, Q.-Y.; Zhao, Y.; Zhan, J.-K.; He, J.-Y.; Li, S.; Liu, Y.-S. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal. Transduct. Target. Ther. 2021, 6, 1–31. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, W.; Wang, W.; Tong, X.; Xia, R.; Fan, J.; Du, J.; Zhang, C.; Shi, X. Platelet-derived exosomes promote neutrophil extracellular trap formation during septic shock. Crit. Care 2020, 24, 1–18. [Google Scholar] [CrossRef]
- Ma, T.; Chen, Y.; Chen, Y.; Meng, Q.; Sun, J.; Shao, L.; Yu, Y.; Huang, H.; Hu, Y.; Yang, Z.; et al. MicroRNA-132, delivered by mesenchymal stem cell-derived exosomes, promote angiogenesis in myocardial infarction. Stem Cells Int. 2018, 2018, 3290372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Galán, A.; Fernández-Messina, L.; Sánchez-Madrid, F. Control of immunoregulatory molecules by miRNAs in T cell activation. Front. Immunol. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Gupta, S.; Rawat, S.; Arora, V.; Kottarath, S.K.; Dinda, A.K.; Vaishnav, P.K.; Nayak, B.; Mohanty, S. An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Stem Cell Res. Ther. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakkar, A.; Nandy, S.B.; Gupta, S.; Bharagava, B.; Airan, B.; Mohanty, S. Adipose tissue derived mesenchymal stem cells are better respondents to TGFβ1 for in vitro generation of cardiomyocyte-like cells. Mol. Cell. Biochem. 2019, 460, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Jiang, J.; Liu, W.; Wang, H.; Zhao, L.; Liu, S.; Li, P.; Zhang, S.; Sun, C.; Wu, Y.; et al. MicroRNA-378 promotes autophagy and inhibits apoptosis in skeletal muscle. Proc. Natl. Acad. Sci. USA 2018, 115, E10849–E10858. [Google Scholar] [CrossRef] [Green Version]
- Simanovich, E.; Brod, V.; Rahat, M.M.; Rahat, M.A. Function of miR-146a-5p in tumor cells as a regulatory switch between cell death and angiogenesis: Macrophage therapy revisited. Front. Immunol. 2018, 8, 1931. [Google Scholar] [CrossRef] [Green Version]
- Boissart, C.; Nissan, X.; Giraud-Triboult, K.; Peschanski, M.; Benchoua, A. MiR-125 potentiates early neural specification of human embryonic stem cells. Development 2012, 139, 1247–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shenoy, A.; Danial, M.; Blelloch, R.H. Let-7 and miR-125 cooperate to prime progenitors for astrogliogenesis. EMBO J. 2015, 34, 1180–1194. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Cheng, W.; Wang, X.; Yang, Z.; Zhou, X.; Pan, C. Overexpression of MicroRNA-145 Ameliorates Astrocyte Injury by Targeting Aquaporin 4 in Cerebral Ischemic Stroke. Biomed. Res. Int. 2017, 2017, 9530951. [Google Scholar] [CrossRef] [Green Version]
- Hegarty, S.V.; Sullivan, A.M.; O’Keeffe, G.W. Inhibition of miR-181a promotes midbrain neuronal growth through a Smad1/5-dependent mechanism: Implications for Parkinson’s disease. Neuronal Signal. 2018, 2, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.F.; Chen, J.C.; Zhang, J.; Xu, J.G. MIR-181a involves in the hippocampus-dependent memory formation via targeting PRKAA1. Sci. Rep. 2017, 7, 1–11. [Google Scholar]
- Wang, Y.-M.; Song, Z.; Qu, Y.; Lu, L.-Q. Down-regulated miR-21 promotes learning-memory recovery after brain injury. Int. J. Clin. Exp. Pathol. 2019, 12, 916–921. [Google Scholar]
- Lundqvist, J.; Andaloussi-Lilja, J.E.; Svensson, C.; Dorfh, H.G.; Forsby, A. Optimisation of culture conditions for differentiation of C17.2 neural stem cells to be used for in vitro toxicity tests. Toxicol. Vitr. 2013, 27, 1565–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgado, A.L.; Podrigues, C.M.P.; Sola, S. MicroRNA-145 Regulates Neural Stem Cell Differentiation Through the Sox2–Lin28/let-7 Signaling Pathway ANA. Stem Cells 2016, 34, 1386–1395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Li, X.; Wang, J.; Song, N.; Zhu, A.; Jia, L. miR-378a Modulates Macrophage Phagocytosis and Differentiation through Targeting CD47-SIRPα Axis in Atherosclerosis. Scand. J. Immunol. 2019, 90, 1–10. [Google Scholar] [CrossRef]
- Gentile, P.; Garcovich, S. Concise review: Adipose-derived stem cells (ASCs) and adipocyte-secreted exosomal microrna (A-SE-miR) modulate cancer growth and promote wound repair. J. Clin. Med. 2019, 8, 855. [Google Scholar] [CrossRef] [Green Version]
- Gentile, P.; Sterodimas, A.; Pizzicannella, J.; Dionisi, L.; De Fazio, D.; Calabrese, C.; Garcovich, S. Systematic review: Allogenic use of stromal vascular fraction (SVF) and decellularized extracellular matrices (ECM) as advanced therapy medicinal products (ATMP) in tissue regeneration. Int. J. Mol. Sci. 2020, 21, 4982. [Google Scholar] [CrossRef] [PubMed]
- Gentile, P.; Scioli, M.G.; Bielli, A.; Orlandi, A.; Cervelli, V. Concise Review: The Use of Adipose-Derived Stromal Vascular Fraction Cells and Platelet Rich Plasma in Regenerative Plastic Surgery. Stem Cells 2017, 35, 117–134. [Google Scholar] [CrossRef]
- Gentile, P.; De Angelis, B.; Pasin, M.; Cervelli, G.; Curcio, C.B.; Floris, M.; Di Pasquali, C.; Bocchini, I.; Balzani, A.; Nicoli, F.; et al. Adipose-derived stromal vascular fraction cells and platelet-rich plasma: Basic and clinical evaluation for cell-based therapies in patients with scars on the face. J. Craniofac. Surg. 2014, 25, 267–272. [Google Scholar] [CrossRef]
- Cervelli, V.; Bocchini, I.; Di Pasquali, C.; De Angelis, B.; Curcio, C.B.; Orlandi, A.; Scioli, M.G.; Tati, E.; Delogu, P.; Gentile, P. P.R.L. Platelet rich lipotransfert: Our experience and current state of art in the combined use of fat and PRP. Biomed. Res. Int. 2013, 2013, 434191. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.S.; Borrelli, M.R.; Lorenz, H.P.; Longaker, M.T.; Wan, D.C. Mesenchymal stromal cells and cutaneous wound healing: A comprehensive review of the background, role, and therapeutic potential. Stem Cells Int. 2018, 2018, 6901983. [Google Scholar] [CrossRef] [Green Version]
- Tak, Y.J.; Lee, S.Y.; Cho, A.R.; Kim, Y.S. A randomized, double-blind, vehicle-controlled clinical study of hair regeneration using adipose-derived stem cell constituent extract in androgenetic alopecia. Stem Cells Transl. Med. 2020, 9, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Gentile, P.; Garcovich, S. Advances in Regenerative Stem Cell Therapy in Androgenic Alopecia and Hair Loss: Wnt Pathway, Growth-Factor, and Mesenchymal Stem Cell Signaling Impact Analysis on Cell Growth and Hair Follicle Development. Cells 2019, 8, 466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentile, P. Autologous cellular method using micrografts of human adipose tissue derived follicle stem cells in androgenic alopecia. Int. J. Mol. Sci. 2019, 20, 3446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentile, P. Breast silicone gel implants versus autologous fat grafting: Biomaterials and bioactive materials in comparison. J. Clin. Med. 2021, 10, 3310. [Google Scholar] [CrossRef] [PubMed]
- Gentile, P.; Kothari, A.; Casella, D.; Calabrese, C. Fat graft enhanced with adipose-Derived stem cells in aesthetic breast augmentation: Clinical, histological, and instrumental evaluation. Aesthetic Surg. J. 2020, 40, 962–977. [Google Scholar] [CrossRef] [PubMed]
- Gentile, P.; Sterodimas, A. Adipose-derived stromal stem cells (ASCs) as a new regenerative immediate therapy combating coronavirus (COVID-19)-induced pneumonia. Expert Opin. Biol. Ther. 2020, 20, 711–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentile, P.; Sterodimas, A.; Pizzicannella, J.; Calabrese, C.; Garcovich, S. Research progress on Mesenchymal Stem Cells (MSCs), adipose-derived mesenchymal stem cells (AD-MSCs), drugs, and vaccines in inhibiting COVID-19 disease. Aging Dis. 2020, 11, 1191–1201. [Google Scholar] [CrossRef]
- Lukomska, B.; Stanaszek, L.; Zuba-Surma, E.; Legosz, P.; Sarzynska, S.; Drela, K. Challenges and Controversies in Human Mesenchymal Stem Cell Therapy. Stem Cells Int. 2019, 2019, 9628536. [Google Scholar] [CrossRef] [Green Version]
- Musiał-Wysocka, A.; Kot, M.; Majka, M. The Pros and Cons of Mesenchymal Stem Cell-Based Therapies. Cell Transplant. 2019, 28, 801–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ragni, E.; Orfei, C.P.; De Luca, P.; Colombini, A.; Viganò, M.; Lugano, G.; Bollati, V.; De Girolamo, L. Identification of miRNA reference genes in extracellular vesicles from adipose derived mesenchymal stem cells for studying osteoarthritis. Int. J. Mol. Sci. 2019, 20, 1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giunti, D.; Marini, C.; Parodi, B.; Usai, C.; Milanese, M.; Bonanno, G.; de Rosbo, N.K.; Uccelli, A. Role of miRNAs shuttled by mesenchymal stem cell-derived small extracellular vesicles in modulating neuroinflammation. Sci. Rep. 2021, 11, 1–17. [Google Scholar] [CrossRef]
- A Canales-Aguirre, A.; E Reza-Zaldivar, E.; Sapiéns, M.A.H.; Mercado, Y.K.G.; Sandoval-Ávila, S.; Gomez-Pinedo, U.; Márquez-Aguirre, A.L.; Vazquez-Mendez, E.; Padilla-Camberos, E. Mesenchymal stem cell-derived exosomes promote neurogenesis and cognitive function recovery in a mouse model of Alzheimer’s disease. Neural Regen. Res. 2019, 14, 1626–1634. [Google Scholar] [CrossRef]
- Hsieh, J.-Y.; Wang, H.-W.; Chang, S.-J.; Liao, K.-H.; Lee, I.-H.; Lin, W.-S.; Wu, C.-H.; Lin, W.-Y.; Cheng, S.-M. Mesenchymal Stem Cells from Human Umbilical Cord Express Preferentially Secreted Factors Related to Neuroprotection, Neurogenesis, and Angiogenesis. PLoS ONE 2013, 8, e72604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Chopp, M.; Meng, Y.; Katakowski, M.; Xin, H.; Mahmood, A.; Xiong, Y. Effect of exosomes derived from multipluripotent mesenchymal stromal cells on functional recovery and neurovascular plasticity in rats after traumatic brain injury. J. Neurosurg. 2015, 122, 856–867. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Silva, M.; Feng, C.; Zhao, S.; Liu, L.; Li, S.; Zhong, J.; Zheng, W. Exosomes derived from human placental mesenchymal stem cells enhanced the recovery of spinal cord injury by activating endogenous neurogenesis. Stem Cell Res. Ther. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Chinnici, C.; Iannolo, G.; Cittadini, E.; Carreca, A.; Nascari, D.; Timoneri, F.; Bella, M.; Cuscino, N.; Amico, G.; Carcione, C.; et al. Extracellular vesicle-derived micrornas of human Wharton’s Jelly mesenchymal stromal cells may activate endogenous VEGF-A to promote angiogenesis. Int. J. Mol. Sci. 2021, 22, 2045. [Google Scholar] [CrossRef]
- Mazini, L.; Rochette, L.; Admou, B.; Amal, S.; Malka, G. Hopes and limits of adipose-derived stem cells (ADSCs) and mesenchymal stem cells (MSCs) in wound healing. Int. J. Mol. Sci. 2020, 21, 1306. [Google Scholar] [CrossRef] [Green Version]
- Beckermann, B.M.; Kallifatidis, G.; Groth, A.; Frommhold, D.; Apel, A.; Mattern, J.; Salnikov, A.V.; Moldenhauer, G.; Wagner, W.; Diehlmann, A.; et al. VEGF expression by mesenchymal stem cells contributes to angiogenesis in pancreatic carcinoma. Br. J. Cancer 2008, 99, 622–631. [Google Scholar] [CrossRef] [Green Version]
- Olejarz, W.; Kubiak-Tomaszewska, G.; Chrzanowska, A.; Lorenc, T. Exosomes in angiogenesis and anti-angiogenic therapy in cancers. Int. J. Mol. Sci. 2020, 21, 5840. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Horwood, N.; Cope, A.; Dazzi, F. The Antiproliferative Effect of Mesenchymal Stem Cells Is a Fundamental Property Shared by All Stromal Cells. J. Immunol. 2007, 179, 2824–2831. [Google Scholar] [CrossRef] [PubMed]
- Tavasolian, F.; Hosseini, A.Z.; Naderi, M. miRNA-146a Improves Immunomodulatory Effects of MSC-derived Exosomes in Rheumatoid Arthritis. Curr. Gene Ther. 2020, 20, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Kong, Y.; Hu, X.; Li, Z.; Li, Y.; Zhong, Y.; Wei, X.; Ling, J. MicroRNA-enriched small extracellular vesicles possess odonto-immunomodulatory properties for modulating the immune response of macrophages and promoting odontogenesis. Stem Cell Res. Ther. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hsu, L.-W.; Huang, K.-T.; Nakano, T.; Chiu, K.-W.; Chen, K.-D.; Goto, S.; Chen, C.-L. MicroRNA-301a inhibition enhances the immunomodulatory functions of adipose-derived mesenchymal stem cells by induction of macrophage M2 polarization. Int. J. Immunopathol. Pharmacol. 2020, 34, 2058738420966092. [Google Scholar] [CrossRef]
- Taghavi-Farahabadi, M.; Mahmoudi, M.; Rezaei, N.; Hashemi, S.M. Wharton’s Jelly Mesenchymal Stem Cells Exosomes and Conditioned Media Increased Neutrophil Lifespan and Phagocytosis Capacity. Immunol. Investig. 2020, 50, 1–16. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Taghavi-Farahabadi, M.; Namaki, S.; Baghaei, K.; Rayzan, E.; Rezaei, N.; Hashemi, S.M. Exosomes derived from mesenchymal stem cells improved function and survival of neutrophils from severe congenital neutropenia patients in vitro. Hum. Immunol. 2019, 80, 990–998. [Google Scholar] [CrossRef]
- Brinkmann, V. Neutrophil Extracellular Traps in the Second Decade. J. Innate Immun. 2018, 10, 414–421. [Google Scholar] [CrossRef]
- Von Köckritz-Blickwede, M.; Nizet, V. Innate immunity turned inside-out: Antimicrobial defense by phagocyte extracellular traps. J. Mol. Med. 2009, 87, 775–783. [Google Scholar] [CrossRef] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soni, N.; Gupta, S.; Rawat, S.; Krishnakumar, V.; Mohanty, S.; Banerjee, A. MicroRNA-Enriched Exosomes from Different Sources of Mesenchymal Stem Cells Can Differentially Modulate Functions of Immune Cells and Neurogenesis. Biomedicines 2022, 10, 69. https://doi.org/10.3390/biomedicines10010069
Soni N, Gupta S, Rawat S, Krishnakumar V, Mohanty S, Banerjee A. MicroRNA-Enriched Exosomes from Different Sources of Mesenchymal Stem Cells Can Differentially Modulate Functions of Immune Cells and Neurogenesis. Biomedicines. 2022; 10(1):69. https://doi.org/10.3390/biomedicines10010069
Chicago/Turabian StyleSoni, Naina, Suchi Gupta, Surender Rawat, Vishnu Krishnakumar, Sujata Mohanty, and Arup Banerjee. 2022. "MicroRNA-Enriched Exosomes from Different Sources of Mesenchymal Stem Cells Can Differentially Modulate Functions of Immune Cells and Neurogenesis" Biomedicines 10, no. 1: 69. https://doi.org/10.3390/biomedicines10010069
APA StyleSoni, N., Gupta, S., Rawat, S., Krishnakumar, V., Mohanty, S., & Banerjee, A. (2022). MicroRNA-Enriched Exosomes from Different Sources of Mesenchymal Stem Cells Can Differentially Modulate Functions of Immune Cells and Neurogenesis. Biomedicines, 10(1), 69. https://doi.org/10.3390/biomedicines10010069