One-Shot Learning for Optical Coherence Tomography Angiography Vessel Segmentation Based on Multi-Scale U2-Net
Abstract
:1. Introduction
- We propose a retinal angiography multi-scale segmentation network (RAMS-Net) for OCTA vessel segmentation under a one-shot learning paradigm, which achieves promising improvement over previous works.
- The INC module is used to extract multi-scale features by expanding the receptive field to preserve the integrity of blood vessels with different sizes in retinal angiography images.
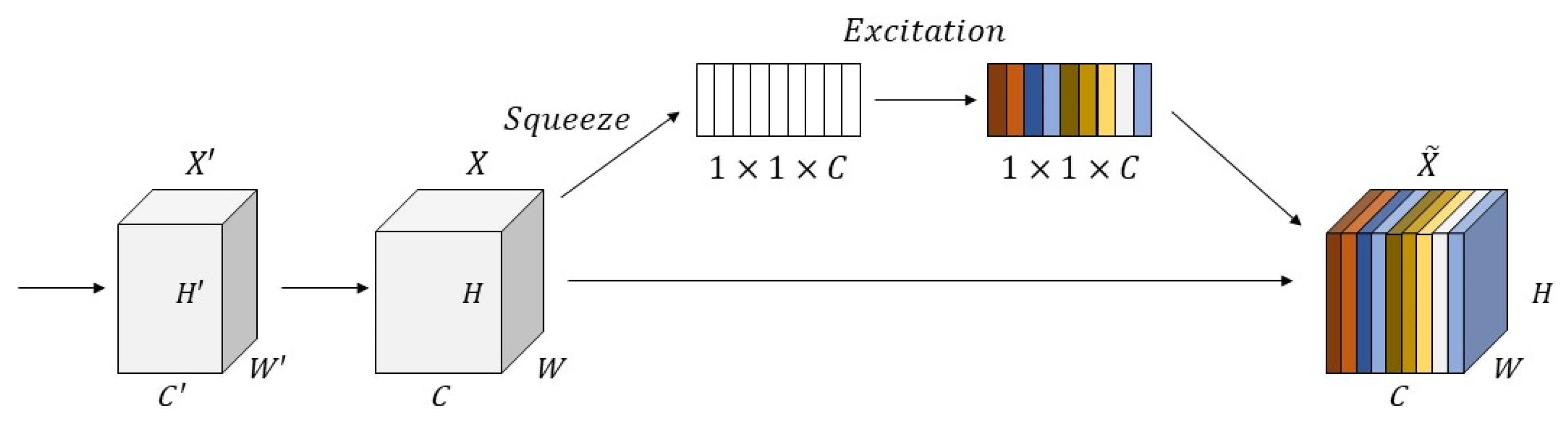
- The SE module is introduced to adjust the weights of each channel adaptively to alleviate ambiguous vessel segmentation under complex noise backgrounds.
2. Materials and Methods
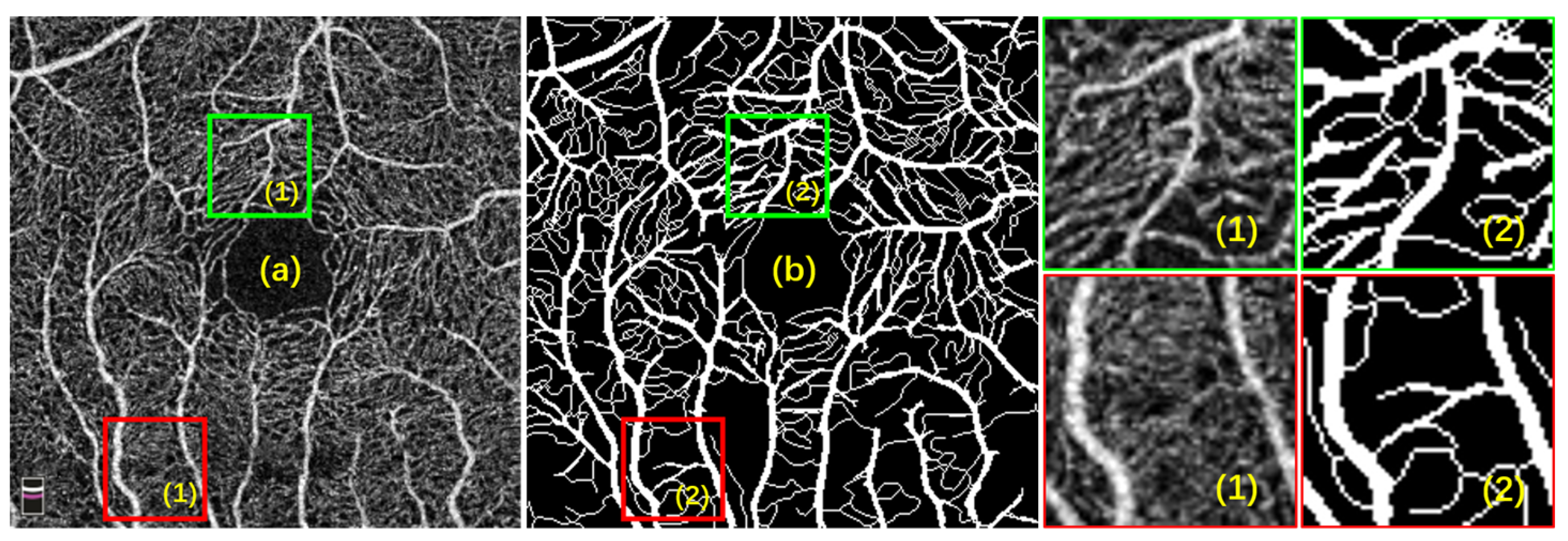
2.1. Dataset
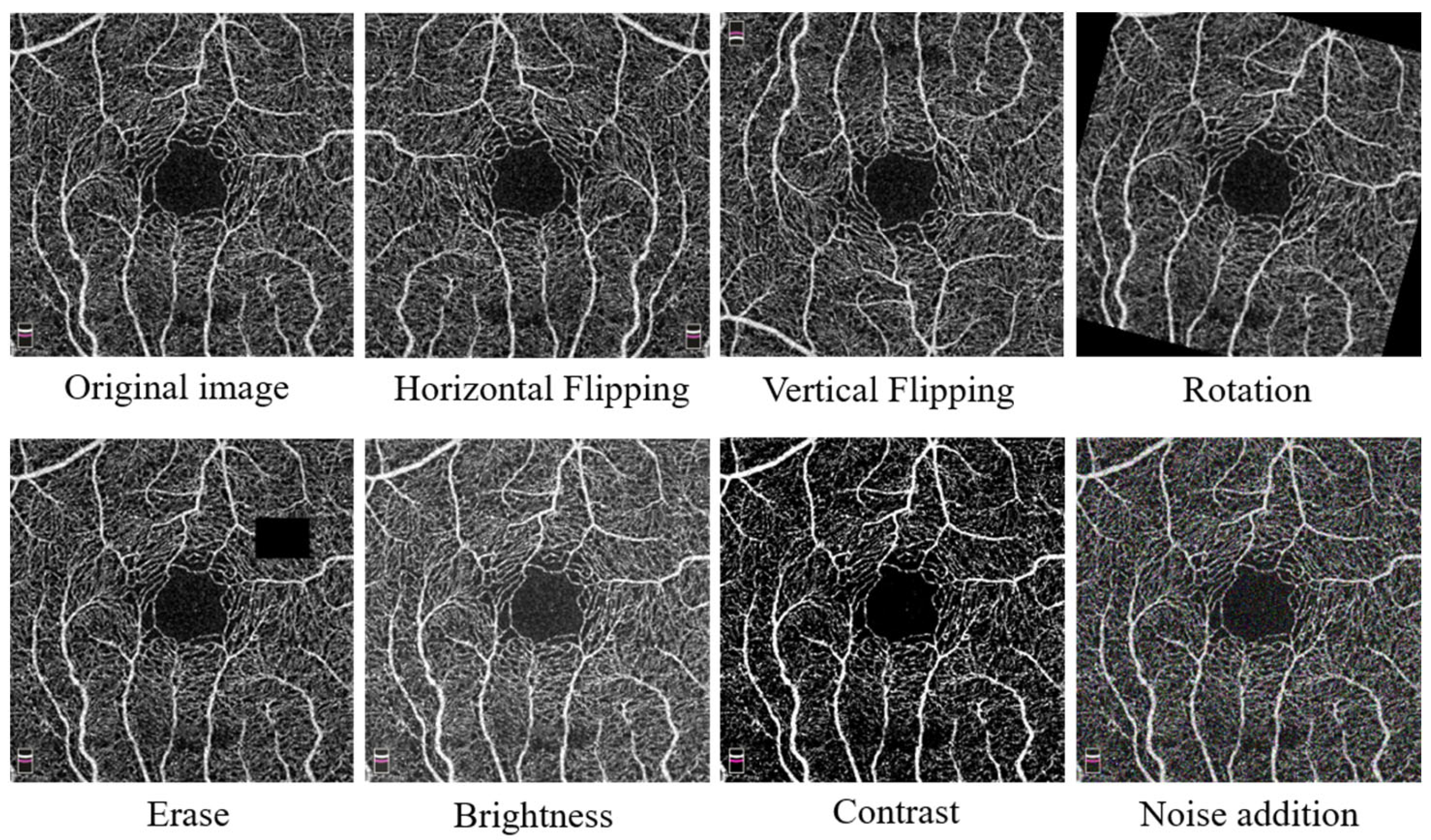
2.2. Data Augmentation
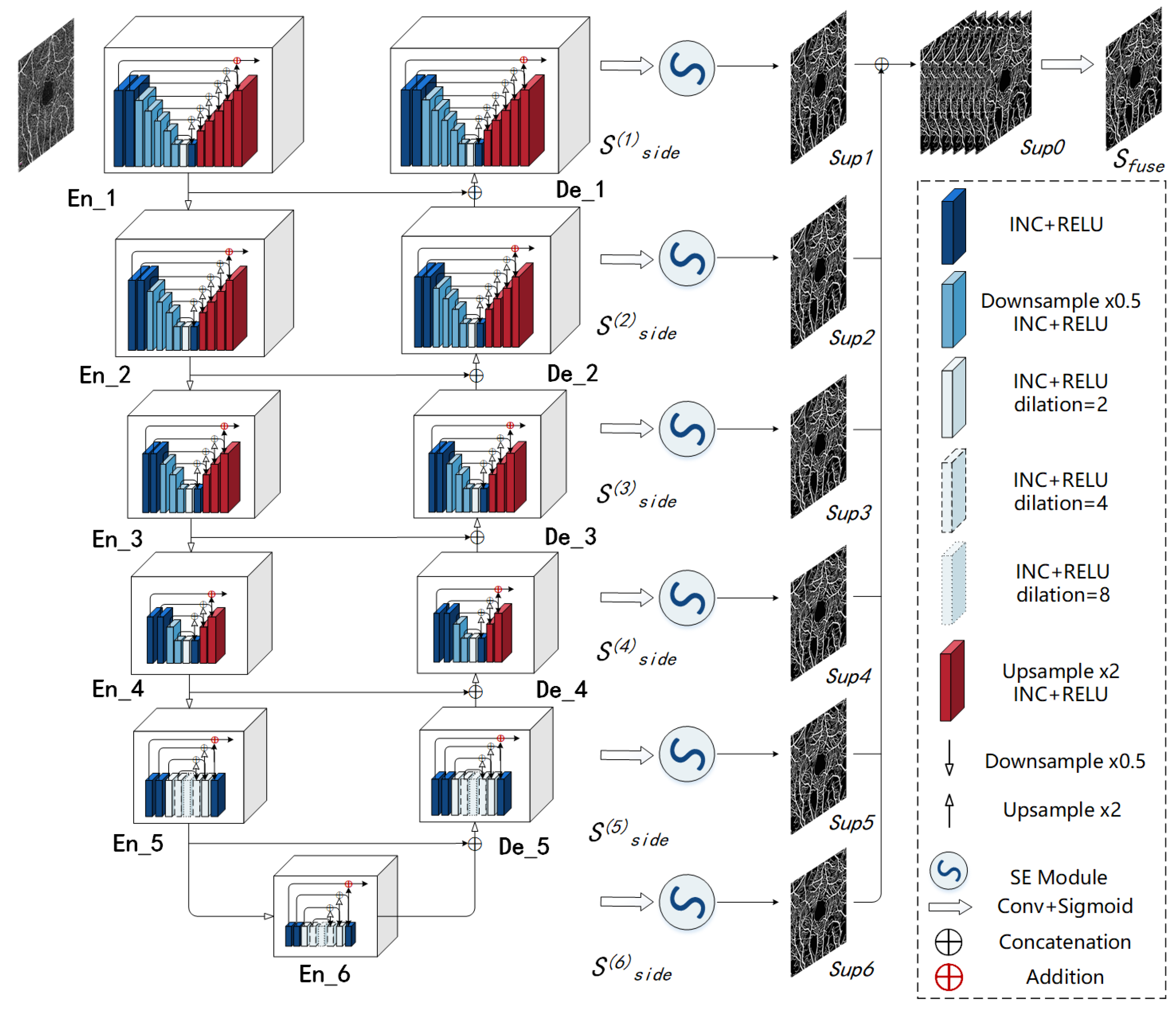
2.3. Retinal Angiography Multi-Scale Segmentation Network (RAMS-Net)
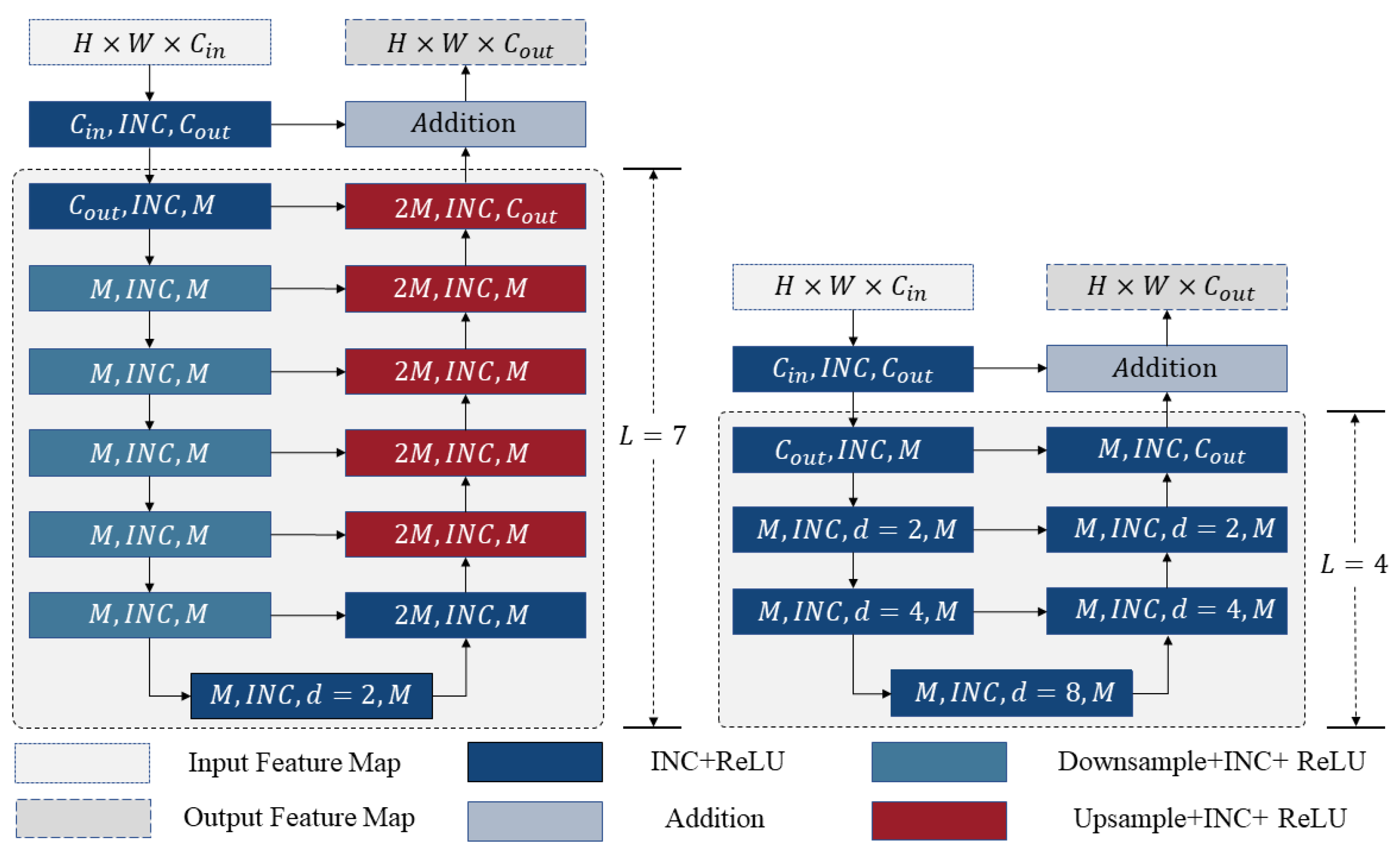
2.3.1. Inception Module
2.3.2. Residual and Inception U-Block (RIU)
2.3.3. Squeeze-and-Excitation Module (SE Module)
2.3.4. Loss Function
3. Results
3.1. Evaluation Metrics
3.2. Implementation Details
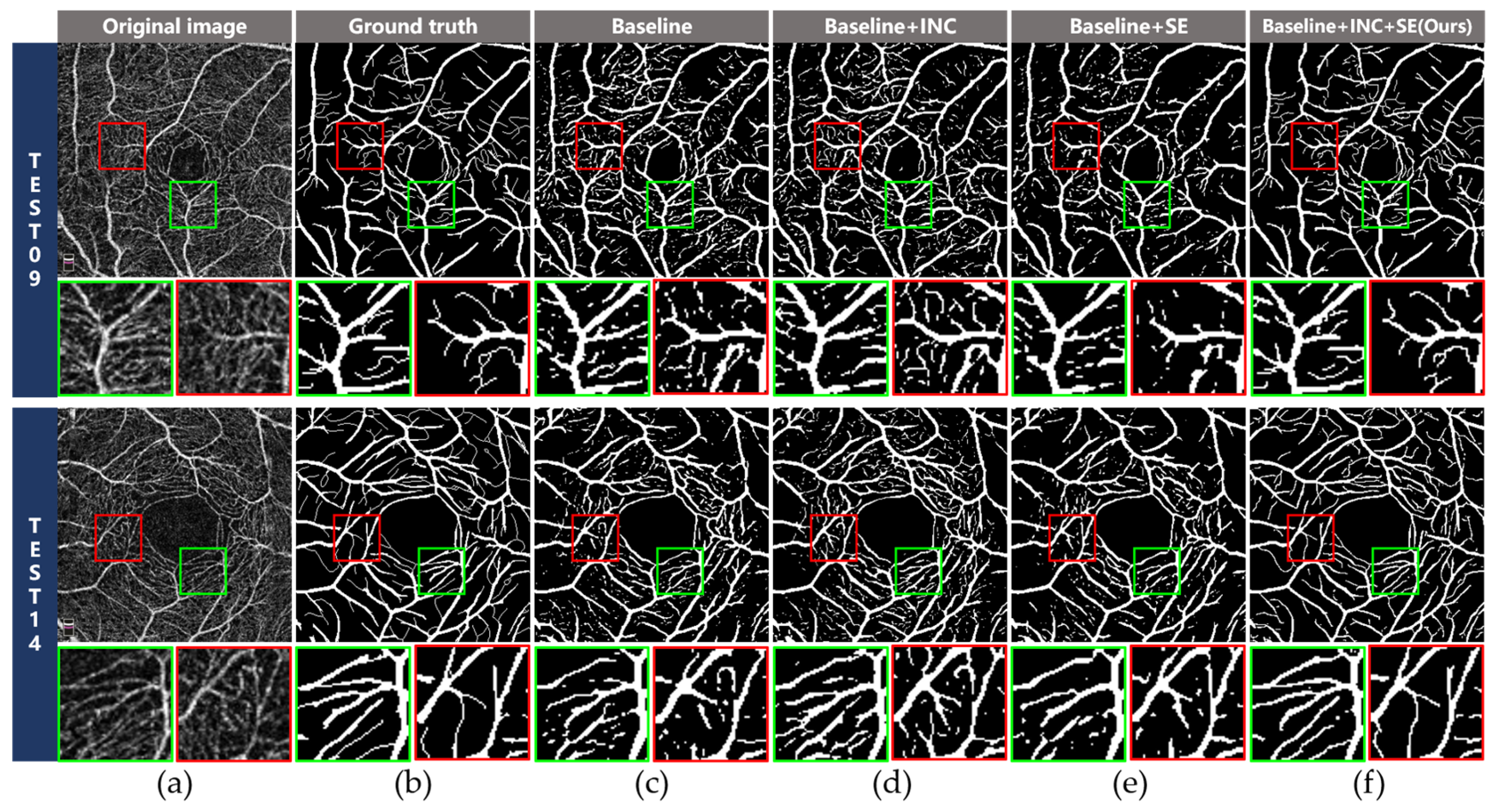
3.3. Ablation Studies
3.3.1. Effectiveness of the INC Module
3.3.2. Effectiveness of the SE Module
3.4. Comparisons with the Other Methods
3.5. Data Augmentation Studies
4. Discussion
4.1. Advantages of RAMS-Net
- Accurate complex vessel segmentation: RAMS-Net outperforms U-Net and U-Net++ in accurately capturing and delineating complex vascular structures, thus minimizing segmentation inaccuracies.
- Multi-scale vascular segmentation: Thanks to the innovative INC module, RAMS-Net excels in detecting small blood vessels, surpassing U2-Net in ensuring the comprehensive segmentation of vessels across a wide range of sizes.
- Effective noise mitigation: Due to the application of the SE module, in contrast to traditional bicubic interpolation and the BM3D algorithm, RAMS-Net effectively mitigates the impact of background noise in retinal angiography images, resulting in improved segmentation accuracy.
- Microvascular connectivity: RAMS-Net effectively minimizes microvascular disconnectivity and incompleteness, contributing to its overall superior performance compared with OCTA-Net.
4.2. Limitations
4.3. Future Improvements and Applications
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Huang, D.; Swanson, E.A.; Lin, C.P.; Schuman, J.S.; Stinson, W.G.; Chang, W.; Hee, M.R.; Flotte, T.; Gregory, K.; Puliafito, C.A.; et al. Optical coherence tomography. Science 1991, 254, 1178–1181. [Google Scholar] [CrossRef]
- Sampson, D.M.; Dubis, A.M.; Chen, F.K.; Zawadzki, R.J.; Sampson, D.D. Towards standardizing retinal optical coherence tomography angiography: A review. Light Sci. Appl. 2022, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; An, L.; Lan, G.; Johnstone, M.; Malchow, D.; Wang, R.K. Extended Imaging Depth to 12 mm for 1050-nm Spectral Domain Optical Coherence Tomography for Imaging the Whole Anterior Segment of the Human Eye at 120-kHz A-scan Rate. J. Biomed. 2013, 18, 016012. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Li, P.; Lan, G.; Malchow, D.; Wang, R.K. High-Resolution 1050 nm Spectral Domain Retinal Optical Coherence Tomography at 120 kHz A-scan Rate with 6.1 mm Imaging Depth. Biomed. Opt. Express 2013, 4, 245–259. [Google Scholar] [CrossRef] [PubMed]
- Spaide, R.F.; Fujimoto, J.G.; Waheed, N.K.; Sadda, S.R.; Staurenghi, G. Optical coherence tomography angiography. Prog. Retin. Eye Res. 2018, 64, 1–55. [Google Scholar] [CrossRef]
- Hwang, T.S.; Jia, Y.; Gao, S.S.; Bailey, S.T.; Lauer, A.K.; Flaxel, C.J.; Wilson, D.J.; Huang, D. Optical coherence tomography angiography features of diabetic retinopathy. Retina 2015, 35, 2371. [Google Scholar] [CrossRef] [PubMed]
- Rao, H.L.; Pradhan, Z.S.; Weinreb, R.N.; Reddy, H.B.; Riyazuddin, M.; Dasari, S.; Palakurthy, M.; Puttaiah, N.K.; Rao, D.A.; Webers, C.A. Regional Comparisons of Optical Coherence Tomography Angiography Vessel Density in Primary Open-Angle Glaucoma. Am. J. Ophthalmol. 2016, 171, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Bailey, S.T.; Wilson, D.J.; Tan, O.; Klein, M.L.; Flaxel, C.J.; Potsaid, B.; Liu, J.J.; Lu, C.D.; Kraus, M.F. Quantitative Optical Coherence Tomography Angiography of Choroidal Neovascularization in Age-Related Macular Degeneration. Ophthalmology 2014, 121, 1435–1444. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.C.; Wang, J.; Hwang, T.S.; Zhang, M.; Gao, S.S.; Pennesi, M.E.; Bailey, B.J.; Lujan, X.; Wang, X.; Wilson, D.J. Plexus-specific detection of retinal vascular pathologic conditions with projection-resolved OCT angiography. Ophthalmol. Retin. 2018, 2, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Vinyals, O.; Blundell, C.; Lillicrap, T.; Wierstra, D. Matching networks for one shot learning. Adv. Neural Inf. Process. Syst. 2016, 29, 3630–3638. [Google Scholar]
- Wu, Y.; Zheng, B.; Chen, J.; Chen, D.Z.; Wu, J. Self-learning and One-shot Learning based Single-slice Annotation for 3D Medical Image Segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2022; pp. 244–254. [Google Scholar]
- Zhang, X.; Wei, Y.; Yang, Y.; Huang, T.S. Sg-one: Similarity guidance network for one-shot semantic segmentation. IEEE Trans. Cybern. 2020, 50, 3855–3865. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lian, C.; Wang, L.; Deng, H.; Fung, S.H.; Nie, D.; Thung, K.H.; Yap, P.T.; Gateno, J.; Xia, J.J.; et al. One-shot generative adversarial learning for MRI segmentation of craniomaxillofacial bony structures. IEEE Trans. Med. Imaging 2019, 39, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Jiang, T.; Zang, Y. Region Growing Method for the Analysis of Functional MRI Data. NeuroImage 2003, 20, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Chudasama, D.; Patel, T.; Joshi, S.; Prajapati, G.I. Image segmentation using morphological operations. Int. J. Comput. Appl. 2015, 117, 16–19. [Google Scholar] [CrossRef]
- Pratondo, A.; Chui, C.K.; Ong, S.H. Integrating machine learning with region-based active contour models in medical image segmentation. J. Visual Commun. Image Represent. 2017, 43, 1–9. [Google Scholar] [CrossRef]
- Dabov, K.; Foi, A.; Katkovnik, V.; Egiazarian, K. Image denoising with block-matching and 3D filtering. Image Process. Algorithms Syst. Neural Netw. Mach. Learn. 2006, 6064, 354–365. [Google Scholar]
- Zhang, H.; Yang, J.; Zhou, K.; Li, F.; Hu, Y.; Zhao, Y.; Zheng, C.; Zhang, X.; Liu, J. Automatic Segmentation and Visualization of Choroid in OCT with Knowledge Infused Deep Learning. IEEE J. Biomed. Health 2020, 24, 3408–3420. [Google Scholar] [CrossRef]
- Kepp, T.; Sudkamp, H.; von der Burchard, C.; Schenke, H.; Koch, P.; Hüttmann, G.; Roider, J.; Heinrich, M.; Handels, H. Segmentation of retinal low-cost optical coherence tomography images using deep learning. SPIE Med. Imaging 2020, 11314, 389–396. [Google Scholar]
- Pekala, M.; Joshi, N.; Liu, T.A.; Bressler, N.M.; DeBuc, D.C.; Burlina, P. Deep learning based retinal OCT segmentation. Comput. Biol. Med. 2019, 114, 103445. [Google Scholar] [CrossRef]
- Yuan, X.; Huang, Y.; An, L.; Qin, J.; Lan, G.; Qiu, H.; Yu, B.; Jia, H.; Ren, S.; Tan, H.; et al. Image Enhancement of Wide-Field Retinal Optical Coherence Tomography Angiography by Super-Resolution Angiogram Reconstruction Generative Adversarial Network. Biomed. Signal Process. Control. 2022, 78, 103957. [Google Scholar] [CrossRef]
- López-Linares, K.; Aranjuelo, N.; Kabongo, L.; Maclair, G.; Lete, N.; Ceresa, M.; Ballester, M.A.G. Fully automatic detection and segmentation of abdominal aortic thrombus in post-operative CTA images using deep convolutional neural networks. Med. Image Anal. 2018, 46, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Zhang, J.; Liu, M.; Wang, L.; Chen, S.; Yuan, P.; Li, J.; Shen, S.G.; Tang, Z.; Chen, K.C.; Xia, J.J.; et al. Context-guided fully convolutional networks for joint craniomaxillofacial bone segmentation and landmark digitization. Med. Image Anal. 2020, 60, 101621. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Proceedings of the 19th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2016), Part II, Athens, Greece, 17–21 October 2016; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 424–432. [Google Scholar]
- Khened, M.; Kollerathu, V.A.; Krishnamurthi, G. Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers. Med. Image Anal. 2019, 51, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015, Proceedings, Part III 18; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Dai, J.; Li, Y.; He, K.; Sun, J. R-FCN: Object Detection via Region-based Fully Convolutional Networks. Adv. Neural Inf. Process. Syst. 2016, 29. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Ktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Rueckert, D. Attention U-Net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Ma, Y.; Hao, H.; Fu, H.; Zhang, J.; Yang, J.; Zhao, Y.; Wang, Z.; Liu, J.; Zheng, Y. ROSE: A retinal OCT-angiography vessel segmentation dataset and new model. IEEE Trans. Med. Imaging 2020, 40, 928–939. [Google Scholar] [CrossRef]
- Qin, X.; Zhang, Z.; Huang, C.; Dehghan, M.; Zaiane, O.R.; Jagersand, M. U2-Net: Going deeper with nested U-structure for salient object detection. Pattern Recognit. 2020, 106, 107404. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Xie, S.; Tu, Z. Holistically-nested edge detection. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 13–16 December 2015; pp. 1395–1403. [Google Scholar]
- Mustofa, F.; Safriandono, A.N.; Muslikh, A.R. Dataset and Feature Analysis for Diabetes Mellitus Classification Using Random Forest. J. Comput. Theor. Appl. 2023, 1, 41–48. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]



| Method | Dice (%) | Acc (%) | Precision (%) | Recall (%) | IOU (%) |
|---|---|---|---|---|---|
| Baseline | 66.51 ± 0.05 | 85.15 ± 0.13 | 58.89 ± 0.25 | 68.39 ± 0.03 | 49.82 ± 0.17 |
| Baseline + INC | 67.79 ± 0.04 | 87.41 ± 0.04 | 61.36 ± 0.02 | 71.29 ± 0.07 | 50.14 ± 0.04 |
| Baseline + SE | 67.09 ± 0.03 | 87.14 ± 0.07 | 60.14 ± 0.09 | 70.86 ± 0.12 | 50.48 ± 0.08 |
| Baseline + INC + SE (ours) | 70.25 ± 0.01 | 89.87 ± 0.02 | 67.51 ± 0.03 | 73.26 ± 0.04 | 54.14 ± 0.01 |
| Method | Dice (%) | Acc (%) | Precision (%) | Recall (%) | IOU (%) |
|---|---|---|---|---|---|
| Bicubic+BM3D | 64.57 ± 0.17 | 85.03 ± 0.19 | 54.63 ± 0.08 | 58.94 ± 0.23 | 47.68 ± 0.05 |
| U-Net | 64.05 ± 0.04 | 78.58 ± 0.11 | 60.21 ± 0.06 | 58.89 ± 0.03 | 47.12 ± 0.10 |
| U-Net++ | 56.85 ± 0.04 | 79.47 ± 0.03 | 54.89 ± 0.05 | 58.93 ± 0.07 | 50.20 ± 0.05 |
| TransUNet | 65.28 ± 0.02 | 79.32 ± 0.06 | 64.53 ± 0.03 | 58.07 ± 0.04 | 48.46 ± 0.16 |
| OCTA-Net | 65.49 ± 0.03 | 79.59 ± 0.11 | 66.64 ± 0.02 | 57.17 ± 0.06 | 48.69 ± 0.04 |
| U2-Net | 66.51 ± 0.12 | 85.15 ± 0.01 | 58.89 ± 0.07 | 68.39 ± 0.04 | 49.82 ± 0.13 |
| RAMS-Net (ours) | 70.25 ± 0.01 | 89.87 ± 0.02 | 67.51 ± 0.03 | 73.26 ± 0.04 | 54.14 ± 0.01 |
| Method | Dice (%) | Acc (%) | Precision (%) | Recall (%) | IOU (%) |
|---|---|---|---|---|---|
| No data augmentation | 54.65 ± 0.04 | 71.14 ± 0.11 | 50.23 ± 0.08 | 60.34 ± 0.03 | 53.35 ± 0.07 |
| Data augmentation | 70.25 ± 0.01 | 89.87 ± 0.02 | 67.51 ± 0.03 | 73.26 ± 0.04 | 54.14 ± 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Guo, S.; Cong, J.; Yang, Y.; Guo, Z.; Gu, B. One-Shot Learning for Optical Coherence Tomography Angiography Vessel Segmentation Based on Multi-Scale U2-Net. Mathematics 2023, 11, 4890. https://doi.org/10.3390/math11244890
Liu S, Guo S, Cong J, Yang Y, Guo Z, Gu B. One-Shot Learning for Optical Coherence Tomography Angiography Vessel Segmentation Based on Multi-Scale U2-Net. Mathematics. 2023; 11(24):4890. https://doi.org/10.3390/math11244890
Chicago/Turabian StyleLiu, Shudong, Shuai Guo, Jia Cong, Yue Yang, Zihui Guo, and Boyu Gu. 2023. "One-Shot Learning for Optical Coherence Tomography Angiography Vessel Segmentation Based on Multi-Scale U2-Net" Mathematics 11, no. 24: 4890. https://doi.org/10.3390/math11244890
APA StyleLiu, S., Guo, S., Cong, J., Yang, Y., Guo, Z., & Gu, B. (2023). One-Shot Learning for Optical Coherence Tomography Angiography Vessel Segmentation Based on Multi-Scale U2-Net. Mathematics, 11(24), 4890. https://doi.org/10.3390/math11244890






