The Potential Global Climate Suitability of Kiwifruit Bacterial Canker Disease (Pseudomonas syringae pv. actinidiae (Psa)) Using Three Modelling Approaches: CLIMEX, Maxent and Multimodel Framework
Abstract
:1. Introduction
2. Materials and Methods
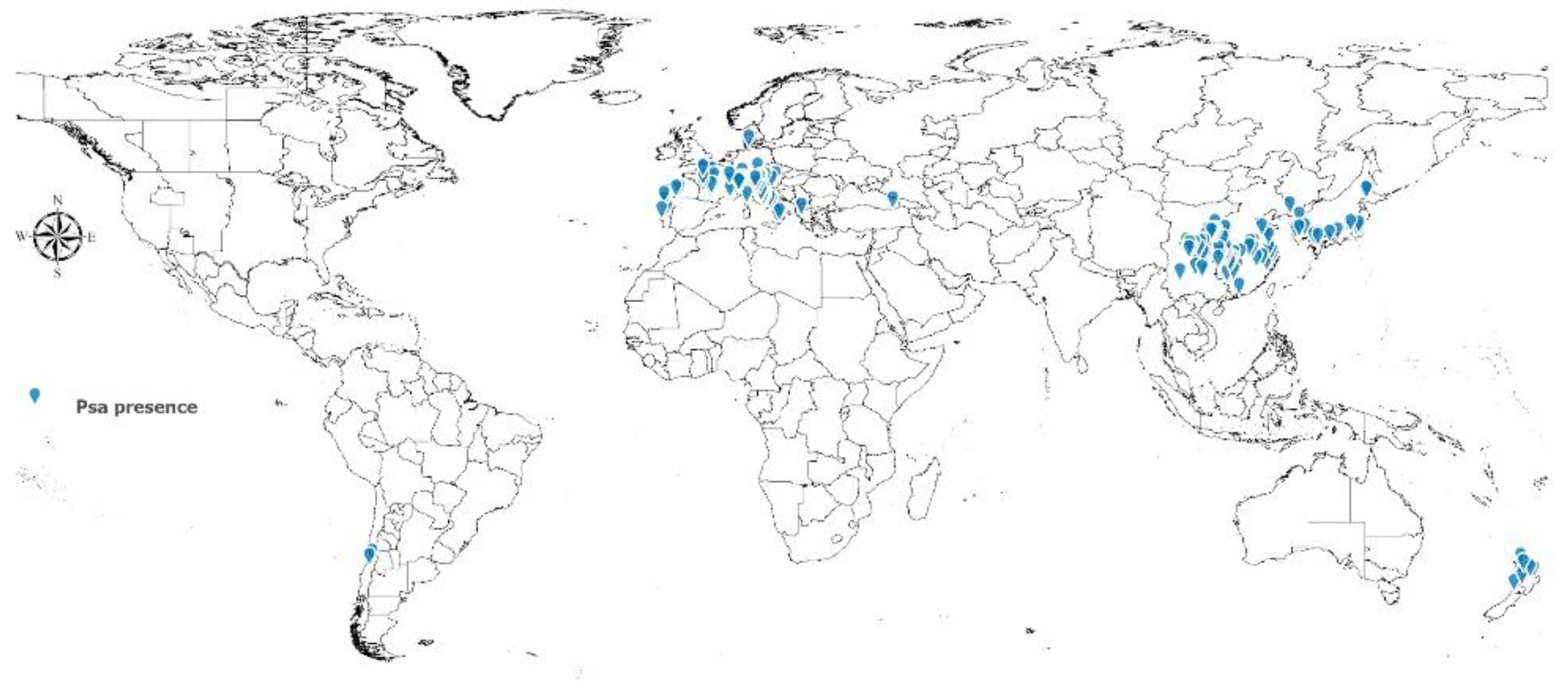
2.1. Psa Presence Data
2.2. Bioclimatic Variables
2.3. Species Distribution Modelling
2.3.1. CLIMEX
2.3.2. MaxEnt
2.3.3. Multimodel Framework
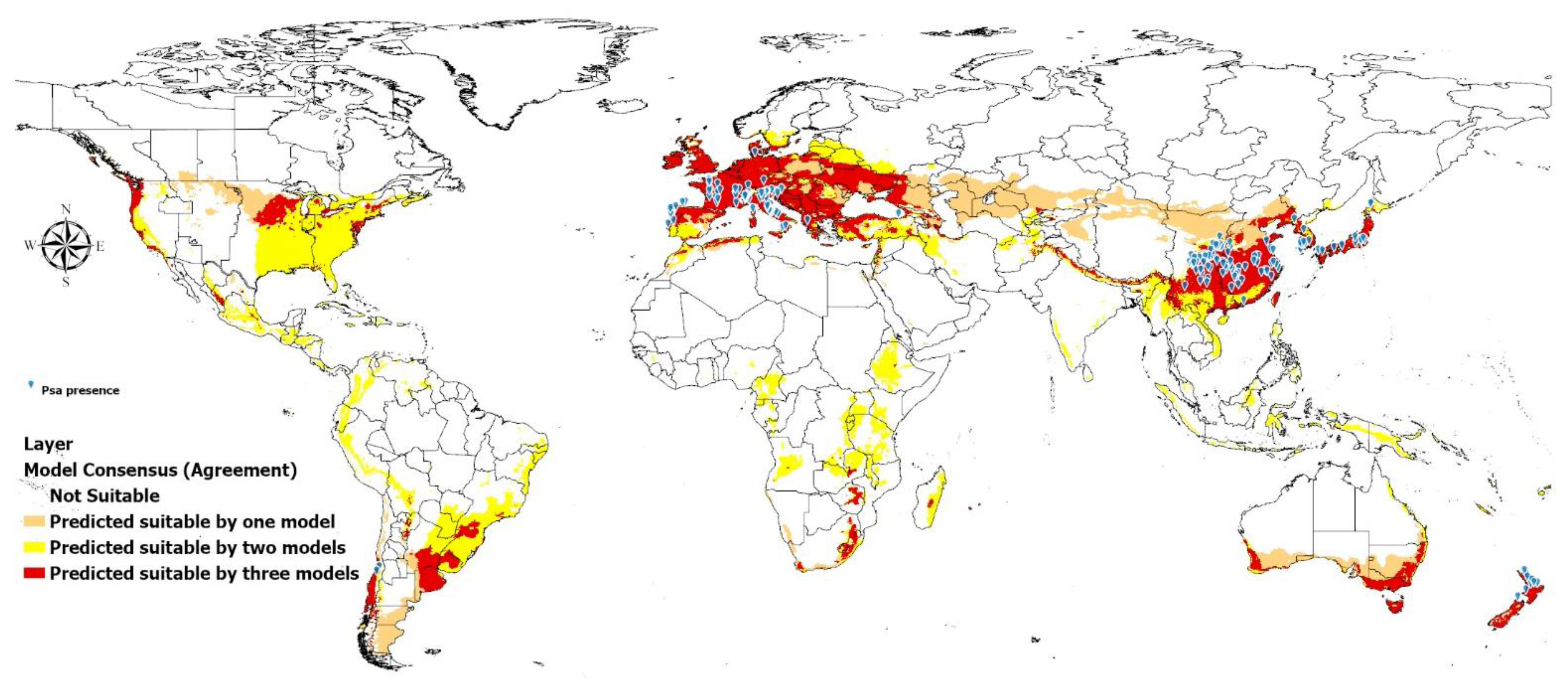
2.4. Model Consensus (Agreement)
3. Results
3.1. Model Performance
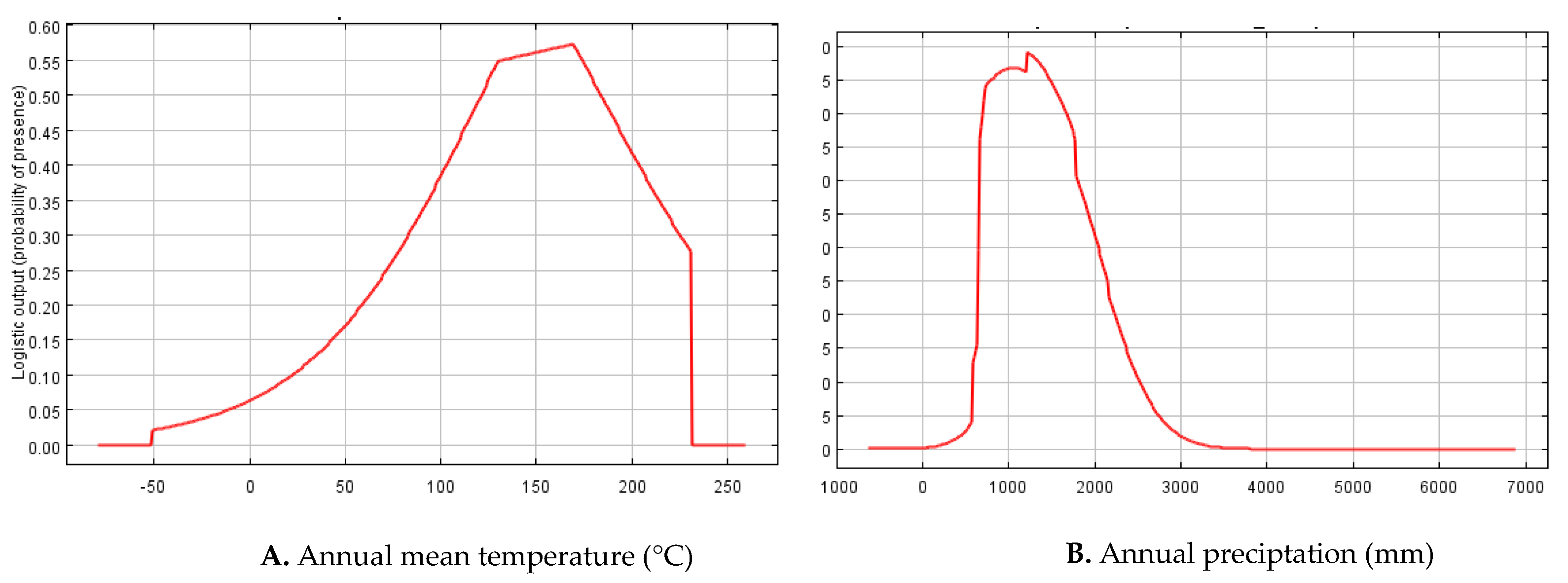
3.2. Predictor (Environmental) Variables
3.3. Potential Distribton
3.4. Model Consensus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Renzi, M.; Copini, P.; Taddei, A.R.; Rossetti, A.; Gallipoli, L.; Mazzaglia, A.; Balestra, G.M. Bacterial canker on kiwifruit in Italy: Anatomical changes in the wood and in the primary infection sites. Phytopathology 2012, 102, 827–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanneste, J.L. The scientific, economic, and social impacts of the New Zealand outbreak of bacterial canker of kiwifruit (Pseudomonas syringae pv. actinidiae). Annu. Rev. Phytopathol. 2017, 55, 377–399. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.H.; Jung, J.S.; Koh, Y.J. Occurrence and epidemics of bacterial canker of kiwifruit in Korea. Plant Pathol. J. 2017, 33, 351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takikawa, Y.; Serizawa, S.; Ichikawa, T.; Tsuyumu, S.; Goto, M. Pseudomonas syringae pv. actinidiae pv. nov.: The causal bacterium of canker of kiwifruit in Japan. Ann. Phytopatb. Soc. Jpn. 1989, 55, 437–444. [Google Scholar]
- Koh, Y.J.; Chung, H.J.; Cha, B.J.; Lee, D.H. Outbreak and spread of bacterial canker in kiwifruit. Plant Pathol. J. 1994, 10, 68–72. [Google Scholar]
- Cheng, H.; Li, Y.; Wan, S.; Zhan, J.; Ping, Q.; Guo, L.; Jiahua, X. Pathogenic indentification of kiwifruit bacterial canker in Anhui. J. Anhui Agric. Univ. 1995, 22, 219–223. [Google Scholar]
- Scortichini, M. Occurrence of Pseudomonas syringae pv. actinidiae on kiwifruit in Italy. Plant Pathol. 1994, 43, 1035–1038. [Google Scholar] [CrossRef]
- Balestra, G.M.; Mazzaglia, A.; Spinelli, R.; Graziani, S.; Quattrucci, A.; Rossetti, A. Cancro batterico su Actinidia chinensis. L’Informatore Agrar. 2008, 38, 7576. [Google Scholar]
- Renzi, M.; Mazzaglia, A.; Balestra, G.M. Widespread distribution of kiwifruit bacterial canker caused by the European Pseudomonas syringae pv. actinidiae genotype in the main production areas of Portugal. Phytopathol. Mediterr. 2012, 51, 402–409. [Google Scholar]
- Balestra, G.M.; Mazzaglia, A.; Quattrucci, A.; Renzi, M.; Ricci, L.; Rossetti, A. Increased spread of bacterial canker of kiwifruit in Italy. Inf. Agrar. 2009, 65, 58–60. [Google Scholar]
- Balestra, G.M.; Renzi, M.; Mazzaglia, A. First report of bacterial canker of Actinidia deliciosa caused by Pseudomonas syringae pv. actinidiae in Portugal. New Dis. Rep. 2010, 22, 10. [Google Scholar] [CrossRef] [Green Version]
- Balestra, G.M.; Renzi, M.; Mazzaglia, A. First report of Pseudomonas syringae pv. actinidiae on kiwifruit plants in Spain. New Dis. Rep. 2011, 24, 10. [Google Scholar] [CrossRef] [Green Version]
- Everett, K.R.; Taylor, R.K.; Romberg, M.K.; Rees-George, J.; Fullerton, R.A.; Vanneste, J.L.; Manning, M.A. First report of Pseudomonas syringae pv. actinidiae causing kiwifruit bacterial canker in New Zealand. Australas. Plant Dis. Notes 2011, 6, 67–71. [Google Scholar] [CrossRef] [Green Version]
- EPPO. First report of Pseudomonas syringae pv. actinidiae in Chile. EPPO Report. Serv. 2011, 3, 55. [Google Scholar]
- Dreo, T.; Pirc, M.; Ravnikar, M.; Žežlina, I.; Poliakoff, F.; Rivoal, C.; Nice, F.; Cunty, A. First report of Pseudomonas syringae pv. actinidiae, the causal agent of bacterial canker of kiwifruit in Slovenia. Plant Dis. 2014, 98, 1578. [Google Scholar] [CrossRef]
- EPPO. First report of Pseudomonas syringae pv. actinidiae in Switzerland. EPPO Rep. Serv. No. 8 2011, 6, 67–71. [Google Scholar]
- Bastas, K.K.; Karakaya, A. First report of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidiae in Turkey. Mol. Plant Pathol. 2012, 13, 631–640. [Google Scholar] [CrossRef]
- Vanneste, J.L.; Poliakoff, F.; Audusseau, C.; Cornish, D.A.; Paillard, S.; Rivoal, C.; Yu, J. First report of Pseudomonas syringae pv. actinidiae, the causal agent of bacterial canker of kiwifruit in France. Mol. Plant Pathol. 2012, 13, 631–640. [Google Scholar] [CrossRef]
- EPPO. First report of Pseudomonas syringae pv. actinidiae in Germany. EPPO Rep. Serv. No. 9. 2013, p. 185, no. 09–2013. Available online: https://gd.eppo.int/reporting/article-2647.2013 (accessed on 5 December 2013).
- Holeva, M.; Glynos, P.; Karafla, C. First report of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidiae in Greece. Plant Dis. 2015, 99, 723. [Google Scholar] [CrossRef]
- Mazzaglia, A.; Turco, S.; Taratufolo, M.; Tatì, M.; Rahi, Y.J.; Gallipoli, L.; Balestra, G. Improved MLVA typing reveals a highly articulated structure in Pseudomonas syringae pv. actinidiae populations. Physiol. Mol. Plant Pathol. 2021, 114, 101636. [Google Scholar] [CrossRef]
- Ciarroni, S.; Gallipoli, L.; Taratufolo, M.C.; Butler, M.I.; Poulter, R.T.; Pourcel, C.; Vergnaud, G.; Balestra, G.M.; Mazzaglia, A. Development of a multiple loci variable number of tandem repeats analysis (MLVA) to unravel the intra-pathovar structure of Pseudomonas syringae pv. actinidiae populations worldwide. PLoS ONE 2015, 10, e0135310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, R.; Liu, P.; Jia, B.; Xue, S.; Wang, X.; Hu, J.; Al Shoffe, Y.; Gallipoli, L.; Mazzaglia, A.; Balestra, G.M. Genetic diversity of Pseudomonas syringae pv. actinidiae strains from different geographic regions in China. Phytopathology 2019, 109, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Cunty, A.; Poliakoff, F.; Rivoal, C.; Cesbron, S.; Fischer-Le Saux, M.; Lemaire, C.; Jacques, M.-A.; Manceau, C.; Vanneste, J. Characterization of Pseudomonas syringae pv. actinidiae (P sa) isolated from France and assignment of Psa biovar 4 to a de novo pathovar: Pseudomonas syringae pv. actinidifoliorum pv. nov. Plant Pathol. 2015, 64, 582–596. [Google Scholar] [CrossRef]
- Froud, K.; Everett, K.; Tyson, J.; Beresford, R.; Cogger, N. Review of the risk factors associated with kiwifruit bacterial canker caused by Pseudomonas syringae pv actinidiae. N. Z. Plant Prot. 2015, 68, 313–327. [Google Scholar] [CrossRef] [Green Version]
- KVH. Kiwifruit Vine Health Psa Statistics Report. Available online: https://kvh.org.nz/protocols-movement-controls/maps-and-stats (accessed on 5 December 2013).
- KVH. Kiwifruit Vine Health Psa Statistics Report. Available online: https://kvh.org.nz/assets/documents/Protocols-and-movement-controls-tab/Maps-and-Stats/KVH-Psa-statistics-06122019.pdf (accessed on 5 December 2021).
- Ushiyama, K. Studies on the epidemics and control of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidiae. Bull. Kanagawa Hortic. Exp. Stn. 1993, 43, 1–76. [Google Scholar]
- Gallipoli, L.; Hu, J.; Pu, L.; Speranza, S.; Mazazaglia, A.; Zhu, L.; Balestra, G.M. Two potential vectors, Nephotettix bipunctatus and Firmiana simplex to spread Pseudomonas syringae pv. actinidiae causing kiwifruit bacterial canker. In Proceedings of the 1st International Symposium on Bacterial Canker of Kiwifruit (Psa), Mt. Maunganui, Tauranga, New Zealand, 19–22 November 2013; p. 73. [Google Scholar]
- Liu, P.; Xue, S.; He, R.; Hu, J.; Wang, X.; Jia, B.; Gallipoli, L.; Mazzaglia, A.; Balestra, G.M.; Zhu, L. Pseudomonas syringae pv. actinidiae isolated from non-kiwifruit plant species in China. Eur. J. Plant Pathol. 2016, 145, 743–754. [Google Scholar] [CrossRef]
- Quattrucci, A.; Renzi, M.; Rossetti, A.; Ricci, L.; Taratufolo, M.C.; Mazzaglia, A.; Balestra, G.M. Cancro batterico del kiwi verde: Nuove strategie di controllo. L’informatore Agrar. 2010, 16, 53–58. [Google Scholar]
- Ferrante, P.; Scortichini, M. Identification of Pseudomonas syringae pv. actinidiae as causal agent of bacterial canker of yellow kiwifruit (Actinidia chinensis Planchon) in central Italy. J. Phytopathol. 2009, 157, 768–770. [Google Scholar] [CrossRef]
- Greer, G.; Saunders, C. The Costs of Psa-V to the New Zealand Kiwifruit Industry and the Wider Community; Agribusiness and Economics Research Unit Report; Lincoln University: Lincoln, New Zealand, 2012. [Google Scholar]
- Birnie, D.; Livesey, A. Lessons learned from the response to Psa-V. Sapere Res. 2014. Available online: https://kvh.org.nz/vdb/document/100538 (accessed on 1 December 2021).
- Serizawa, S.; Ichikawa, T.; Takikawa, Y.; Tsuyumu, S.; Goto, M. Occurrence of bacterial canker of kiwifruit in Japan: Description of symptoms, isolation of the pathogen and screening of bactericides. Ann. Phytopathol. Soc. Jpn. 1989, 55, 427–436. [Google Scholar] [CrossRef] [Green Version]
- Donati, I.; Mauri, S.; Buriani, G.; Cellini, A.; Spinelli, F. Role of Metcalfa pruinosa as a vector for Pseudomonas syringae pv. actinidiae. Plant Pathol. J. 2017, 33, 554. [Google Scholar] [CrossRef] [PubMed]
- Serizawa, S.; Ichikawa, T. Epidemiology of bacterial canker of kiwifruit. 4. Optimum temperature for disease development on new canes. Ann. Phytopathol. Soc. Jpn. 1993, 59, 694–701. [Google Scholar] [CrossRef]
- Serizawa, S.; Ichikawa, T. Epidemiology of bacterial canker of kiwifruit, 3: The seasonal changes of bacterial population in lesions and of its exudation from lesion. Ann. Phytopathol. Soc. Jpn. 1993, 59, 46. [Google Scholar] [CrossRef]
- Beresford, R.; Tyson, J.; Henshall, W. Development and validation of an infection risk model for bacterial canker of kiwifruit, using a multiplication and dispersal concept for forecasting bacterial diseases. Phytopathology 2017, 107, 184–191. [Google Scholar] [CrossRef]
- EPPO. Final Pest Risk Analysis for Pseudomonas Syringae pv. Actinidiae 12-17928; European and Mediterranean Plant Protection Organization: Paris, France, 2012; p. 50. [Google Scholar]
- Yao, L.I.; Heyuan, C.; Shumiao, F.; Zihua, Q. Prevalent forecast of kiwifruit bacterial canker caused by Pseudomonas syringae pv. actinidiae. Chin. J. Appl. Ecol. 2001, 3, 355–358. [Google Scholar]
- Tyson, J.; Rees-George, J.; Curtis, C.; Manning, M.; Fullerton, R. Survival of Pseudomonas syringae pv. actinidiae on the orchard floor over winter. N. Z. Plant Prot. 2012, 65, 25–28. [Google Scholar] [CrossRef] [Green Version]
- Narouei Khandan, H.A.; Worner, S.P.; Jones, E.E.; Villjanen-Rollinson, S.L.H.; Gallipoli, L.; Mazzaglia, A.; Balestra, G.M. Predicting the potential global distribution of Pseudomonas syringae pv. actinidiae (Psa). N. Z. Plant Prot. 2013, 66, 184–193. [Google Scholar]
- Wang, R.; Li, Q.; He, S.; Liu, Y.; Wang, M.; Jiang, G. Modeling and mapping the current and future distribution of Pseudomonas syringae pv. actinidiae under climate change in China. PLoS ONE 2018, 13, e0192153. [Google Scholar] [CrossRef] [Green Version]
- Qin, Z.; Zhang, J.; Jiang, Y.; Wang, R.; Wu, R. Predicting the potential distribution of Pseudomonas syringae pv. actinidiae in China using ensemble models. Plant Pathol. 2020, 69, 120–131. [Google Scholar] [CrossRef]
- Do, K.S.; Chung, B.N.; Joa, J.H. D-PSA-K: A model for estimating the accumulated potential damage on kiwifruit canes caused by bacterial canker during the growing and overwintering seasons. Plant Pathol. J. 2016, 32, 537. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.H.; Son, K.; Koh, Y. Adaptation of the New Zealand Psa Risk Model for forecasting kiwifruit bacterial canker in Korea. Plant Pathol. 2018, 67, 1208–1219. [Google Scholar] [CrossRef]
- Kim, K.-H.; Koh, Y.J. An integrated modeling approach for predicting potential epidemics of bacterial blossom blight in kiwifruit under climate change. Plant Pathol. J. 2019, 35, 459. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-H.; Koh, Y.J. Development of a Maryblyt-based forecasting model for kiwifruit bacterial blossom blight. Res. Plant Dis. 2015, 21, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Narouei-Khandan, H.; Worner, S.; Viljanen, S.; van Bruggen, A.; Jones, E. Projecting the suitability of global and local habitats for myrtle rust (Austropuccinia psidii) using model consensus. Plant Pathol. 2020, 69, 17–27. [Google Scholar] [CrossRef]
- Narouei-Khandan, H.A.; Halbert, S.E.; Worner, S.P.; van Bruggen, A.H. Global climate suitability of citrus huanglongbing and its vector, the Asian citrus psyllid, using two correlative species distribution modeling approaches, with emphasis on the USA. Eur. J. Plant Pathol. 2016, 144, 655–670. [Google Scholar] [CrossRef]
- Kriticos, D.J.; Morin, L.; Leriche, A.; Anderson, R.C.; Caley, P. Combining a climatic niche model of an invasive fungus with its host species distributions to identify risks to natural assets: Puccinia psidii Sensu Lato in Australia. PLoS ONE 2013, 8, e64479. [Google Scholar] [CrossRef] [Green Version]
- Kriticos, D.J.; Webber, B.L.; Leriche, A.; Ota, N.; Macadam, I.; Bathols, J.; Scott, J.K. CliMond: Global high-resolution historical and future scenario climate surfaces for bioclimatic modelling. Methods Ecol. Evol. 2012, 3, 53–64. [Google Scholar] [CrossRef]
- Hutchinson, M.; Xu, T.; Houlder, D.; Nix, H.; McMahon, J. ANUCLIM 6.0 User’s Guide; Fenner School of Environment and Society, Australian National University: Canberra, Austrilia, 2009. [Google Scholar]
- Elith, J. Predicting Distributions of Invasive Species. 2013. Available online: http://arxiv.org/abs/1312.0851 (accessed on 1 December 2021).
- Stephens, A.E.; Kriticos, D.J.; Leriche, A. The current and future potential geographical distribution of the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Bull. Entomol Res. 2007, 97, 369–378. [Google Scholar] [CrossRef]
- Beddow, J.M.; Hurley, T.M.; Kriticos, D.J.; Pardey, P.G. Measuring the Global Occurrence and Probabilistic Consequences of Wheat Stem Rust. St. Paul, MN, HarvestChoice, April 2013. pp. 1–24. Available online: https://www.instepp.umn.edu/products/measuring-global-occurrence-and-probabilistic-consequences-wheat-stem-rust (accessed on 1 December 2021).
- Venette, R.C.; Cohen, S.D. Potential climatic suitability for establishment of Phytophthora ramorum within the contiguous United States. For. Ecol. Manage. 2006, 231, 18–26. [Google Scholar] [CrossRef]
- Pattison, R.R.; Mack, R.N. Potential distribution of the invasive tree Triadica sebifera (Euphorbiaceae) in the United States: Evaluating CLIMEX predictions with field trials. Glob. Change Biol. 2008, 14, 813–826. [Google Scholar] [CrossRef]
- Poutsma, J.; Loomans, A.J.M.; Aukema, B.; Heijerman, T. Predicting the potential geographical distribution of the harlequin ladybird, Harmonia axyridis, using the CLIMEX model. BioControl 2008, 53, 103–125. [Google Scholar] [CrossRef]
- Senaratne, K.A.D.; Palmer, W.A.; Sutherst, R.W. Use of CLIMEX modelling to identify prospective areas for exploration to find new biological control agents for prickly acacia. Aust. J. Entomol. 2006, 45, 298–302. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef] [Green Version]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Merow, C.; Smith, M.J.; Silander, J.A. A practical guide to MaxEnt for modeling species’ distributions: What it does, and why inputs and settings matter. Ecography 2013, 36, 1–12. [Google Scholar] [CrossRef]
- Syfert, M.M.; Smith, M.J.; Coomes, D.A. The effects of sampling bias and model complexity on the predictive performance of MaxEnt species distribution models. PLoS ONE 2013, 8, e55158. [Google Scholar] [CrossRef]
- Worner, S.P.; Ikeda, T.; Leday, G.; Zealand, N.; Joy, M. Surveillance Tools for Freshwater Invertebrates. MAF Biosecurity Tech. Pap. 2010. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.459.1082&rep=rep1&type=pdf (accessed on 1 December 2021).
- Radosavljevic, A.; Anderson, R.P. Making better Maxent models of species distributions: Complexity, overfitting and evaluation. J. Biogeogr. 2013, 41, 629–643. [Google Scholar] [CrossRef]
- Biosecurity Australia. Final Pest risk Analysis Report for Pseudomonas Syringae pv. Actinidiae Associated with Actinidia (Kiwifruit)Propagative Material; Department of Agriculture, Fisheries and Forestry: Canberra, Australia, 2011. Available online: https://www.awe.gov.au/biosecurity-trade/policy/risk-analysis/memos/2011/baa_2011-22-final. (accessed on 1 December 2021).
- Wolpert, D.H. Stacked generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Sutherst, R.W.; Maywald, G.F.; Kriticos, D.J. CLIMEX Version 3: User’s Guide. South Yarra: Hearne Scientific Software. 2007. Available online: http://hdl.handle.net/102.100.100/127529?index=1 (accessed on 1 December 2021).
- Donati, I.; Cellini, A.; Sangiorgio, D.; Vanneste, J.L.; Scortichini, M.; Balestra, G.M.; Spinelli, F. Pseudomonas syringae pv. actinidiae: Ecology, infection dynamics and disease epidemiology. Microb. Ecol. 2020, 80, 81–102. [Google Scholar] [CrossRef]
- Chapman, J.; Taylor, R.; Weir, B.; Romberg, M.; Vanneste, J.; Luck, J.; Alexander, B. Phylogenetic relationships among global populations of Pseudomonas syringae pv. actinidiae. Phytopathology 2012, 102, 1034–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazarei, M.; Mostofipour, P. First report of bacterial canker of kiwifruit in Iran. Plant Pathol. 1994, 43, 1055–1056. [Google Scholar] [CrossRef]
- Araújo, M.B.; Whittaker, R.J.; Ladle, R.J.; Erhard, M. Reducing uncertainty in projections of extinction risk from climate change. Glob. Ecol. Biogeogr. 2005, 14, 529–538. [Google Scholar] [CrossRef]
- Watling, J.I.; Brandt, L.A.; Bucklin, D.N.; Fujisaki, I.; Mazzotti, F.J.; Romanach, S.S.; Speroterra, C. Performance metrics and variance partitioning reveal sources of uncertainty in species distribution models. Ecol. Model. 2015, 309, 48–59. [Google Scholar] [CrossRef]
- Narouei-Khandan, H.A.; Harmon, C.L.; Harmon, P.; Olmstead, J.; Zelenev, V.V.; van der Werf, W.; Worner, S.P.; Senay, S.D.; van Bruggen, A.H.C. Potential global and regional geographic distribution of Phomopsis vaccinii on Vaccinium species projected by two species distribution models. Eur. J. Plant Pathol. 2017, 148, 919–930. [Google Scholar] [CrossRef]
- van Bruggen, A.; West, J.; Van der Werf, W.; Potting, R.; Gardi, C.; Koufakis, I.; Zelenev, V.; Narouei-Khandan, H.; Schilder, A.; Harmon, P. Input data needed for a risk model for the entry, establishment and spread of a pathogen (Phomopsis vaccinii) of blueberries and cranberries in the EU. Ann. Appl. Biol. 2018, 172, 126–147. [Google Scholar] [CrossRef]
- Araujo, M.B.; New, M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 2007, 22, 42–47. [Google Scholar] [CrossRef]
- Webber, B.L.; Yates, C.J.; Le Maitre, D.C.; Scott, J.K.; Kriticos, D.J.; Ota, N.; McNeill, A.; Le Roux, J.J.; Midgley, G.F. Modelling horses for novel climate courses: Insights from projecting potential distributions of native and alien Australian acacias with correlative and mechanistic models. Divers. Distrib. 2011, 17, 978–1000. [Google Scholar] [CrossRef]
- Ward, D.F. Modelling the potential geographic distribution of invasive ant species in New Zealand. Biol. Invasions 2007, 9, 723–735. [Google Scholar] [CrossRef]
- Pereira, C.; Costa, P.; Pinheiro, L.; Balcão, V.M.; Almeida, A. Kiwifruit bacterial canker: An integrative view focused on biocontrol strategies. Planta 2021, 253, 1–20. [Google Scholar] [CrossRef]


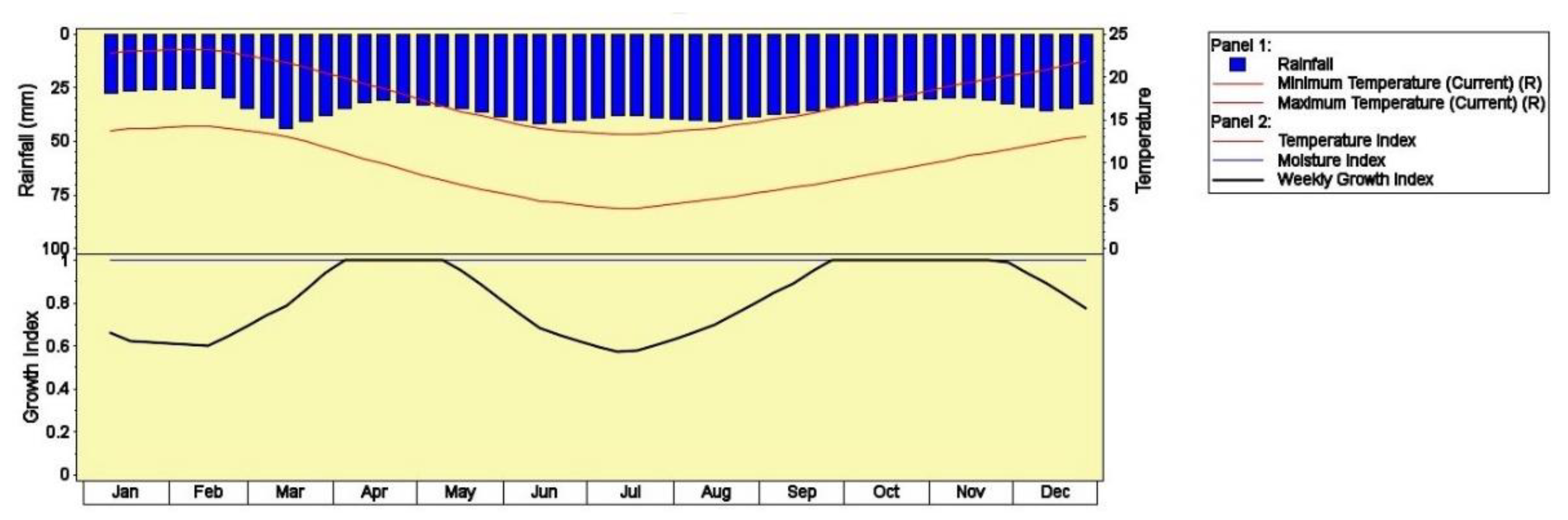
| Index | Parameters | Current Study | EPPO | Unit |
|---|---|---|---|---|
| SM0 | Lower soil moisture threshold | 0.75 | 0.2 | - |
| SM1 | Lower optimum soil moisture | 0.9 | 0.5 | - |
| SM2 | Upper optimum soil moisture | 2.2 | 1.5 | - |
| SM3 | Upper soil moisture threshold | 3 | 2.5 | - |
| DV0 | Lower temperature threshold | 5 | 10 | °C |
| DV1 | Lower optimum temperature | 12 | 20 | °C |
| DV2 | Upper optimum temperature | 20 | 25 | °C |
| DV3 | Upper temperature threshold | 28 | 30 | °C |
| TTCS | Cold stress temperature threshold | 5 | 2 | °C |
| THCS | Cold stress temperature rate | −0.00005 | −0.01 | Week−1 |
| DTCS | Cold stress degree-day threshold | 15 | - | °C |
| DHCS | Cold stress degree-day rate | −0.0001 | - | Week−1 |
| TTHS | Heat Stress Temperature threshold | 30 | - | °C |
| THHS | Heat Stress Temperature rate | 0.0005 | - | Week−1 |
| SDMS | Dry Stress Threshold | 0.2 | 0.15 | Week−1 |
| HDS | Dry Stress Rate | −0.005 | −0.02 | Week−1 |
| SMWS | Wet stress threshold (1–10) | 2 | - | - |
| HWS | Wet stress rate | 0.001 | - | Week−1 |
| Variable | Percent Contribution | Permutation Importance |
|---|---|---|
| Annual_Precipitation | 26.7 | 5.6 |
| An_Mean_Tem | 19 | 79.1 |
| Min_Tem_Coldest_Month | 15.9 | 3.4 |
| Mean_Tem_Coldest_Quarter | 15.8 | 0.3 |
| Mean_Diurnal_Range | 10.5 | 6.4 |
| Max_Tem_Warmest_Month | 8.6 | 1.9 |
| Temperature_Seasonality | 2.2 | 1 |
| Precipitation_Coldest_Quarter | 1.1 | 1.6 |
| Precipitation_Wettest_Quarter | 0.1 | 0.5 |
| Precipitation_Wettest_Month | 0 | 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narouei-Khandan, H.A.; Worner, S.P.; Viljanen, S.L.H.; van Bruggen, A.H.C.; Balestra, G.M.; Jones, E. The Potential Global Climate Suitability of Kiwifruit Bacterial Canker Disease (Pseudomonas syringae pv. actinidiae (Psa)) Using Three Modelling Approaches: CLIMEX, Maxent and Multimodel Framework. Climate 2022, 10, 14. https://doi.org/10.3390/cli10020014
Narouei-Khandan HA, Worner SP, Viljanen SLH, van Bruggen AHC, Balestra GM, Jones E. The Potential Global Climate Suitability of Kiwifruit Bacterial Canker Disease (Pseudomonas syringae pv. actinidiae (Psa)) Using Three Modelling Approaches: CLIMEX, Maxent and Multimodel Framework. Climate. 2022; 10(2):14. https://doi.org/10.3390/cli10020014
Chicago/Turabian StyleNarouei-Khandan, Hossein A., Susan P. Worner, Suvi L. H. Viljanen, Ariena H. C. van Bruggen, Giorgio M. Balestra, and Eirian Jones. 2022. "The Potential Global Climate Suitability of Kiwifruit Bacterial Canker Disease (Pseudomonas syringae pv. actinidiae (Psa)) Using Three Modelling Approaches: CLIMEX, Maxent and Multimodel Framework" Climate 10, no. 2: 14. https://doi.org/10.3390/cli10020014







