Insights into the Roles of Epigenetic Modifications in Ferroptosis
Abstract
:Simple Summary
Abstract
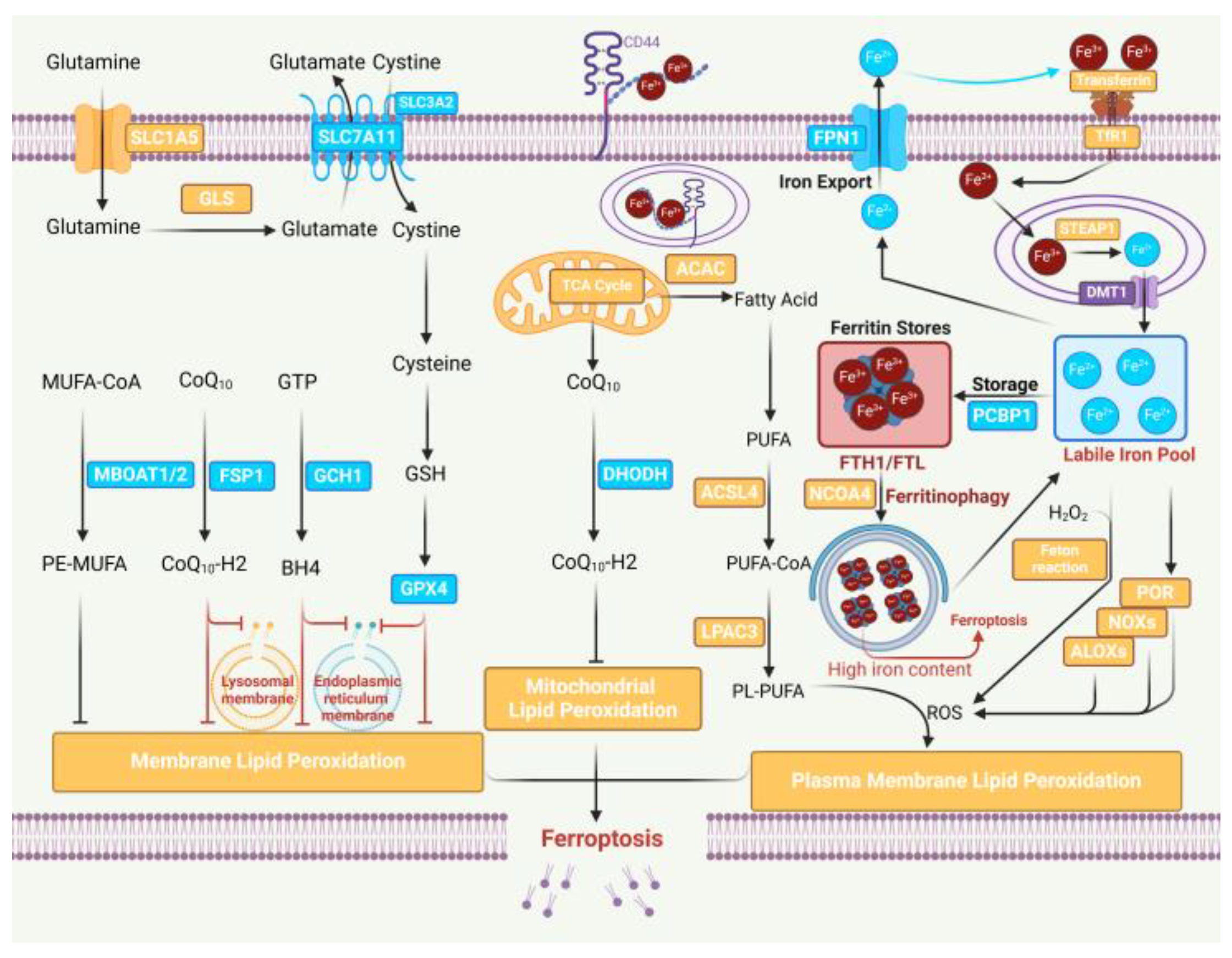
1. Introduction
2. Epigenetic Modification
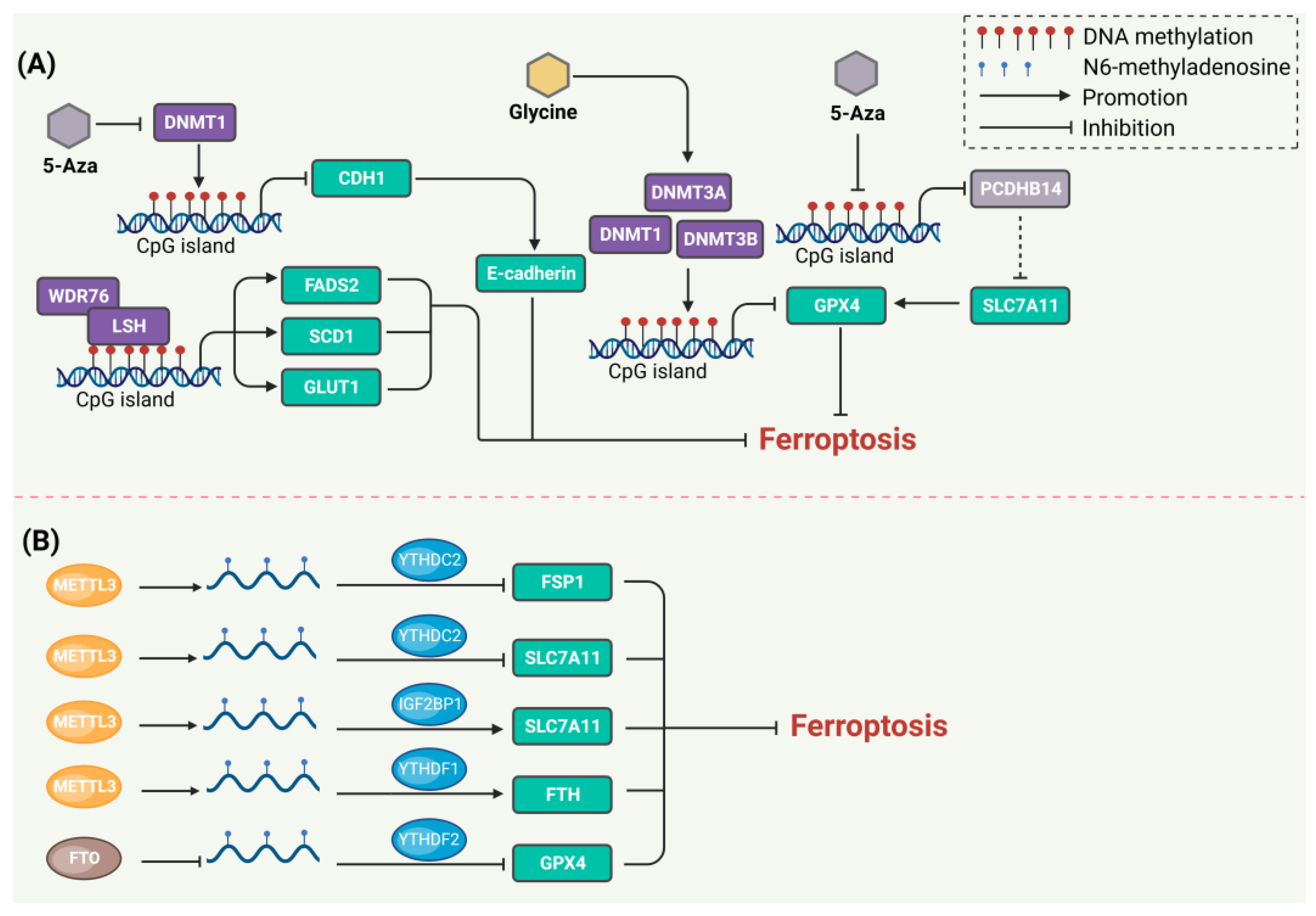
3. DNA Methylation Regulates Ferroptosis
4. RNA Methylation Regulates Ferroptosis
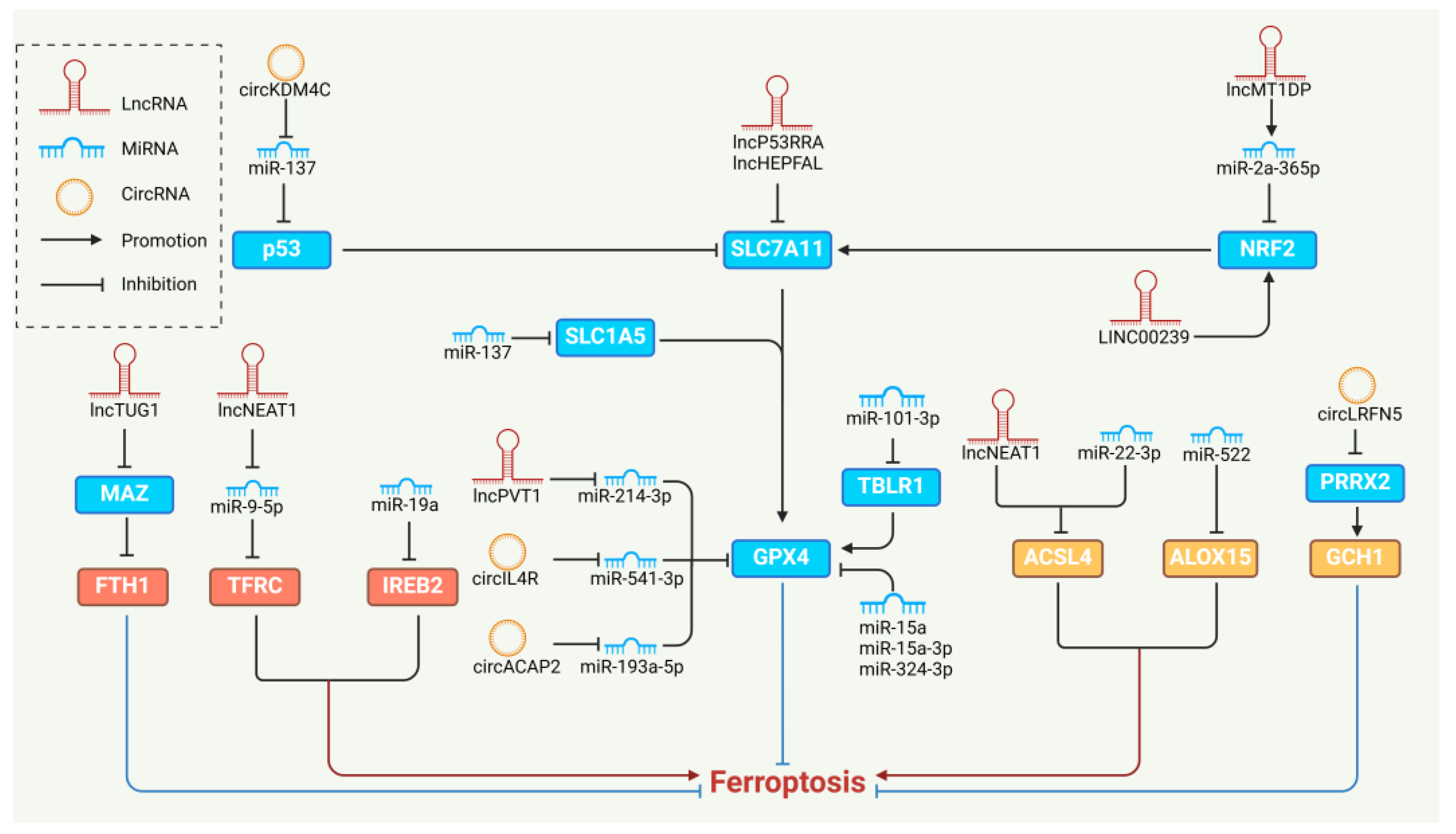
5. NcRNAs Regulating Ferroptosis
5.1. MiRNAs and Ferroptosis
5.2. LncRNAs and Ferroptosis
5.3. CircRNAs and Ferroptosis
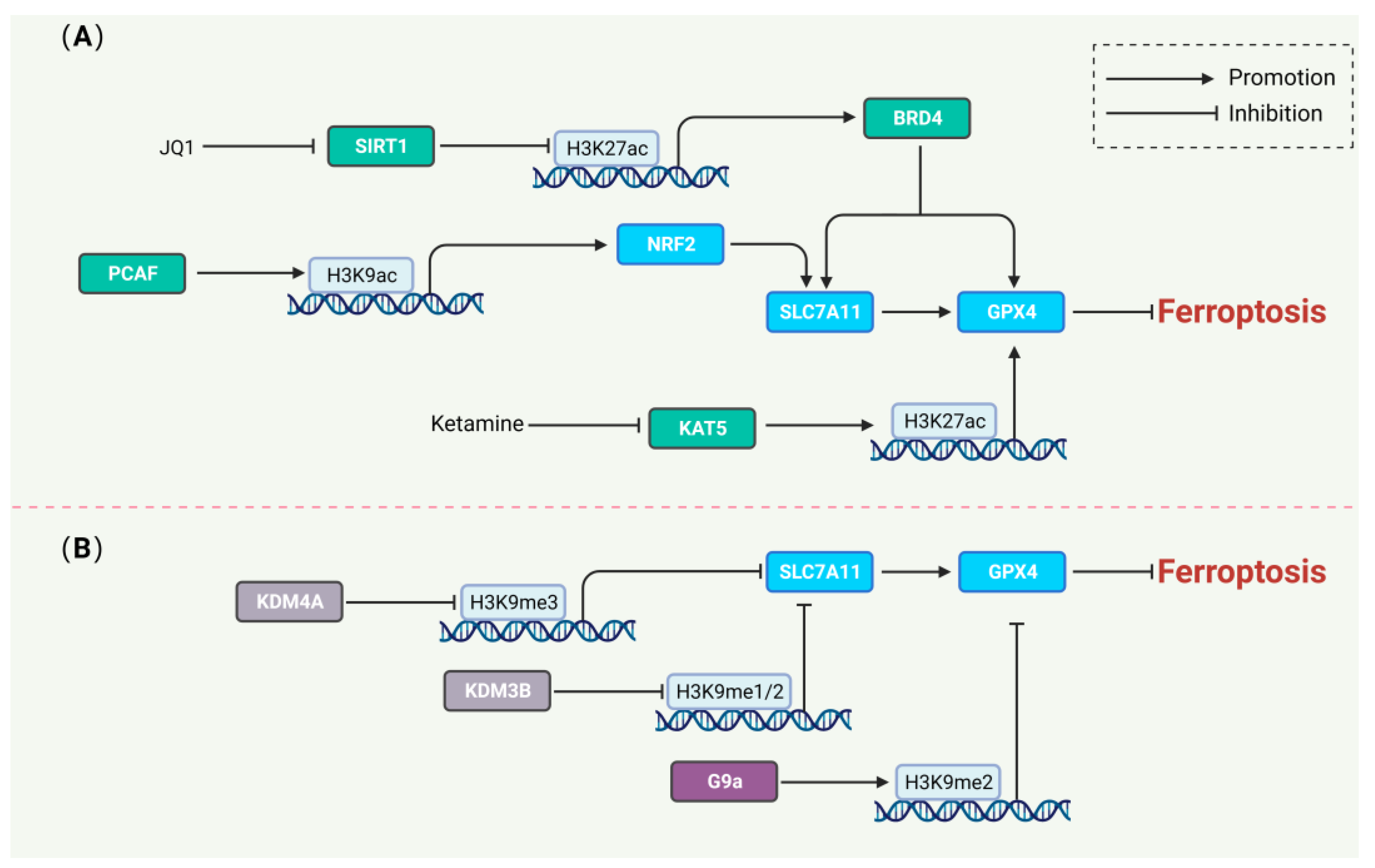
6. Histone Modifications Regulating Ferroptosis
6.1. Histone Acetylation and Ferroptosis
6.2. Histone Methylation and Ferroptosis
7. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cao, F.; Yin, H.L.; Huang, Z.J.; Lin, Z.T.; Mao, N.; Sun, B.; Wang, G. Ferroptosis: Past, present and future. Cell Death Dis. 2020, 11, 88. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Jiang, L.; Kon, N.; Li, T.; Wang, S.J.; Su, T.; Hibshoosh, H.; Baer, R.; Gu, W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015, 520, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Winter, G.E.; Musavi, L.S.; Lee, E.D.; Snijder, B.; Rebsamen, M.; Superti-Furga, G.; Stockwell, B.R. Human Haploid Cell Genetics Reveals Roles for Lipid Metabolism Genes in Nonapoptotic Cell Death. ACS Chem. Biol. 2015, 10, 1604–1609. [Google Scholar] [CrossRef]
- Nakamura, T.; Hipp, C.; Santos Dias Mourão, A.; Borggräfe, J.; Aldrovandi, M.; Henkelmann, B.; Wanninger, J.; Mishima, E.; Lytton, E.; Emler, D.; et al. Phase separation of FSP1 promotes ferroptosis. Nature 2023, 619, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Mishima, E.; Ito, J.; Wu, Z.; Nakamura, T.; Wahida, A.; Doll, S.; Tonnus, W.; Nepachalovich, P.; Eggenhofer, E.; Aldrovandi, M.; et al. A non-canonical vitamin K cycle is a potent ferroptosis suppressor. Nature 2022, 608, 778–783. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Zhang, Y.; Lou, H.; Ou, Z.; Liu, J.; Duan, W.; Wang, H.; Ge, Y.; Min, J.; Wang, F.; et al. GPX4 and vitamin E cooperatively protect hematopoietic stem and progenitor cells from lipid peroxidation and ferroptosis. Cell Death Dis. 2021, 12, 706. [Google Scholar] [CrossRef]
- Bersuker, K.; Hendricks, J.M.; Li, Z.; Magtanong, L.; Ford, B.; Tang, P.H.; Roberts, M.A.; Tong, B.; Maimone, T.J.; Zoncu, R.; et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 2019, 575, 688–692. [Google Scholar] [CrossRef]
- Kraft, V.A.N.; Bezjian, C.T.; Pfeiffer, S.; Ringelstetter, L.; Müller, C.; Zandkarimi, F.; Merl-Pham, J.; Bao, X.; Anastasov, N.; Kössl, J.; et al. GTP Cyclohydrolase 1/Tetrahydrobiopterin Counteract Ferroptosis through Lipid Remodeling. ACS Cent. Sci. 2020, 6, 41–53. [Google Scholar] [CrossRef]
- Mao, C.; Liu, X.; Zhang, Y.; Lei, G.; Yan, Y.; Lee, H.; Koppula, P.; Wu, S.; Zhuang, L.; Fang, B.; et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature 2021, 593, 586–590. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, J.; Wu, S.; Fleishman, J.S.; Li, Y.; Xu, Y.; Zou, W.; Wang, J.; Feng, Y.; Chen, J.; et al. Targeting epigenetic and posttranslational modifications regulating ferroptosis for the treatment of diseases. Signal Transduct. Target. Ther. 2023, 8. [Google Scholar] [CrossRef]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef]
- Dangol, S.; Chen, Y.; Hwang, B.K.; Jwa, N.S. Iron- and Reactive Oxygen Species-Dependent Ferroptotic Cell Death in Rice-Magnaporthe oryzae Interactions. Plant Cell 2019, 31, 189–209. [Google Scholar] [CrossRef] [PubMed]
- Canizal-García, M.; Olmos-Orizaba, B.E.; Moreno-Jiménez, M.; Calderón-Cortés, E.; Saavedra-Molina, A.; Cortés-Rojo, C. Glutathione peroxidase 2 (Gpx2) preserves mitochondrial function and decreases ROS levels in chronologically aged yeast. Free Radic. Res. 2021, 55, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Kagan, V.E.; Bayir, H.; Pagnussat, G.C.; Head, B.; Traber, M.G.; Stockwell, B.R. Regulation of lipid peroxidation and ferroptosis in diverse species. Genes. Dev. 2018, 32, 602–619. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.; Lamberson, C.R.; Xu, L.; To, R.; Tsui, H.S.; Shmanai, V.V.; Bekish, A.V.; Awad, A.M.; Marbois, B.N.; Cantor, C.R.; et al. Small amounts of isotope-reinforced polyunsaturated fatty acids suppress lipid autoxidation. Free Radic. Biol. Med. 2012, 53, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Nuszkiewicz, J.; Sutkowy, P.; Wróblewski, M.; Pawłowska, M.; Wesołowski, R.; Wróblewska, J.; Woźniak, A. Links between Vitamin K, Ferroptosis and SARS-CoV-2 Infection. Antioxidants 2023, 12, 733. [Google Scholar] [CrossRef] [PubMed]
- Bayır, H.; Dixon, S.J.; Tyurina, Y.Y.; Kellum, J.A.; Kagan, V.E. Ferroptotic mechanisms and therapeutic targeting of iron metabolism and lipid peroxidation in the kidney. Nat. Rev. Nephrol. 2023, 19, 315–336. [Google Scholar] [CrossRef] [PubMed]
- Forcina, G.C.; Dixon, S.J. GPX4 at the Crossroads of Lipid Homeostasis and Ferroptosis. Proteomics 2019, 19, e1800311. [Google Scholar] [CrossRef] [PubMed]
- Lei, G.; Zhuang, L.; Gan, B. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer 2022, 22, 381–396. [Google Scholar] [CrossRef]
- Gao, M.; Monian, P.; Pan, Q.; Zhang, W.; Xiang, J.; Jiang, X. Ferroptosis is an autophagic cell death process. Cell Res. 2016, 26, 1021–1032. [Google Scholar] [CrossRef]
- Giorgio, M.; Trinei, M.; Migliaccio, E.; Pelicci, P.G. Hydrogen peroxide: A metabolic by-product or a common mediator of ageing signals? Nat. Rev. Mol. Cell Biol. 2007, 8, 722–728. [Google Scholar] [CrossRef]
- Tang, D.; Chen, X.; Kang, R.; Kroemer, G. Ferroptosis: Molecular mechanisms and health implications. Cell Res. 2021, 31, 107–125. [Google Scholar] [CrossRef]
- Lorent, J.H.; Levental, K.R.; Ganesan, L.; Rivera-Longsworth, G.; Sezgin, E.; Doktorova, M.; Lyman, E.; Levental, I. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat. Chem. Biol. 2020, 16, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Minikes, A.M.; Jiang, X. Ferroptosis at the intersection of lipid metabolism and cellular signaling. Mol. Cell 2022, 82, 2215–2227. [Google Scholar] [CrossRef] [PubMed]
- Aldrovandi, M.; Fedorova, M.; Conrad, M. Juggling with lipids, a game of Russian roulette. Trends Endocrinol. Metab. TEM 2021, 32, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Doll, S.; Proneth, B.; Tyurina, Y.Y.; Panzilius, E.; Kobayashi, S.; Ingold, I.; Irmler, M.; Beckers, J.; Aichler, M.; Walch, A.; et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017, 13, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Kagan, V.E.; Mao, G.; Qu, F.; Angeli, J.P.; Doll, S.; Croix, C.S.; Dar, H.H.; Liu, B.; Tyurin, V.A.; Ritov, V.B.; et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017, 13, 81–90. [Google Scholar] [CrossRef]
- Beharier, O.; Tyurin, V.A.; Goff, J.P.; Guerrero-Santoro, J.; Kajiwara, K.; Chu, T.; Tyurina, Y.Y.; St Croix, C.M.; Wallace, C.T.; Parry, S.; et al. PLA2G6 guards placental trophoblasts against ferroptotic injury. Proc. Natl. Acad. Sci. USA 2020, 117, 27319–27328. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.Y.; Tyurin, V.A.; Mikulska-Ruminska, K.; Shrivastava, I.H.; Anthonymuthu, T.S.; Zhai, Y.J.; Pan, M.H.; Gong, H.B.; Lu, D.H.; Sun, J.; et al. Phospholipase iPLA(2)β averts ferroptosis by eliminating a redox lipid death signal. Nat. Chem. Biol. 2021, 17, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Kang, R.; Kroemer, G.; Tang, D. Broadening horizons: The role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 2021, 18, 280–296. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ran, Q.; Duan, Q.; Jin, J.; Wang, Y.; Yu, L.; Wang, C.; Zhu, Z.; Chen, X.; Weng, L.; et al. 7-Dehydrocholesterol dictates ferroptosis sensitivity. Nature 2024, 626, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Freitas, F.P.; Alborzinia, H.; Dos Santos, A.F.; Nepachalovich, P.; Pedrera, L.; Zilka, O.; Inague, A.; Klein, C.; Aroua, N.; Kaushal, K.; et al. 7-Dehydrocholesterol is an endogenous suppressor of ferroptosis. Nature 2024, 626, 401–410. [Google Scholar] [CrossRef]
- Ji, J.; Jing, A.; Geng, T.; Ma, X.; Liu, W.; Liu, B. Editorial: Protein modifications in epigenetic dysfunctional diseases: Mechanisms and potential therapeutic strategies. Front. Cell Dev. Biol. 2023, 11, 1216637. [Google Scholar] [CrossRef]
- Esteve-Puig, R.; Bueno-Costa, A.; Esteller, M. Writers, readers and erasers of RNA modifications in cancer. Cancer Lett. 2020, 474, 127–137. [Google Scholar] [CrossRef]
- Wang, D.; Xie, D.; Zhang, J.; Cai, B.; Yang, B.; Zhou, L.; Huang, X. Comprehensive analysis of the coding and non-coding RNA transcriptome expression profiles of hippocampus tissue in tx-J animal model of Wilson’s disease. Sci. Rep. 2023, 13, 9252. [Google Scholar] [CrossRef]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef]
- Frazzi, R.; Auffray, C.; Ferrari, A.; Filippini, P.; Rutella, S.; Cesario, A. Integrative systems medicine approaches to identify molecular targets in lymphoid malignancies. J. Transl. Med. 2016, 14, 252. [Google Scholar] [CrossRef]
- Kutay, H.; Klepper, C.; Wang, B.; Hsu, S.H.; Datta, J.; Yu, L.; Zhang, X.; Majumder, S.; Motiwala, T.; Khan, N.; et al. Reduced susceptibility of DNA methyltransferase 1 hypomorphic (Dnmt1N/+) mice to hepatic steatosis upon feeding liquid alcohol diet. PloS ONE 2012, 7, e41949. [Google Scholar] [CrossRef] [PubMed]
- Hossain, K.; Suzuki, T.; Hasibuzzaman, M.M.; Islam, M.S.; Rahman, A.; Paul, S.K.; Tanu, T.; Hossain, S.; Saud, Z.A.; Rahman, M.; et al. Chronic exposure to arsenic, LINE-1 hypomethylation, and blood pressure: A cross-sectional study in Bangladesh. Environ. Health Glob. Access Sci. Source 2017, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Buysschaert, I.; Schmidt, T.; Roncal, C.; Carmeliet, P.; Lambrechts, D. Genetics, epigenetics and pharmaco-(epi)genomics in angiogenesis. J. Cell. Mol. Med. 2008, 12, 2533–2551. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Zhang, J.; Zhang, C.; Shi, Y.; Zhao, B.; Jiao, A.; Chen, B. Using Illumina Infinium HumanMethylation 450K BeadChip to explore genome-wide DNA methylation profiles in a human hepatocellular carcinoma cell line. Mol. Med. Rep. 2018, 18, 4446–4456. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Yu, H.; Huang, X.; Shen, J.; Xiao, G.; Chen, L.; Wang, H.; Xing, L.; Chen, D. Current understanding of osteoarthritis pathogenesis and relevant new approaches. Bone Res. 2022, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Song, S.; Liu, J.; Zhang, L.; Guo, X.; Lu, J.; Li, L.; Yang, C.; Fu, Q.; Zeng, B. Epigenetic regulation of programmed cell death in hypoxia-induced pulmonary arterial hypertension. Front. Immunol. 2023, 14, 1206452. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Fan, B.Y.; Sun, T.; Wang, X.X.; Li, J.J.; Zhang, J.P.; Gu, G.J.; Shen, W.Y.; Liu, D.R.; Wei, Z.J.; et al. Bioinformatics analysis of ferroptosis in spinal cord injury. Neural Regen. Res. 2023, 18, 626–633. [Google Scholar] [CrossRef] [PubMed]
- Xi, C.; Pang, J.; Zhi, W.; Chang, C.S.; Siddaramappa, U.; Shi, H.; Horuzsko, A.; Pace, B.S.; Zhu, X. Nrf2 sensitizes ferroptosis through l-2-hydroxyglutarate-mediated chromatin modifications in sickle cell disease. Blood 2023, 142, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Skvortsova, K.; Stirzaker, C.; Taberlay, P. The DNA methylation landscape in cancer. Essays Biochem. 2019, 63, 797–811. [Google Scholar] [CrossRef]
- Park, H.; Cho, B.; Kim, J. Rad50 mediates DNA demethylation to establish pluripotent reprogramming. Exp. Mol. Med. 2020, 52, 1116–1127. [Google Scholar] [CrossRef]
- Yang, M.; Luo, H.; Yi, X.; Wei, X.; Jiang, D.S. The epigenetic regulatory mechanisms of ferroptosis and its implications for biological processes and diseases. MedComm 2023, 4, e267. [Google Scholar] [CrossRef]
- Ling, H.; Li, M.; Yang, C.; Sun, S.; Zhang, W.; Zhao, L.; Xu, N.; Zhang, J.; Shen, Y.; Zhang, X.; et al. Glycine increased ferroptosis via SAM-mediated GPX4 promoter methylation in rheumatoid arthritis. Rheumatology 2022, 61, 4521–4534. [Google Scholar] [CrossRef]
- Zeng, Z.; Li, T.; Liu, X.; Ma, Y.; Luo, L.; Wang, Z.; Zhao, Z.; Li, H.; He, X.; Zeng, H.; et al. DNA dioxygenases TET2 deficiency promotes cigarette smoke induced chronic obstructive pulmonary disease by inducing ferroptosis of lung epithelial cell. Redox Biol. 2023, 67, 102916. [Google Scholar] [CrossRef]
- Pontel, L.B.; Bueno-Costa, A.; Morellato, A.E.; Carvalho Santos, J.; Roué, G.; Esteller, M. Acute lymphoblastic leukemia necessitates GSH-dependent ferroptosis defenses to overcome FSP1-epigenetic silencing. Redox Biol. 2022, 55, 102408. [Google Scholar] [CrossRef]
- Lee, J.; You, J.H.; Kim, M.S.; Roh, J.L. Epigenetic reprogramming of epithelial-mesenchymal transition promotes ferroptosis of head and neck cancer. Redox Biol. 2020, 37, 101697. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ouyang, L.; Mao, C.; Chen, Y.; Li, T.; Liu, N.; Wang, Z.; Lai, W.; Zhou, Y.; Cao, Y.; et al. PCDHB14 promotes ferroptosis and is a novel tumor suppressor in hepatocellular carcinoma. Oncogene 2022, 41, 3570–3583. [Google Scholar] [CrossRef] [PubMed]
- Spruijt, C.G.; Gnerlich, F.; Smits, A.H.; Pfaffeneder, T.; Jansen, P.W.; Bauer, C.; Munzel, M.; Wagner, M.; Muller, M.; Khan, F.; et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 2013, 152, 1146–1159. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Mao, C.; Yang, R.; Yan, B.; Shi, Y.; Liu, X.; Lai, W.; Liu, Y.; Wang, X.; Xiao, D.; et al. EGLN1/c-Myc Induced Lymphoid-Specific Helicase Inhibits Ferroptosis through Lipid Metabolic Gene Expression Changes. Theranostics 2017, 7, 3293–3305. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Li, Z.; Xie, J.; Wu, Z.; Huang, Y.; Xie, H.; Xu, H.; Huang, Y.; Zhou, H. Methylation Drives SLC2A1 Transcription and Ferroptosis Process Decreasing Autophagy Pressure in Colon Cancer. J. Oncol. 2022, 2022, 9077424. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Li, H.; Zhan, H.; Liu, Y.; Li, X.; Huang, Y.; Wang, L.; Zhang, F.; Li, Y. Alterations of m6A RNA methylation regulators contribute to autophagy and immune infiltration in primary Sjögren’s syndrome. Front. Immunol. 2022, 13, 949206. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Huai, Q.L.; Sun, S.J.; Guo, L.; Xue, X.M.; Song, P.; Ying, J.M.; Gao, Y.B.; Gao, S.G.; He, J. Development and validation of m6A RNA methylation regulators-based signature in lung adenocarcinoma. Chin. Med. J. 2021, 134, 2128–2130. [Google Scholar] [CrossRef] [PubMed]
- He, P.C.; Wei, J.; Dou, X.; Harada, B.T.; Zhang, Z.; Ge, R.; Liu, C.; Zhang, L.S.; Yu, X.; Wang, S.; et al. Exon architecture controls mRNA m(6)A suppression and gene expression. Science 2023, 379, 677–682. [Google Scholar] [CrossRef]
- Shen, M.; Li, Y.; Wang, Y.; Shao, J.; Zhang, F.; Yin, G.; Chen, A.; Zhang, Z.; Zheng, S. N(6)-methyladenosine modification regulates ferroptosis through autophagy signaling pathway in hepatic stellate cells. Redox Biol. 2021, 47, 102151. [Google Scholar] [CrossRef]
- Zhuang, S.; Ma, Y.; Zeng, Y.; Lu, C.; Yang, F.; Jiang, N.; Ge, J.; Ju, H.; Zhong, C.; Wang, J.; et al. METTL14 promotes doxorubicin-induced cardiomyocyte ferroptosis by regulating the KCNQ1OT1-miR-7-5p-TFRC axis. Cell Biol. Toxicol. 2021. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, X.; Yu, K.; Xu, X.; Chen, T.; Shi, Y.; Wang, Y.; Qiu, S.; Guo, S.; Cui, J.; et al. Targeting SLC3A2 subunit of system X(C)(-) is essential for m(6)A reader YTHDC2 to be an endogenous ferroptosis inducer in lung adenocarcinoma. Free Radic. Biol. Med. 2021, 168, 25–43. [Google Scholar] [CrossRef]
- Ma, L.; Chen, T.; Zhang, X.; Miao, Y.; Tian, X.; Yu, K.; Xu, X.; Niu, Y.; Guo, S.; Zhang, C.; et al. The m(6)A reader YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing SLC7A11-dependent antioxidant function. Redox Biol. 2021, 38, 101801. [Google Scholar] [CrossRef]
- Sun, S.; Gao, T.; Pang, B.; Su, X.; Guo, C.; Zhang, R.; Pang, Q. RNA binding protein NKAP protects glioblastoma cells from ferroptosis by promoting SLC7A11 mRNA splicing in an m(6)A-dependent manner. Cell Death Dis. 2022, 13, 73. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Yu, H.; Yuan, Z.; Ye, T.; Hu, B. ALKBH5 decreases SLC7A11 expression by erasing m6A modification and promotes the ferroptosis of colorectal cancer cells. Clin. Transl. Oncol. 2023, 25, 2265–2276. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; He, J.; Sun, G.; Huang, N.; Bian, Z.; Xu, C.; Zhang, Y.; Cui, Z.; Xu, W.; Sun, F.; et al. The N6-methyladenosine modification enhances ferroptosis resistance through inhibiting SLC7A11 mRNA deadenylation in hepatoblastoma. Clin. Transl. Med. 2022, 12, e778. [Google Scholar] [CrossRef]
- Li, N.; Yi, X.; He, Y.; Huo, B.; Chen, Y.; Zhang, Z.; Wang, Q.; Li, Y.; Zhong, X.; Li, R.; et al. Targeting Ferroptosis as a Novel Approach to Alleviate Aortic Dissection. Int. J. Biol. Sci. 2022, 18, 4118–4134. [Google Scholar] [CrossRef]
- Zhang, G.; Mi, W.; Wang, C.; Li, J.; Zhang, Y.; Liu, N.; Jiang, M.; Jia, G.; Wang, F.; Yang, G.; et al. Targeting AKT induced Ferroptosis through FTO/YTHDF2-dependent GPX4 m6A methylation up-regulating and degradating in colorectal cancer. Cell Death Discov. 2023, 9, 457. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, X.; Li, L.; Gao, Z.; Su, X.; Ji, M.; Liu, J. N(6)-methyladenosine METTL3 promotes cervical cancer tumorigenesis and Warburg effect through YTHDF1/HK2 modification. Cell Death Dis. 2020, 11, 911. [Google Scholar] [CrossRef]
- He, Y.; Wang, W.; Xu, X.; Yang, B.; Yu, X.; Wu, Y.; Wang, J. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through mediating Bcl2 stability via Ythdf1-mediated m(6)A modification. Bone 2022, 154, 116182. [Google Scholar] [CrossRef]
- Xu, Y.; Lv, D.; Yan, C.; Su, H.; Zhang, X.; Shi, Y.; Ying, K. METTL3 promotes lung adenocarcinoma tumor growth and inhibits ferroptosis by stabilizing SLC7A11 m(6)A modification. Cancer Cell Int. 2022, 22, 11. [Google Scholar] [CrossRef]
- Diao, H.; Tan, H.; Hu, Y.; Wang, R.; Cai, P.; Huang, B.; Shao, X.; Yan, M.; Yin, C.; Zhang, Y. The m(6)A Reader YTHDF1 Promotes Lung Carcinoma Progression via Regulating Ferritin Mediate Ferroptosis in an m(6)A-Dependent Manner. Pharmaceuticals 2023, 16, 185. [Google Scholar] [CrossRef]
- Zhu, R.; Gao, C.; Feng, Q.; Guan, H.; Wu, J.; Samant, H.; Yang, F.; Wang, X. Ferroptosis-related genes with post-transcriptional regulation mechanisms in hepatocellular carcinoma determined by bioinformatics and experimental validation. Ann. Transl. Med. 2022, 10, 1390. [Google Scholar] [CrossRef]
- Liu, J.; Ren, Z.; Yang, L.; Zhu, L.; Li, Y.; Bie, C.; Liu, H.; Ji, Y.; Chen, D.; Zhu, M.; et al. The NSUN5-FTH1/FTL pathway mediates ferroptosis in bone marrow-derived mesenchymal stem cells. Cell Death Discov. 2022, 8, 99. [Google Scholar] [CrossRef]
- Qu, Z.; Adelson, D.L. Evolutionary conservation and functional roles of ncRNA. Front. Genet. 2012, 3, 205. [Google Scholar] [CrossRef]
- Kim, T.; Reitmair, A. Non-Coding RNAs: Functional Aspects and Diagnostic Utility in Oncology. Int. J. Mol. Sci. 2013, 14, 4934–4968. [Google Scholar] [CrossRef]
- Ling, H.; Vincent, K.; Pichler, M.; Fodde, R.; Berindan-Neagoe, I.; Slack, F.J.; Calin, G.A. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 2015, 34, 5003–5011. [Google Scholar] [CrossRef]
- Almeida, M.I.; Reis, R.M.; Calin, G.A. MicroRNA history: Discovery, recent applications, and next frontiers. Mutat. Res. 2011, 717, 1–8. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, D. The Pattern of microRNA Binding Site Distribution. Genes 2017, 8, 296. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, D.; Chen, X.; Li, J.; Li, L.; Bian, Z.; Sun, F.; Lu, J.; Yin, Y.; Cai, X.; et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell 2010, 39, 133–144. [Google Scholar] [CrossRef]
- Yin, Y.; Cai, X.; Chen, X.; Liang, H.; Zhang, Y.; Li, J.; Wang, Z.; Chen, X.; Zhang, W.; Yokoyama, S.; et al. Tumor-secreted miR-214 induces regulatory T cells: A major link between immune evasion and tumor growth. Cell Res. 2014, 24, 1164–1180. [Google Scholar] [CrossRef]
- Santamaria, X.; Taylor, H. MicroRNA and gynecological reproductive diseases. Fertil. Steril. 2014, 101, 1545–1551. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Lee, W. Role of MiR-325-3p in the Regulation of CFL2 and Myogenic Differentiation of C2C12 Myoblasts. Cells 2021, 10, 2725. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Xing, J.; Liu, H.; Jiang, W.; Wang, L. LncRNA-Encoded Peptide: Functions and Predicting Methods. Front. Oncol. 2020, 10, 622294. [Google Scholar] [CrossRef] [PubMed]
- Jonas, K.; Calin, G.A.; Pichler, M. RNA-Binding Proteins as Important Regulators of Long Non-Coding RNAs in Cancer. Int. J. Mol. Sci. 2020, 21, 2969. [Google Scholar] [CrossRef] [PubMed]
- Bartonicek, N.; Maag, J.L.; Dinger, M.E. Long noncoding RNAs in cancer: Mechanisms of action and technological advancements. Mol. Cancer 2016, 15, 43. [Google Scholar] [CrossRef]
- Bartolomei, M.S.; Zemel, S.; Tilghman, S.M. Parental imprinting of the mouse H19 gene. Nature 1991, 351, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.; Ballabio, A.; Rupert, J.L.; Lafreniere, R.G.; Grompe, M.; Tonlorenzi, R.; Willard, H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991, 349, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Chinnaiyan, A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef] [PubMed]
- Teppan, J.; Barth, D.A.; Prinz, F.; Jonas, K.; Pichler, M.; Klec, C. Involvement of Long Non-Coding RNAs (lncRNAs) in Tumor Angiogenesis. Non-Coding RNA 2020, 6, 42. [Google Scholar] [CrossRef]
- Balihodzic, A.; Barth, D.A.; Prinz, F.; Pichler, M. Involvement of Long Non-Coding RNAs in Glucose Metabolism in Cancer. Cancers 2021, 13, 977. [Google Scholar] [CrossRef] [PubMed]
- Ashekyan, O.; Abdallah, S.; Shoukari, A.A.; Chamandi, G.; Choubassy, H.; Itani, A.R.S.; Alwan, N.; Nasr, R. Spotlight on Exosomal Non-Coding RNAs in Breast Cancer: An In Silico Analysis to Identify Potential lncRNA/circRNA-miRNA-Target Axis. Int. J. Mol. Sci. 2022, 23, 8351. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Liu, C.X.; Chen, L.L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef]
- Verduci, L.; Tarcitano, E.; Strano, S.; Yarden, Y.; Blandino, G. CircRNAs: Role in human diseases and potential use as biomarkers. Cell Death Dis. 2021, 12, 468. [Google Scholar] [CrossRef]
- Gong, H.; Gao, M.; Lin, Y.; Liu, J.; Hu, Z.; Liu, J. TUG1/MAZ/FTH1 Axis Attenuates the Antiglioma Effect of Dihydroartemisinin by Inhibiting Ferroptosis. Oxidative Med. Cell. Longev. 2022, 2022, 7843863. [Google Scholar] [CrossRef]
- Wei, X.B.; Jiang, W.Q.; Zeng, J.H.; Huang, L.Q.; Ding, H.G.; Jing, Y.W.; Han, Y.L.; Li, Y.C.; Chen, S.L. Exosome-Derived lncRNA NEAT1 Exacerbates Sepsis-Associated Encephalopathy by Promoting Ferroptosis Through Regulating miR-9-5p/TFRC and GOT1 Axis. Mol. Neurobiol. 2022, 59, 1954–1969. [Google Scholar] [CrossRef]
- Fan, H.; Ai, R.; Mu, S.; Niu, X.; Guo, Z.; Liu, L. MiR-19a suppresses ferroptosis of colorectal cancer cells by targeting IREB2. Bioengineered 2022, 13, 12021–12029. [Google Scholar] [CrossRef] [PubMed]
- He, G.N.; Bao, N.R.; Wang, S.; Xi, M.; Zhang, T.H.; Chen, F.S. Ketamine Induces Ferroptosis of Liver Cancer Cells by Targeting lncRNA PVT1/miR-214-3p/GPX4. Drug Des. Dev. Ther. 2021, 15, 3965–3978. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Niu, G.; Yi, H.; Li, Q.; Wu, Z.; Wang, J.; Yang, J.; Li, B.; Peng, Y.; Liang, Y.; et al. Nanomedicine promotes ferroptosis to inhibit tumour proliferation in vivo. Redox Biol. 2021, 42, 101908. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.H.; Wu, D.M.; Li, L.; Liu, T.; Zhang, T.; Li, J.; Yu, Y.; He, M.; Zhao, Y.Y.; Han, R.; et al. miR-324-3p reverses cisplatin resistance by inducing GPX4-mediated ferroptosis in lung adenocarcinoma cell line A549. Biochem. Biophys. Res. Commun. 2021, 549, 54–60. [Google Scholar] [CrossRef]
- Hou, Y.; Cai, S.; Yu, S.; Lin, H. Metformin induces ferroptosis by targeting miR-324-3p/GPX4 axis in breast cancer. Acta Biochim. Et. Biophys. Sin. 2021, 53, 333–341. [Google Scholar] [CrossRef]
- Liu, L.; Yao, H.; Zhou, X.; Chen, J.; Chen, G.; Shi, X.; Wu, G.; Zhou, G.; He, S. MiR-15a-3p regulates ferroptosis via targeting glutathione peroxidase GPX4 in colorectal cancer. Mol. Carcinog. 2022, 61, 301–310. [Google Scholar] [CrossRef]
- Xu, P.; Wang, Y.; Deng, Z.; Tan, Z.; Pei, X. MicroRNA-15a promotes prostate cancer cell ferroptosis by inhibiting GPX4 expression. Oncol. Lett. 2022, 23, 67. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, T.; Liu, R.; Ning, T.; Yang, H.; Liu, D.; Zhang, Q.; Lin, D.; Ge, S.; Bai, M.; et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer 2020, 19, 43. [Google Scholar] [CrossRef]
- Scalise, M.; Pochini, L.; Console, L.; Losso, M.A.; Indiveri, C. The Human SLC1A5 (ASCT2) Amino Acid Transporter: From Function to Structure and Role in Cell Biology. Front. Cell Dev. Biol. 2018, 6, 96. [Google Scholar] [CrossRef]
- Luo, M.; Wu, L.; Zhang, K.; Wang, H.; Zhang, T.; Gutierrez, L.; O’Connell, D.; Zhang, P.; Li, Y.; Gao, T.; et al. miR-137 regulates ferroptosis by targeting glutamine transporter SLC1A5 in melanoma. Cell Death Differ. 2018, 25, 1457–1472. [Google Scholar] [CrossRef]
- Yuan, Y.; Mei, Z.; Qu, Z.; Li, G.; Yu, S.; Liu, Y.; Liu, K.; Shen, Z.; Pu, J.; Wang, Y.; et al. Exosomes secreted from cardiomyocytes suppress the sensitivity of tumor ferroptosis in ischemic heart failure. Signal Transduct. Target. Ther. 2023, 8, 121. [Google Scholar] [CrossRef]
- Zhang, B.; Bao, W.; Zhang, S.; Chen, B.; Zhou, X.; Zhao, J.; Shi, Z.; Zhang, T.; Chen, Z.; Wang, L.; et al. LncRNA HEPFAL accelerates ferroptosis in hepatocellular carcinoma by regulating SLC7A11 ubiquitination. Cell Death Dis. 2022, 13, 734. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, Q.; Kang, W.; Yu, C. Long non-coding RNA LINC00472 suppresses hepatocellular carcinoma cell proliferation, migration and invasion through miR-93-5p/PDCD4 pathway. Clin. Res. Hepatol. Gastroenterol. 2019, 43, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Koppula, P.; Zhuang, L.; Gan, B. Cystine transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021, 12, 599–620. [Google Scholar] [CrossRef] [PubMed]
- Gai, C.; Liu, C.; Wu, X.; Yu, M.; Zheng, J.; Zhang, W.; Lv, S.; Li, W. MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in non-small cell lung cancer cells. Cell Death Dis. 2020, 11, 751. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Gao, X.; Wu, N.; Jin, Y.; Zhou, H.; Wang, W.; Liu, H.; Chu, Y.; Cao, J.; Jiang, M.; et al. Long noncoding RNA LINC00239 inhibits ferroptosis in colorectal cancer by binding to Keap1 to stabilize Nrf2. Cell Death Dis. 2022, 13, 742. [Google Scholar] [CrossRef]
- Wu, H.; Liu, A. Long non-coding RNA NEAT1 regulates ferroptosis sensitivity in non-small-cell lung cancer. J. Int. Med. Res. 2021, 49, 300060521996183. [Google Scholar] [CrossRef]
- Luo, W.; Wang, J.; Xu, W.; Ma, C.; Wan, F.; Huang, Y.; Yao, M.; Zhang, H.; Qu, Y.; Ye, D.; et al. LncRNA RP11-89 facilitates tumorigenesis and ferroptosis resistance through PROM2-activated iron export by sponging miR-129-5p in bladder cancer. Cell Death Dis. 2021, 12, 1043. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.H.; Huang, J.J.; Zu, P.; Liu, J.; Gao, X.; Du, J.W.; Li, Y.F. CircKDM4C upregulates P53 by sponging hsa-let-7b-5p to induce ferroptosis in acute myeloid leukemia. Environ. Toxicol. 2021, 36, 1288–1302. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Zhou, L.; Yang, G.; Meng, F.; Wan, Y.; Wang, L.; Zhang, L. CircIL4R facilitates the tumorigenesis and inhibits ferroptosis in hepatocellular carcinoma by regulating the miR-541-3p/GPX4 axis. Cell Biol. Int. 2020, 44, 2344–2356. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhao, J.; Li, R.; Liu, Y.; Zhou, L.; Wang, C.; Lv, C.; Gao, L.; Cui, D. CircLRFN5 inhibits the progression of glioblastoma via PRRX2/GCH1 mediated ferroptosis. J. Exp. Clin. Cancer Res. 2022, 41, 307. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, L.; Yang, Z.; Wen, D.; Hu, Z. Circular RNA circACAP2 Suppresses Ferroptosis of Cervical Cancer during Malignant Progression by miR-193a-5p/GPX4. J. Oncol. 2022, 2022, 5228874. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhu, S.; Wang, P.; Wang, J.; Huang, J.; Wang, T.; Guo, L.; Liang, D.; Meng, Q.; Pan, H. Regulators of epigenetic change in ferroptosis-associated cancer (Review). Oncol. Rep. 2022, 48. [Google Scholar] [CrossRef]
- Schonberg, D.L.; Miller, T.E.; Wu, Q.; Flavahan, W.A.; Das, N.K.; Hale, J.S.; Hubert, C.G.; Mack, S.C.; Jarrar, A.M.; Karl, R.T.; et al. Preferential Iron Trafficking Characterizes Glioblastoma Stem-like Cells. Cancer Cell 2015, 28, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Basuli, D.; Tesfay, L.; Deng, Z.; Paul, B.; Yamamoto, Y.; Ning, G.; Xian, W.; McKeon, F.; Lynch, M.; Crum, C.P.; et al. Iron addiction: A novel therapeutic target in ovarian cancer. Oncogene 2017, 36, 4089–4099. [Google Scholar] [CrossRef]
- Müller, S.; Sindikubwabo, F.; Cañeque, T.; Lafon, A.; Versini, A.; Lombard, B.; Loew, D.; Wu, T.D.; Ginestier, C.; Charafe-Jauffret, E.; et al. CD44 regulates epigenetic plasticity by mediating iron endocytosis. Nat. Chem. 2020, 12, 929–938. [Google Scholar] [CrossRef]
- Mai, T.T.; Hamaï, A.; Hienzsch, A.; Cañeque, T.; Müller, S.; Wicinski, J.; Cabaud, O.; Leroy, C.; David, A.; Acevedo, V.; et al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat. Chem. 2017, 9, 1025–1033. [Google Scholar] [CrossRef]
- Solier, S.; Müller, S.; Cañeque, T.; Versini, A.; Mansart, A.; Sindikubwabo, F.; Baron, L.; Emam, L.; Gestraud, P.; Pantoș, G.D.; et al. A druggable copper-signalling pathway that drives inflammation. Nature 2023, 617, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20, 245. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Kim, S.; Son, M.; Kim, M.; Koh, E.S.; Shin, S.J.; Park, C.W.; Kim, H.S. Inhibition of p300/CBP-Associated Factor Attenuates Renal Tubulointerstitial Fibrosis through Modulation of NF-kB and Nrf2. Int. J. Mol. Sci. 2019, 20, 1554. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, W.; Zhang, X.; Wu, F.; Sun, D.; Wang, Z. Ketamine suppresses proliferation and induces ferroptosis and apoptosis of breast cancer cells by targeting KAT5/GPX4 axis. Biochem. Biophys. Res. Commun. 2021, 585, 111–116. [Google Scholar] [CrossRef]
- Sui, S.; Zhang, J.; Xu, S.; Wang, Q.; Wang, P.; Pang, D. Ferritinophagy is required for the induction of ferroptosis by the bromodomain protein BRD4 inhibitor (+)-JQ1 in cancer cells. Cell Death Dis. 2019, 10, 331. [Google Scholar] [CrossRef]
- Rothammer, N.; Woo, M.S.; Bauer, S.; Binkle-Ladisch, L.; Di Liberto, G.; Egervari, K.; Wagner, I.; Haferkamp, U.; Pless, O.; Merkler, D.; et al. G9a dictates neuronal vulnerability to inflammatory stress via transcriptional control of ferroptosis. Sci. Adv. 2022, 8, eabm5500. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Wang, H.; Zhang, C.; Wang, M.; Yang, Y.; Xu, X.; Hu, Z. Histone demethylase KDM3B protects against ferroptosis by upregulating SLC7A11. FEBS Open Bio 2020, 10, 637–643. [Google Scholar] [CrossRef]
- Chen, M.; Jiang, Y.; Sun, Y. KDM4A-mediated histone demethylation of SLC7A11 inhibits cell ferroptosis in osteosarcoma. Biochem. Biophys. Res. Commun. 2021, 550, 77–83. [Google Scholar] [CrossRef]
- Cao, Z.; Liu, X.; Zhang, W.; Zhang, K.; Pan, L.; Zhu, M.; Qin, H.; Zou, C.; Wang, W.; Zhang, C.; et al. Biomimetic Macrophage Membrane-Camouflaged Nanoparticles Induce Ferroptosis by Promoting Mitochondrial Damage in Glioblastoma. ACS Nano 2023, 17, 23746–23760. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Zhu, J.; Wu, J.; Thompson, C.B.; Jiang, X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 31189–31197. [Google Scholar] [CrossRef]
- Mao, C.; Wang, X.; Liu, Y.; Wang, M.; Yan, B.; Jiang, Y.; Shi, Y.; Shen, Y.; Liu, X.; Lai, W.; et al. A G3BP1-Interacting lncRNA Promotes Ferroptosis and Apoptosis in Cancer via Nuclear Sequestration of p53. Cancer Res. 2018, 78, 3484–3496. [Google Scholar] [CrossRef]
| Gene/Metabolite | Humans | Mice | Plants | Yeast | Bacteria |
|---|---|---|---|---|---|
| GPX4 | Inhibitor [3] | Inhibitor | Inhibitor [14] | Presence of homologue [15] | Not found [16] |
| SLC7A11 | Inhibitor | Inhibitor | Unknown | Unknown | Unknown |
| Glutathione | Synthetic substrate for GPX4 | Synthetic substrate for GPX4 | Works with ascorbate to suppress ROS | Antioxidant | Antioxidant |
| PUFAs | Essential for ferroptosis | Essential for ferroptosis | Essential for ferroptosis | Driving ferroptosis in CoQ-deficient yeast [17] | Not essential for ferroptosis |
| ACSL4 | Promoter | Promoter | Present but unclearly function | Unknown | Unknown |
| Lipid peroxidation | Detected in ferroptosis | Detected in ferroptosis | Detected in ferroptosis | Detected in ferroptosis | Not typically observed |
| Vitamin E | Inhibitor | Inhibitor | Produced by many plants | Unknown | Unknown |
| Vitamin K | Inhibitor [18] | Inhibitor | Produced by many plants | Unknown | Unknown |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, J.; Lyu, H.; Ouyang, Q.; Shi, H.; Zhang, R.; Xiao, S.; Guo, D.; Zhang, Q.; Chen, X.-Z.; Zhou, C.; et al. Insights into the Roles of Epigenetic Modifications in Ferroptosis. Biology 2024, 13, 122. https://doi.org/10.3390/biology13020122
Kong J, Lyu H, Ouyang Q, Shi H, Zhang R, Xiao S, Guo D, Zhang Q, Chen X-Z, Zhou C, et al. Insights into the Roles of Epigenetic Modifications in Ferroptosis. Biology. 2024; 13(2):122. https://doi.org/10.3390/biology13020122
Chicago/Turabian StyleKong, Jinghua, Hao Lyu, Qian Ouyang, Hao Shi, Rui Zhang, Shuai Xiao, Dong Guo, Qi Zhang, Xing-Zhen Chen, Cefan Zhou, and et al. 2024. "Insights into the Roles of Epigenetic Modifications in Ferroptosis" Biology 13, no. 2: 122. https://doi.org/10.3390/biology13020122





