Base Flipping in Open Complex Formation at Bacterial Promoters
Abstract
:1. Introduction
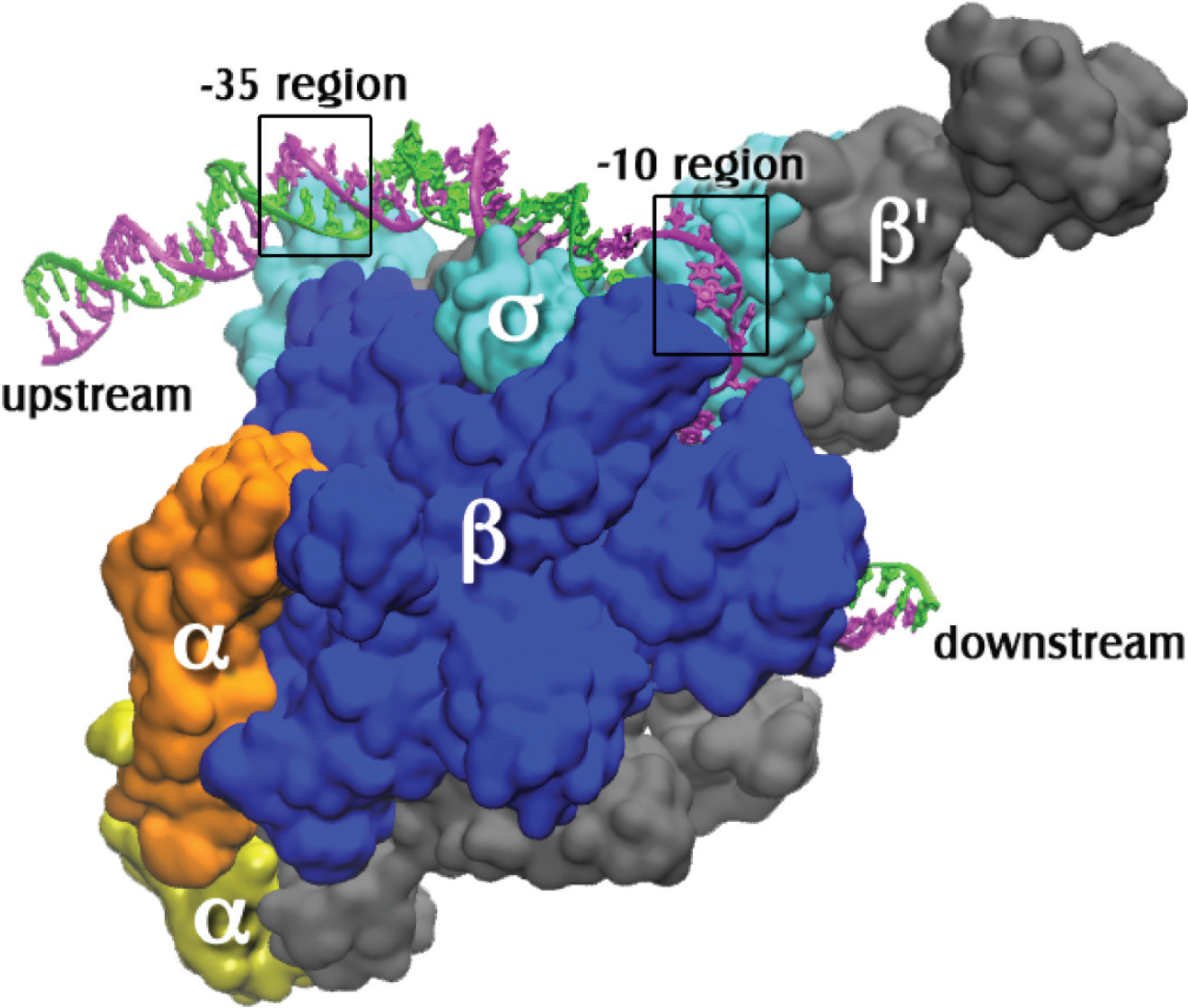

2. History
3. The Closed Complex and Other Intermediates
4. The Importance of −11A for Promoter DNA Melting
5. Flipping of Other Bases of the Non-Template Strand
6. Mechanism of Strand Opening
7. Structural and Functional Properties of Open Complexes
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- Saecker, R.M.; Record, M.T., Jr.; deHaseth, P.L. Mechanism of bacterial transcription initiation: RNA polymerase—Promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011, 412, 754–771. [Google Scholar] [CrossRef] [PubMed]
- Ebright, R.H. RNA polymerase: Structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol. 2000, 304, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Lane, W.J.; Darst, S.A. Molecular evolution of multisubunit RNA polymerases: Sequence analysis. J. Mol. Biol. 2010, 395, 671–685. [Google Scholar] [CrossRef] [PubMed]
- Lane, W.J.; Darst, S.A. Molecular evolution of multisubunit RNA polymerases: Structural analysis. J. Mol. Biol. 2010, 395, 686–704. [Google Scholar] [CrossRef] [PubMed]
- Feklistov, A.; Sharon, B.D.; Darst, S.A.; Gross, C.A. Bacterial sigma factors: A historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014, 68, 357–376. [Google Scholar] [CrossRef] [PubMed]
- Decker, K.B.; Hinton, D.M. Transcription regulation at the core: Similarities among bacterial, archaeal, and eukaryotic RNA polymerases. Annu. Rev. Microbiol. 2013, 67, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Gruber, T.M.; Gross, C.A. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003, 57, 441–466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Feng, Y.; Chatterjee, S.; Tuske, S.; Ho, M.X.; Arnold, E.; Ebright, R.H. Structural basis of transcription initiation. Science 2012, 338, 1076–1080. [Google Scholar] [CrossRef] [PubMed]
- Hudson, B.P.; Quispe, J.; Lara-Gonzalez, S.; Kim, Y.; Berman, H.M.; Arnold, E.; Ebright, R.H.; Lawson, C.L. Three-dimensional EM structure of an intact activator-dependent transcription initiation complex. Proc. Natl. Acad. Sci. USA 2009, 106, 19830–19835. [Google Scholar] [CrossRef] [PubMed]
- Hsu, L.M. Promoter clearance and escape in prokaryotes. Biochim. Biophys. Acta 2002, 1577, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Siebenlist, U.; Gilbert, W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc. Natl. Acad. Sci. USA 1980, 77, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Kirkegaard, K.; Buc, H.; Spassky, A.; Wang, J.C. Mapping of single-stranded regions in duplex DNA at the sequence level: Single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc. Natl. Acad. Sci. USA 1983, 80, 2544–2548. [Google Scholar] [CrossRef] [PubMed]
- Sasse-Dwight, S.; Gralla, J.D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J. Biol. Chem. 1989, 264, 8074–8081. [Google Scholar] [PubMed]
- Murakami, K.S.; Masuda, S.; Campbell, E.A.; Muzzin, O.; Darst, S.A. Structural basis of transcription initiation: An RNA polymerase holoenzyme-DNA complex. Science 2002, 296, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Basu, R.S.; Warner, B.A.; Molodtsov, V.; Pupov, D.; Esyunina, D.; Fernandez-Tornero, C.; Kulbachinskiy, A.; Murakami, K.S. Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J. Biol. Chem. 2014, 289, 24549–24559. [Google Scholar] [CrossRef] [PubMed]
- Chamberlin, M.J. The selectivity of transcription. Annu. Rev. Biochem. 1974, 43, 721–775. [Google Scholar] [CrossRef] [PubMed]
- Saucier, J.M.; Wang, J.C. Angular alteration of the DNA helix by E. coli RNA polymerase. Nat. New Biol. 1972, 239, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Siebenlist, U.; Simpson, R.B.; Gilbert, W. E. coli RNA polymerase interacts homologously with two different promoters. Cell 1980, 20, 269–281. [Google Scholar] [CrossRef] [PubMed]
- DeHaseth, P.L.; Zupancic, M.L.; Record, M.T., Jr. RNA polymerase-promoter interactions: The comings and goings of RNA polymerase. J. Bacteriol. 1998, 180, 3019–3025. [Google Scholar] [PubMed]
- Lonetto, M.; Gribskov, M.; Gross, C.A. The σ70 family: Sequence conservation and evolutionary relationships. J. Bacteriol. 1992, 174, 3843–3849. [Google Scholar] [PubMed]
- Campbell, E.A.; Muzzin, O.; Chlenov, M.; Sun, J.L.; Olson, C.A.; Weinman, O.; Trester-Zedlitz, M.L.; Darst, S.A. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell 2002, 9, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Juang, Y.L.; Helmann, J.D. A promoter melting region in the primary sigma factor of Bacillus subtilis. Identification of functionally important aromatic amino acids. J. Mol. Biol. 1994, 235, 1470–1488. [Google Scholar] [CrossRef] [PubMed]
- Fenton, M.S.; Lee, S.J.; Gralla, J.D. Escherichia coli promoter opening and −10 recognition: Mutational analysis of σ70. EMBO J. 2000, 19, 1130–1137. [Google Scholar] [CrossRef] [PubMed]
- Panaghie, G.; Aiyar, S.E.; Bobb, K.L.; Hayward, R.S.; deHaseth, P.L. Aromatic amino acids in region 2.3 of Escherichia coli σ70 participate collectively in the formation of an RNA polymerase-promoter open complex. J. Mol. Biol. 2000, 299, 1217–1230. [Google Scholar] [CrossRef] [PubMed]
- Tomsic, M.; Tsujikawa, L.; Panaghie, G.; Wang, Y.; Azok, J.; deHaseth, P.L. Different roles for basic and aromatic amino acids in conserved region 2 of Escherichia coli σ70 in the nucleation and maintenance of the single-stranded DNA bubble in open RNA polymerase-promoter complexes. J. Biol. Chem. 2001, 276, 31891–31896. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gralla, J.D. Promoter opening via a DNA fork junction binding activity. Proc. Natl. Acad. Sci. USA 1998, 95, 11655–11660. [Google Scholar] [CrossRef] [PubMed]
- Helmann, J.D.; deHaseth, P.L. Protein-nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry 1999, 38, 5959–5967. [Google Scholar] [CrossRef] [PubMed]
- Heyduk, E.; Kuznedelov, K.; Severinov, K.; Heyduk, T. A consensus adenine at position −11 of the nontemplate strand of bacterial promoter is important for nucleation of promoter melting. J. Biol. Chem. 2006, 281, 12362–12369. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.M.; Lee, H.J.; Roy, S.; Adhya, S. A “master” in base unpairing during isomerization of a promoter upon RNA polymerase binding. Proc. Natl. Acad. Sci. USA 2001, 98, 14849–14852. [Google Scholar] [CrossRef] [PubMed]
- Tsujikawa, L.; Strainic, M.G.; Watrob, H.; Barkley, M.D.; deHaseth, P.L. RNA polymerase alters the mobility of an A-residue crucial to polymerase-induced melting of promoter DNA. Biochemistry 2002, 41, 15334–15341. [Google Scholar] [CrossRef] [PubMed]
- Matlock, D.L.; Heyduk, T. Sequence determinants for the recognition of the fork junction DNA containing the −10 region of promoter DNA by E. coli RNA polymerase. Biochemistry 2000, 39, 12274–12283. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Lim, H.M.; Adhya, S. An unsubstituted C2 hydrogen of adenine is critical and sufficient at the −11 position of a promoter to signal base pair deformation. J. Biol. Chem. 2004, 279, 16899–16902. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Lim, H.M.; Liu, M.; Adhya, S. Asynchronous basepair openings in transcription initiation: CRP enhances the rate-limiting step. EMBO J. 2004, 23, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.J.; Bjornson, K.P.; Sowers, L.C.; deHaseth, P.L. Spectroscopic determination of open complex formation at promoters for Escherichia coli RNA polymerase. Biochemistry 1997, 36, 8005–8012. [Google Scholar] [CrossRef]
- Fedoriw, A.M.; Liu, H.; Anderson, V.E.; deHaseth, P.L. Equilibrium and kinetic parameters of the sequence-specific interaction of Escherichia coli RNA polymerase with nontemplate strand oligodeoxyribonucleotides. Biochemistry 1998, 37, 11971–11979. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, L.A.; Gries, T.J.; Saecker, R.M.; Record, M.T., Jr.; Harris, M.E.; deHaseth, P.L. Evidence for a tyrosine-adenine stacking interaction and for a short-lived open intermediate subsequent to initial binding of Escherichia coli RNA polymerase to promoter DNA. J. Mol. Biol. 2009, 385, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Liebert, K.; Hermann, A.; Schlickenrieder, M.; Jeltsch, A. Stopped-flow and mutational analysis of base flipping by the Escherichia coli Dam DNA-(adenine-N6)-methyltransferase. J. Mol. Biol. 2004, 341, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Tamulaitis, G.; Zaremba, M.; Szczepanowski, R.H.; Bochtler, M.; Siksnys, V. Nucleotide flipping by restriction enzymes analyzed by 2-aminopurine steady-state fluorescence. Nucleic Acids Res. 2007, 35, 4792–4799. [Google Scholar] [CrossRef] [PubMed]
- Shultzaberger, R.K.; Chen, Z.; Lewis, K.A.; Schneider, T.D. Anatomy of Escherichia coli σ70 promoters. Nucleic Acids Res. 2007, 35, 771–788. [Google Scholar] [CrossRef] [PubMed]
- Feklistov, A.; Darst, S.A. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell 2011, 147, 1257–1269. [Google Scholar] [CrossRef] [PubMed]
- Campagne, S.; Marsh, M.E.; Capitani, G.; Vorholt, J.A.; Allain, F.H. Structural basis for −10 promoter element melting by environmentally induced sigma factors. Nat. Struct. Mol. Biol. 2014, 21, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.W.; Roberts, J.W. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell 1996, 86, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Fenton, M.S.; Gralla, J.D. Function of the bacterial TATAAT −10 element as single-stranded DNA during RNA polymerase isomerization. Proc. Natl. Acad. Sci. USA 2001, 98, 9020–9025. [Google Scholar] [CrossRef] [PubMed]
- Mekler, V.; Severinov, K. Cooperativity and interaction energy threshold effects in recognition of the −10 promoter element by bacterial RNA polymerase. Nucleic Acids Res. 2013, 41, 7276–7285. [Google Scholar] [CrossRef] [PubMed]
- Heyduk, E.; Heyduk, T. Next generation sequencing-based parallel analysis of melting kinetics of 4096 variants of a bacterial promoter. Biochemistry 2014, 53, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Darst, S.A.; Thirumalai, D. Promoter melting triggered by bacterial RNA polymerase occurs in three steps. Proc. Natl. Acad. Sci. USA 2010, 107, 12523–12528. [Google Scholar] [CrossRef] [PubMed]
- Severinov, K.; Darst, S.A. A mutant RNA polymerase that forms unusual open promoter complexes. Proc. Natl. Acad. Sci. USA 1997, 94, 13481–13486. [Google Scholar] [CrossRef] [PubMed]
- Rogozina, A.; Zaychikov, E.; Buckle, M.; Heumann, H.; Sclavi, B. DNA melting by RNA polymerase at the T7A1 promoter precedes the rate-limiting step at 37 degrees C and results in the accumulation of an off-pathway intermediate. Nucleic Acids Res. 2009, 37, 5390–5404. [Google Scholar] [CrossRef] [PubMed]
- Zaychikov, E.; Denissova, L.; Meier, T.; Gotte, M.; Heumann, H. Influence of Mg2+ and temperature on formation of the transcription bubble. J. Biol. Chem. 1997, 272, 2259–2267. [Google Scholar] [CrossRef] [PubMed]
- Mekler, V.; Pavlova, O.; Severinov, K. The interaction of Escherichia coli RNA polymerase σ70 subunit with promoter elements in the context of free σ70, RNA polymerase holoenzyme, and the b'-σ70 complex. J. Biol. Chem. 2011, 286, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Gries, T.J.; Kontur, W.S.; Capp, M.W.; Saecker, R.M.; Record, M.T., Jr. One-step DNA melting in the RNA polymerase cleft opens the initiation bubble to form an unstable open complex. Proc. Natl. Acad. Sci. USA 2010, 107, 10418–10423. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, L.A.; Karpen, M.E.; deHaseth, P.L. Threonine 429 of Escherichia coli σ70 is a key participant in promoter DNA melting by RNA polymerase. J. Mol. Biol. 2008, 376, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Saecker, R.M.; Tsodikov, O.V.; McQuade, K.L.; Schlax, P.E., Jr.; Capp, M.W.; Record, M.T., Jr. Kinetic studies and structural models of the association of E. coli σ70 RNA polymerase with the λPR promoter: Large scale conformational changes in forming the kinetically significant intermediates. J. Mol. Biol. 2002, 319, 649–671. [Google Scholar] [CrossRef] [PubMed]
- Robb, N.C.; Cordes, T.; Hwang, L.C.; Gryte, K.; Duchi, D.; Craggs, T.D.; Santoso, Y.; Weiss, S.; Ebright, R.H.; Kapanidis, A.N. The transcription bubble of the RNA polymerase-promoter open complex exhibits conformational heterogeneity and millisecond-scale dynamics: Implications for transcription start-site selection. J. Mol. Biol. 2013, 425, 875–885. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karpen, M.E.; DeHaseth, P.L. Base Flipping in Open Complex Formation at Bacterial Promoters. Biomolecules 2015, 5, 668-678. https://doi.org/10.3390/biom5020668
Karpen ME, DeHaseth PL. Base Flipping in Open Complex Formation at Bacterial Promoters. Biomolecules. 2015; 5(2):668-678. https://doi.org/10.3390/biom5020668
Chicago/Turabian StyleKarpen, Mary E., and Pieter L. DeHaseth. 2015. "Base Flipping in Open Complex Formation at Bacterial Promoters" Biomolecules 5, no. 2: 668-678. https://doi.org/10.3390/biom5020668





