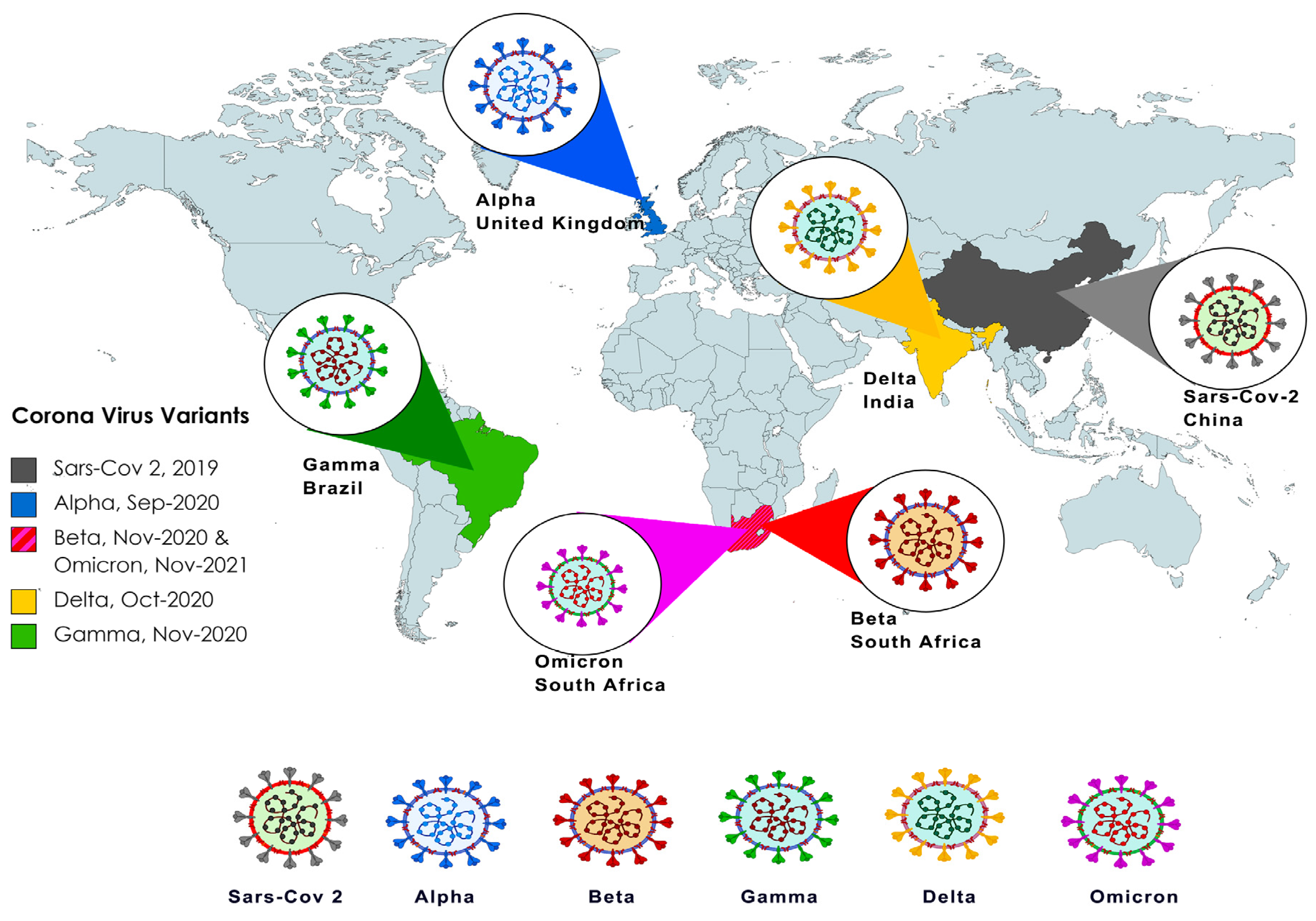
From Alpha to Omicron: How Different Variants of Concern of the SARS-Coronavirus-2 Impacted the World
Abstract
:Simple Summary
Abstract
1. Introduction
2. Alpha (B.1.1.7 Lineage)
2.1. Genetic Origin and Genetic Makeup
2.2. Epidemiology and Morbidity
2.3. Response to Treatment/Vaccination
3. Beta (B.1.351 Lineage)
3.1. Genetic Origin and Genetic Makeup
3.2. Epidemiology and Morbidity
3.3. Response to Treatment/Vaccination
4. Gamma (P.1 Lineage)
4.1. Genetic Origin and Genetic Makeup
4.2. Epidemiology and Morbidity
4.3. Response to Treatment/Vaccination
5. Delta (B.1.617.2 Lineage)
5.1. Genetic Origin and Genetic Makeup
5.2. Epidemiology and Morbidity
5.3. Response to Treatment/Vaccination
6. Omicron (B.1.1.529 Lineage)
6.1. Genetic Origin and Genetic Makeup
Omicron Sub-Lineages (BA.1, BA.1.1, BA.2, BA.4, BA.5, BQ.1, BQ.1.1, and XBB.1.5)
6.2. Epidemiology and Morbidity
6.3. Response to Treatment/Vaccination
7. Global Health Response and Future Variant Prediction
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Xu, Y.; Bao, L.; Zhang, L.; Yu, P.; Qu, Y.; Zhu, H.; Zhao, W.; Han, Y.; Qin, C. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses 2019, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.R.; Yin, W.C.; Jiang, Y.; Xu, H.E. Structure genomics of SARS-CoV-2 and its Omicron variant: Drug design templates for COVID-19. Acta Pharmacol. Sin. 2022, 43, 3021–3033. [Google Scholar] [CrossRef]
- Bai, C.; Zhong, Q.; Gao, G.F. Overview of SARS-CoV-2 genome-encoded proteins. Sci. China Life Sci. 2022, 65, 280–294. [Google Scholar] [CrossRef]
- Nelson, C.W.; Ardern, Z.; Goldberg, T.L.; Meng, C.; Kuo, C.H.; Ludwig, C.; Kolokotronis, S.O.; Wei, X. Dynamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic. eLife 2020, 9, e59633. [Google Scholar] [CrossRef]
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef]
- Papanikolaou, V.; Chrysovergis, A.; Ragos, V.; Tsiambas, E.; Katsinis, S.; Manoli, A.; Papouliakos, S.; Roukas, D.; Mastronikolis, S.; Peschos, D.; et al. From delta to Omicron: S1-RBD/S2 mutation/deletion equilibrium in SARS-CoV-2 defined variants. Gene 2022, 814, 146134. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Zhao, R.; Gao, L.J.; Gao, X.F.; Wang, D.P.; Cao, J.M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Lu, G.; Wang, Q.; Gao, G.F. Bat-to-human: Spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015, 23, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Kemp, S.A.; Papa, G.; Datir, R.; Ferreira, I.; Marelli, S.; Harvey, W.T.; Lytras, S.; Mohamed, A.; Gallo, G.; et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021, 35, 109292. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021, 27, 717–726. [Google Scholar] [CrossRef]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; de Oliveira, T.; Vermeulen, M.; van der Berg, K.; et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021, 27, 622–625. [Google Scholar] [CrossRef]
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A.; et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021, 592, 616–622. [Google Scholar] [CrossRef]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.D.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J. Med. Virol. 2022, 94, 1641–1649. [Google Scholar] [CrossRef]
- Liu, Y.; Rocklöv, J. The reproductive number of the Delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J. Travel Med. 2021, 28, taab124. [Google Scholar] [CrossRef] [PubMed]
- Ladhani, S.N. COVID-19 vaccination for children aged 5-11 years. Lancet 2022, 400, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Karim, S.S.A.; Karim, Q.A. Omicron SARS-CoV-2 variant: A new chapter in the COVID-19 pandemic. Lancet 2021, 398, 2126–2128. [Google Scholar] [CrossRef] [PubMed]
- Ingraham, N.E.; Ingbar, D.H. The omicron variant of SARS-CoV-2: Understanding the known and living with unknowns. Clin. Transl. Med. 2021, 11, e685. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Krüger, N.; Schulz, S.; Cossmann, A.; Rocha, C.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Moldenhauer, A.S.; Winkler, M.S.; et al. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell 2022, 185, 447–456.e11. [Google Scholar] [CrossRef]
- VanBlargan, L.A.; Errico, J.M.; Halfmann, P.J.; Zost, S.J.; Crowe, J.E., Jr.; Purcell, L.A.; Kawaoka, Y.; Corti, D.; Fremont, D.H.; Diamond, M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022, 28, 490–495. [Google Scholar] [CrossRef]
- Scobie, H.M.; Johnson, A.G.; Suthar, A.B.; Severson, R.; Alden, N.B.; Balter, S.; Bertolino, D.; Blythe, D.; Brady, S.; Cadwell, B.; et al. Monitoring Incidence of COVID-19 Cases, Hospitalizations, and Deaths, by Vaccination Status—13 U.S. Jurisdictions, 4 April–17 July 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1284–1290. [Google Scholar] [CrossRef]
- Higham, A.; Singh, D. Increased ACE2 Expression in Bronchial Epithelium of COPD Patients who are Overweight. Obes. (Silver Spring) 2020, 28, 1586–1589. [Google Scholar] [CrossRef]
- McAlinden, K.D.; Lu, W.; Ferdowsi, P.V.; Myers, S.; Markos, J.; Larby, J.; Chia, C.; Weber, H.C.; Haug, G.; Eapen, M.S.; et al. Electronic Cigarette Aerosol Is Cytotoxic and Increases ACE2 Expression on Human Airway Epithelial Cells: Implications for SARS-CoV-2 (COVID-19). J. Clin. Med. 2021, 10, 1028. [Google Scholar] [CrossRef] [PubMed]
- Salka, K.; Abutaleb, K.; Chorvinsky, E.; Thiruvengadam, G.; Arroyo, M.; Gomez, J.L.; Gutierrez, M.J.; Pillai, D.K.; Jaiswal, J.K.; Nino, G. IFN Stimulates ACE2 Expression in Pediatric Airway Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 2021, 64, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Wark, P.A.B.; Pathinayake, P.S.; Kaiko, G.; Nichol, K.; Ali, A.; Chen, L.; Sutanto, E.N.; Garratt, L.W.; Sohal, S.S.; Lu, W.; et al. ACE2 expression is elevated in airway epithelial cells from older and male healthy individuals but reduced in asthma. Respirology 2021, 26, 442–451. [Google Scholar] [CrossRef]
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020, 14, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Duchene, S.; Featherstone, L.; Haritopoulou-Sinanidou, M.; Rambaut, A.; Lemey, P.; Baele, G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020, 6, veaa061. [Google Scholar] [CrossRef] [PubMed]
- Walensky, R.P.; Walke, H.T.; Fauci, A.S. SARS-CoV-2 Variants of Concern in the United States-Challenges and Opportunities. JAMA 2021, 325, 1037–1038. [Google Scholar] [CrossRef] [PubMed]
- WHO. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 27 June 2023).
- Rahimi, A.; Mirzazadeh, A.; Tavakolpour, S. Genetics and genomics of SARS-CoV-2: A review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics 2021, 113 Pt 2, 1221–1232. [Google Scholar] [CrossRef]
- Winger, A.; Caspari, T. The Spike of Concern-The Novel Variants of SARS-CoV-2. Viruses 2021, 13, 1002. [Google Scholar] [CrossRef]
- Braga, L.; Ali, H.; Secco, I.; Chiavacci, E.; Neves, G.; Goldhill, D.; Penn, R.; Jimenez-Guardeño, J.M.; Ortega-Prieto, A.M.; Bussani, R.; et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature 2021, 594, 88–93. [Google Scholar] [CrossRef]
- Bussani, R.; Schneider, E.; Zentilin, L.; Collesi, C.; Ali, H.; Braga, L.; Volpe, M.C.; Colliva, A.; Zanconati, F.; Berlot, G.; et al. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine 2020, 61, 103104. [Google Scholar] [CrossRef]
- Loconsole, D.; Centrone, F.; Morcavallo, C.; Campanella, S.; Accogli, M.; Sallustio, A.; Peccarisi, D.; Stufano, A.; Lovreglio, P.; Chironna, M. Changing Features of COVID-19: Characteristics of Infections with the SARS-CoV-2 Delta (B.1.617.2) and Alpha (B.1.1.7) Variants in Southern Italy. Vaccines 2021, 9, 1354. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Jarvis, C.I.; Edmunds, W.J.; Jewell, N.P.; Diaz-Ordaz, K.; Keogh, R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature 2021, 593, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Benton, D.J.; Wrobel, A.G.; Roustan, C.; Borg, A.; Xu, P.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2021, 118, e2022586118. [Google Scholar] [CrossRef] [PubMed]
- Juri, T.; Kihira, S.; Higuchi, R.; Yanagawa, T.; Koike, M. Ultrasonographic evaluation of the development of the brain sulcus. No Hattatsu 1986, 18, 322–323. [Google Scholar]
- Ulrich, L.; Halwe, N.J.; Taddeo, A.; Ebert, N.; Schön, J.; Devisme, C.; Trüeb, B.S.; Hoffmann, B.; Wider, M.; Fan, X.; et al. Enhanced fitness of SARS-CoV-2 variant of concern Alpha but not Beta. Nature 2022, 602, 307–313. [Google Scholar] [CrossRef]
- Tian, F.; Tong, B.; Sun, L.; Shi, S.; Zheng, B.; Wang, Z.; Dong, X.; Zheng, P. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. eLife 2021, 10, e69091. [Google Scholar] [CrossRef]
- Radvak, P.; Kwon, H.J.; Kosikova, M.; Ortega-Rodriguez, U.; Xiang, R.; Phue, J.N.; Shen, R.F.; Rozzelle, J.; Kapoor, N.; Rabara, T.; et al. SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nat. Commun. 2021, 12, 6559. [Google Scholar] [CrossRef]
- Jassat, W.; Abdool Karim, S.S.; Mudara, C.; Welch, R.; Ozougwu, L.; Groome, M.J.; Govender, N.; von Gottberg, A.; Wolter, N.; Wolmarans, M.; et al. Clinical severity of COVID-19 in patients admitted to hospital during the omicron wave in South Africa: A retrospective observational study. Lancet Glob. Health 2022, 10, e961–e969. [Google Scholar] [CrossRef]
- Chemaitelly, H.; Yassine, H.M.; Benslimane, F.M.; Al Khatib, H.A.; Tang, P.; Hasan, M.R.; Malek, J.A.; Coyle, P.; Ayoub, H.H.; Al Kanaani, Z.; et al. mRNA-1273 COVID-19 vaccine effectiveness against the B.1.1.7 and B.1.351 variants and severe COVID-19 disease in Qatar. Nat. Med. 2021, 27, 1614–1621. [Google Scholar] [CrossRef]
- Skelly, D.T.; Harding, A.C.; Gilbert-Jaramillo, J.; Knight, M.L.; Longet, S.; Brown, A.; Adele, S.; Adland, E.; Brown, H.; Tipton, T.; et al. Two doses of SARS-CoV-2 vaccination induce robust immune responses to emerging SARS-CoV-2 variants of concern. Nat. Commun. 2021, 12, 5061. [Google Scholar] [CrossRef]
- Abu-Raddad, L.J.; Chemaitelly, H.; Butt, A.A. Effectiveness of the BNT162b2 COVID-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021, 385, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Mahase, E. COVID-19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ 2021, 372, n296. [Google Scholar] [CrossRef] [PubMed]
- da Silva, J.C.; Félix, V.B.; Leão, S.; Trindade-Filho, E.M.; Scorza, F.A. New Brazilian variant of the SARS-CoV-2 (P1/Gamma) of COVID-19 in Alagoas state. Braz. J. Infect. Dis. 2021, 25, 101588. [Google Scholar] [CrossRef] [PubMed]
- Giovanetti, M.; Fonseca, V.; Wilkinson, E.; Tegally, H.; San, E.J.; Althaus, C.L.; Xavier, J.; Nanev Slavov, S.; Viala, V.L.; Ranieri Jerônimo Lima, A.; et al. Replacement of the Gamma by the Delta variant in Brazil: Impact of lineage displacement on the ongoing pandemic. Virus Evol. 2022, 8, veac024. [Google Scholar] [CrossRef]
- Wang, P.; Casner, R.G.; Nair, M.S.; Wang, M.; Yu, J.; Cerutti, G.; Liu, L.; Kwong, P.D.; Huang, Y.; Shapiro, L.; et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe 2021, 29, 747–751.e4. [Google Scholar] [CrossRef]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I.; et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef]
- Kannan, S.R.; Spratt, A.N.; Cohen, A.R.; Naqvi, S.H.; Chand, H.S.; Quinn, T.P.; Lorson, C.L.; Byrareddy, S.N.; Singh, K. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J. Autoimmun. 2021, 124, 102715. [Google Scholar] [CrossRef]
- Zhang, J.; Cai, Y.; Xiao, T.; Lu, J.; Peng, H.; Sterling, S.M.; Walsh, R.M., Jr.; Rits-Volloch, S.; Zhu, H.; Woosley, A.N.; et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 2021, 372, 525–530. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e9. [Google Scholar] [CrossRef]
- Gobeil, S.M.; Janowska, K.; McDowell, S.; Mansouri, K.; Parks, R.; Stalls, V.; Kopp, M.F.; Manne, K.; Li, D.; Wiehe, K.; et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science 2021, 373, eabi6226. [Google Scholar] [CrossRef]
- Saville, J.W.; Mannar, D.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Demers, J.P.; Zhou, S.; Tuttle, K.S.; Sekirov, I.; Kim, A.; et al. Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants. Nat. Commun. 2022, 13, 742. [Google Scholar] [CrossRef] [PubMed]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Campbell, F.; Archer, B.; Laurenson-Schafer, H.; Jinnai, Y.; Konings, F.; Batra, N.; Pavlin, B.; Vandemaele, K.; Van Kerkhove, M.D.; Jombart, T.; et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill 2021, 26, 2100509. [Google Scholar] [CrossRef]
- Chavda, V.P.; Apostolopoulos, V. Global impact of delta plus variant and vaccination. Expert Rev. Vaccines 2022, 21, 597–600. [Google Scholar] [CrossRef] [PubMed]
- Fiolet, T.; Kherabi, Y.; MacDonald, C.J.; Ghosn, J.; Peiffer-Smadja, N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: A narrative review. Clin. Microbiol. Infect. 2022, 28, 202–221. [Google Scholar] [CrossRef]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O’Connell, A.M.; et al. COVID-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022, 386, 1532–1546. [Google Scholar] [CrossRef]
- Crocker, T.F.; Brown, L.; Lam, N.; Wray, F.; Knapp, P.; Forster, A. Information provision for stroke survivors and their carers. Cochrane Database Syst. Rev. 2021, 11, Cd001919. [Google Scholar] [CrossRef]
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of COVID-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef]
- Wang, L.; Cheng, G. Sequence analysis of the emerging SARS-CoV-2 variant Omicron in South Africa. J. Med. Virol. 2022, 94, 1728–1733. [Google Scholar] [CrossRef]
- He, X.; Hong, W.; Pan, X.; Lu, G.; Wei, X. SARS-CoV-2 Omicron variant: Characteristics and prevention. MedComm (2020) 2021, 2, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, S.; Sarkar, R. Mutational and phylogenetic analyses of the two lineages of the Omicron variant. J. Med. Virol. 2022, 94, 1777–1779. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Karuppanan, K.; Subramaniam, G. Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: A comparative sequence and structural-based computational assessment. J. Med. Virol. 2022, 94, 4780–4791. [Google Scholar] [CrossRef] [PubMed]
- Nutalai, R.; Zhou, D.; Tuekprakhon, A.; Ginn, H.M.; Supasa, P.; Liu, C.; Huo, J.; Mentzer, A.J.; Duyvesteyn, H.M.E.; Dijokaite-Guraliuc, A.; et al. Potent cross-reactive antibodies following Omicron breakthrough in vaccinees. Cell 2022, 185, 2116–2131.e18. [Google Scholar] [CrossRef]
- Bazargan, M.; Elahi, R.; Esmaeilzadeh, A. OMICRON: Virology, immunopathogenesis, and laboratory diagnosis. J. Gene Med. 2022, 24, e3435. [Google Scholar] [CrossRef]
- Yamasoba, D.; Kimura, I.; Nasser, H.; Morioka, Y.; Nao, N.; Ito, J.; Uriu, K.; Tsuda, M.; Zahradnik, J.; Shirakawa, K.; et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell 2022, 185, 2103–2115.e19. [Google Scholar] [CrossRef]
- Li, L.; Liao, H.; Meng, Y.; Li, W.; Han, P.; Liu, K.; Wang, Q.; Li, D.; Zhang, Y.; Wang, L.; et al. Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1. Cell 2022, 185, 2952–2960.e10. [Google Scholar] [CrossRef]
- Hirotsu, Y.; Maejima, M.; Shibusawa, M.; Natori, Y.; Nagakubo, Y.; Hosaka, K.; Sueki, H.; Mochizuki, H.; Tsutsui, T.; Kakizaki, Y.; et al. Classification of Omicron BA.1, BA.1.1, and BA.2 sublineages by TaqMan assay consistent with whole genome analysis data. Int. J. Infect. Dis. 2022, 122, 486–491. [Google Scholar] [CrossRef]
- Iuliano, A.D.; Brunkard, J.M.; Boehmer, T.K.; Peterson, E.; Adjei, S.; Binder, A.M.; Cobb, S.; Graff, P.; Hidalgo, P.; Panaggio, M.J.; et al. Trends in Disease Severity and Health Care Utilization During the Early Omicron Variant Period Compared with Previous SARS-CoV-2 High Transmission Periods—United States, December 2020-January 2022. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 146–152. [Google Scholar] [CrossRef]
- Evans, J.P.; Zeng, C.; Qu, P.; Faraone, J.; Zheng, Y.M.; Carlin, C.; Bednash, J.S.; Zhou, T.; Lozanski, G.; Mallampalli, R.; et al. Neutralization of SARS-CoV-2 Omicron sub-lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe 2022, 30, 1093–1102.e3. [Google Scholar] [CrossRef]
- Kannan, S.R.; Spratt, A.N.; Sharma, K.; Goyal, R.; Sönnerborg, A.; Apparsundaram, S.; Lorson, C.L.; Byrareddy, S.N.; Singh, K. Complex Mutation Pattern of Omicron BA.2: Evading Antibodies without Losing Receptor Interactions. Int. J. Mol. Sci. 2022, 23, 5534. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Yisimayi, A.; Jian, F.; Song, W.; Xiao, T.; Wang, L.; Du, S.; Wang, J.; Li, Q.; Chen, X.; et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022, 608, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Tuekprakhon, A.; Nutalai, R.; Dijokaite-Guraliuc, A.; Zhou, D.; Ginn, H.M.; Selvaraj, M.; Liu, C.; Mentzer, A.J.; Supasa, P.; Duyvesteyn, H.M.E.; et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 2022, 185, 2422–2433.e13. [Google Scholar] [CrossRef]
- Callaway, E. What Omicron’s BA.4 and BA.5 variants mean for the pandemic. Nature 2022, 606, 848–849. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Yamasoba, D.; Tamura, T.; Nao, N.; Suzuki, T.; Oda, Y.; Mitoma, S.; Ito, J.; Nasser, H.; Zahradnik, J.; et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell 2022, 185, 3992–4007.e16. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. Coronavirus variant XBB.1.5 rises in the United States—Is it a global threat? Nature 2023, 613, 222–223. [Google Scholar] [CrossRef]
- Vitiello, A.; Ferrara, F.; Auti, A.M.; Di Domenico, M.; Boccellino, M. Advances in the Omicron variant development. J. Intern. Med. 2022, 292, 81–90. [Google Scholar] [CrossRef]
- Taylor, C.A.; Whitaker, M.; Anglin, O.; Milucky, J.; Patel, K.; Pham, H.; Chai, S.J.; Alden, N.B.; Yousey-Hindes, K.; Anderson, E.J.; et al. COVID-19-Associated Hospitalizations Among Adults During SARS-CoV-2 Delta and Omicron Variant Predominance, by Race/Ethnicity and Vaccination Status—COVID-NET, 14 States, July 2021-January 2022. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 466–473. [Google Scholar] [CrossRef]
- Shuai, H.; Chan, J.F.; Hu, B.; Chai, Y.; Yuen, T.T.; Yin, F.; Huang, X.; Yoon, C.; Hu, J.C.; Liu, H.; et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 2022, 603, 693–699. [Google Scholar] [CrossRef]
- Khandia, R.; Singhal, S.; Alqahtani, T.; Kamal, M.A.; El-Shall, N.A.; Nainu, F.; Desingu, P.A.; Dhama, K. Emergence of SARS-CoV-2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ. Res. 2022, 209, 112816. [Google Scholar] [CrossRef]
- Leshem, E.; Gonen, T.; Hoffman, T.; Barsisat, A.; Kreiss, Y.; Regev-Yochay, G. Low rate of transmission to triple-vaccinated contacts of an imported case of SARS-CoV-2 omicron infection: A contact tracing study in Israel. J. Travel Med. 2022, 29, taac003. [Google Scholar] [CrossRef]
- Sigal, A.; Milo, R.; Jassat, W. Estimating disease severity of Omicron and Delta SARS-CoV-2 infections. Nat. Rev. Immunol. 2022, 22, 267–269. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, Y.; Zhao, Y.; He, D. Reduction in the infection fatality rate of Omicron variant compared with previous variants in South Africa. Int. J. Infect. Dis. 2022, 120, 146–149. [Google Scholar] [CrossRef]
- Adamoski, D.; Baura, V.A.; Rodrigues, A.C.; Royer, C.A.; Aoki, M.N.; Tschá, M.K.; Bonatto, A.C.; Wassem, R.; Nogueira, M.B.; Raboni, S.M.; et al. SARS-CoV-2 Delta and Omicron Variants Surge in Curitiba, Southern Brazil, and Its Impact on Overall COVID-19 Lethality. Viruses 2022, 14, 809. [Google Scholar] [CrossRef]
- Pierce, C.A.; Herold, K.C.; Herold, B.C.; Chou, J.; Randolph, A.; Kane, B.; McFarland, S.; Gurdasani, D.; Pagel, C.; Hotez, P.; et al. COVID-19 and children. Science 2022, 377, 1144–1149. [Google Scholar] [CrossRef]
- Lewnard, J.A.; Hong, V.X.; Patel, M.M.; Kahn, R.; Lipsitch, M.; Tartof, S.Y. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in Southern California. Nat. Med. 2022, 28, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.C.; Shirk, P.; Lambrou, A.S.; Hassell, N.; Zheng, X.Y.; Payne, A.B.; Ali, A.R.; Batra, D.; Caravas, J.; Chau, R.; et al. Genomic Surveillance for SARS-CoV-2 Variants: Circulation of Omicron Lineages—United States, January 2022-May 2023. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 651–656. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Technical Advisory Group on Virus Evolution (TAG-VE). 2023. Available online: https://www.who.int/docs/default-source/coronaviruse/11jan2023_xbb15_rapid_risk_assessment.pdf (accessed on 1 June 2023).
- Chenchula, S.; Amerneni, K.C.; Ghanta, M.K.; Padmavathi, R.; Chandra, M.B.; Adusumilli, M.B.; Chavan, M.; Mudda, S.; Gupta, R.; Lakhawat, B. Clinical virology and effect of COVID-19 vaccination and monoclonal antibodies against highly infectious SARS-CoV-2 omicron sub variant BF.7 (BA.5.2.1.7): A systematic review. Virology 2023, 584, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.G.; Amin, A.B.; Ali, A.R.; Hoots, B.; Cadwell, B.L.; Arora, S.; Avoundjian, T.; Awofeso, A.O.; Barnes, J.; Bayoumi, N.S.; et al. COVID-19 Incidence and Death Rates Among Unvaccinated and Fully Vaccinated Adults with and Without Booster Doses During Periods of Delta and Omicron Variant Emergence—25 U.S. Jurisdictions, April 4–December 25, 2021. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.; Wang, X.; He, X.; Zhao, X.; Zhang, Y.; Jiang, Y.; Li, M.; Cui, Y.; Chen, Y.; Qiao, R.; et al. Antibody evasion of SARS-CoV-2 Omicron BA.1, BA.1.1, BA.2, and BA.3 sub-lineages. Cell Host Microbe 2022, 30, 1077–1083.e4. [Google Scholar] [CrossRef]
- Thompson, M.G.; Natarajan, K.; Irving, S.A.; Rowley, E.A.; Griggs, E.P.; Gaglani, M.; Klein, N.P.; Grannis, S.J.; DeSilva, M.B.; Stenehjem, E.; et al. Effectiveness of a Third Dose of mRNA Vaccines Against COVID-19-Associated Emergency Department and Urgent Care Encounters and Hospitalizations Among Adults During Periods of Delta and Omicron Variant Predominance—VISION Network, 10 States, August 2021-–January 2022. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Chenchula, S.; Karunakaran, P.; Sharma, S.; Chavan, M. Current evidence on efficacy of COVID-19 booster dose vaccination against the Omicron variant: A systematic review. J. Med. Virol. 2022, 94, 2969–2976. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Song, S.; Guo, H.; Zhou, X.; Fan, Q.; Liu, W.; Cheng, L.; Ge, X.; Ju, B.; Zhang, Z. A fourth dose of Omicron RBD vaccine enhances broad neutralization against SARS-CoV-2 variants including BA.1 and BA.2 in vaccinated mice. J. Med. Virol. 2022, 94, 3992–3997. [Google Scholar] [CrossRef]
- Miller, J.; Hachmann, N.P.; Collier, A.Y.; Lasrado, N.; Mazurek, C.R.; Patio, R.C.; Powers, O.; Surve, N.; Theiler, J.; Korber, B.; et al. Substantial Neutralization Escape by SARS-CoV-2 Omicron Variants BQ.1.1 and XBB.1. N. Engl. J. Med. 2023, 388, 662–664. [Google Scholar] [CrossRef] [PubMed]
- Barbosa-Camacho, F.J.; Romero-Limón, O.M.; Ibarrola-Peña, J.C.; Almanza-Mena, Y.L.; Pintor-Belmontes, K.J.; Sánchez-López, V.A.; Chejfec-Ciociano, J.M.; Guzmán-Ramírez, B.G.; Sapién-Fernández, J.H.; Guzmán-Ruvalcaba, M.J.; et al. Depression, anxiety, and academic performance in COVID-19: A cross-sectional study. BMC Psychiatry 2022, 22, 443. [Google Scholar] [CrossRef]
- Panchal, U.; Salazar de Pablo, G.; Franco, M.; Moreno, C.; Parellada, M.; Arango, C.; Fusar-Poli, P. The impact of COVID-19 lockdown on child and adolescent mental health: Systematic review. Eur. Child. Adolesc. Psychiatry 2023, 32, 1151–1177. [Google Scholar] [CrossRef]
- Mugabe, P.A.; Renkamp, T.M.; Rybak, C.; Mbwana, H.; Gordon, C.; Sieber, S.; Löhr, K. Governing COVID-19: Analyzing the effects of policy responses on food systems in Tanzania. Agric. Food Secur. 2022, 11, 47. [Google Scholar] [CrossRef]
- Bhargava, A.; Shewade, H.D. The potential impact of the COVID-19 response related lockdown on TB incidence and mortality in India. Indian J. Tuberc. 2020, 67, S139–S146. [Google Scholar] [CrossRef]
- Arbel, R.; Pliskin, J. Vaccinations versus Lockdowns to Prevent COVID-19 Mortality. Vaccines 2022, 10, 1347. [Google Scholar] [CrossRef]
- Borri, N.; Drago, F.; Santantonio, C.; Sobbrio, F. The “Great Lockdown”: Inactive workers and mortality by COVID-19. Health Econ. 2021, 30, 2367–2382. [Google Scholar] [CrossRef]
- CDC. CDC Strategy for Global Response to COVID-19 (2020–2023). 10 November 2022. Available online: https://www.cdc.gov/coronavirus/2019-ncov/global-covid-19/global-response-strategy.html#:~:text=public%20health%20emergencies.-,Goals,global%20public%20health%20leadership%3B%20and (accessed on 9 September 2023).
- Otto, S.P.; Day, T.; Arino, J.; Colijn, C.; Dushoff, J.; Li, M.; Mechai, S.; Van Domselaar, G.; Wu, J.; Earn, D.J.D.; et al. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 2021, 31, R918–R929. [Google Scholar] [CrossRef]
- Magazine, N.; Zhang, T.; Wu, Y.; McGee, M.C.; Veggiani, G.; Huang, W. Mutations and Evolution of the SARS-CoV-2 Spike Protein. Viruses 2022, 14, 640. [Google Scholar] [CrossRef]
- Newcomb, K.; Bilal, S.; Michael, E. Combining predictive models with future change scenarios can produce credible forecasts of COVID-19 futures. PLoS ONE 2022, 17, e0277521. [Google Scholar] [CrossRef] [PubMed]
- Obermeyer, F.; Jankowiak, M.; Barkas, N.; Schaffner, S.F.; Pyle, J.D.; Yurkovetskiy, L.; Bosso, M.; Park, D.J.; Babadi, M.; MacInnis, B.L.; et al. Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness. Science 2022, 376, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Maher, M.C.; Bartha, I.; Weaver, S.; di Iulio, J.; Ferri, E.; Soriaga, L.; Lempp, F.A.; Hie, B.L.; Bryson, B.; Berger, B.; et al. Predicting the mutational drivers of future SARS-CoV-2 variants of concern. Sci. Transl. Med. 2022, 14, eabk3445. [Google Scholar] [CrossRef]
- Giovanetti, M.; Benedetti, F.; Campisi, G.; Ciccozzi, A.; Fabris, S.; Ceccarelli, G.; Tambone, V.; Caruso, A.; Angeletti, S.; Zella, D.; et al. Evolution patterns of SARS-CoV-2: Snapshot on its genome variants. Biochem. Biophys. Res. Commun. 2021, 538, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Subhadra, B.; Agrawal, R.; Pal, V.K.; Chenine, A.L.; Mattathil, J.G.; Singh, A. Significant Broad-Spectrum Antiviral Activity of Bi121 against Different Variants of SARS-CoV-2. Viruses 2023, 15, 129. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Zhang, L.; Rocha, C.; Sidarovich, A.; Kempf, A.; Schulz, S.; Cossmann, A.; Manger, B.; Baier, E.; Tampe, B.; et al. Comparable neutralisation evasion of SARS-CoV-2 omicron subvariants BA.1, BA.2, and BA.3. Lancet Infect. Dis. 2022, 22, 766–767. [Google Scholar] [CrossRef]
- Rössler, A.; Netzl, A.; Knabl, L.; Schäfer, H.; Wilks, S.H.; Bante, D.; Falkensammer, B.; Borena, W.; von Laer, D.; Smith, D.J.; et al. BA.2 and BA.5 omicron differ immunologically from both BA.1 omicron and pre-omicron variants. Nat. Commun. 2022, 13, 7701. [Google Scholar] [CrossRef]
- Morens, D.M.; Taubenberger, J.K.; Fauci, A.S. Universal Coronavirus Vaccines—An Urgent Need. N. Engl. J. Med. 2022, 386, 297–299. [Google Scholar] [CrossRef]


| Variant | Key Mutations | Infectivity & Transmissibility | Response to Treatment | References Cited |
|---|---|---|---|---|
| Alpha (B.1.1.7) | N501Y, H69/V70, P681H, Y144 | Increased transmissibility and virulence | Reduced efficacy of monoclonal antibodies and vaccines | [13,14,15] |
| Beta (B.1.351) | N501Y, E484K, K417N, L18F, D80A, D215G, A701V | Increased transmissibility | Reduced efficacy of monoclonal antibodies and vaccines | [16,17,18,19] |
| Gamma (P.1 lineage) | K417T, E484K, N501Y | Increased transmissibility and virulence | Reduced efficacy of monoclonal antibodies and vaccines, but previous infection provided between 54 to 79% protection | [20] |
| Delta (B.1.617) | R158G, L452R, T478K, D614G P681R, D950N | Increased infectivity and transmissibility compared to previous variants through the association of infected persons having a higher viral RNA load. | Treatment of monoclonal antibodies casirivimab, imdevimab and sotrovimab were associated with decreased hospitalizations and mortalities. Vaccination efficacy proved 80% effective at preventing symptomatic delta infection >240 after second vaccine dose. | [21,22,23] |
| Omicron (B.1.1.529) | Δ69–70 deletion, T95I, G142D/ Δ143–145 deletion, K417N, T478K, N501Y, N655Y, N679K, and P681H | Higher transmissibility with reduced case fatality compared to other variants. | Vaccination was ineffective at protection from symptomatic infection. Efficacious treatment modality was still largely restricted to glucocorticoids. | [24,25] [26,27] [28] [29] |
| Subvariants (BA.1, BA.1.1, BA.2, BA.4, BA.5, BQ.1, BQ.1.1, XBB.1.5) BA2.75 BA.4 and BA.5 XBB.1.5 XBB.1.16 BQ.1 and BQ.1.1 | >60 mutations >50% mutations reside in the spike gene (~30 substitutions, 6 residue deletions, 3 residue insertions); other mutations dominate the RBD and NTD. W152R, F157L, I210V, G257S, D339H, G446S, N460K, Q493 Δ69–70 deletion, L452R, F486V, and R493Q N460K, S486P, F490S E180V, T478R, F486P K444T, L452R, N460K, and F486V Y273H (NSP12), N268S (NSP13) | High infectivity and transmissibility; most dominant strain. BA.4 and BA.5 more infectious than BA.2. Antibody evasion and enhanced viral fitness. | Insufficient neutralization of Omicron antibodies from current COVID-19 vaccines. Enhanced neutralization resistance Resistant to the monoclonal antibody drug therapy |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andre, M.; Lau, L.-S.; Pokharel, M.D.; Ramelow, J.; Owens, F.; Souchak, J.; Akkaoui, J.; Ales, E.; Brown, H.; Shil, R.; et al. From Alpha to Omicron: How Different Variants of Concern of the SARS-Coronavirus-2 Impacted the World. Biology 2023, 12, 1267. https://doi.org/10.3390/biology12091267
Andre M, Lau L-S, Pokharel MD, Ramelow J, Owens F, Souchak J, Akkaoui J, Ales E, Brown H, Shil R, et al. From Alpha to Omicron: How Different Variants of Concern of the SARS-Coronavirus-2 Impacted the World. Biology. 2023; 12(9):1267. https://doi.org/10.3390/biology12091267
Chicago/Turabian StyleAndre, Mickensone, Lee-Seng Lau, Marissa D. Pokharel, Julian Ramelow, Florida Owens, Joseph Souchak, Juliet Akkaoui, Evan Ales, Harry Brown, Rajib Shil, and et al. 2023. "From Alpha to Omicron: How Different Variants of Concern of the SARS-Coronavirus-2 Impacted the World" Biology 12, no. 9: 1267. https://doi.org/10.3390/biology12091267





