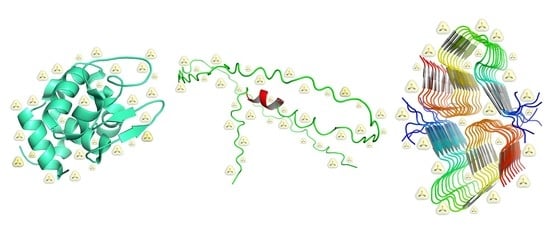
The Action of Chemical Denaturants: From Globular to Intrinsically Disordered Proteins
Abstract
:Simple Summary
Abstract
1. Introduction
2. Molecular Mechanism(s) of Chemical Denaturation of Globular Proteins
3. Denatured and Native States in the Polymer Physics Perspective
4. Intrinsically Disordered Proteins: Conformational Preferences in Different Contexts
Effects of Chemical Denaturants on IDPs
5. Effects of Chemical Denaturants on Amyloid-Like Aggregates
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tompa, P. Intrinsically Unstructured Proteins Evolve by Repeat Expansion. BioEssays 2003, 25, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically Disordered Proteins and Their Environment: Effects of Strong Denaturants, Temperature, PH, Counter Ions, Membranes, Binding Partners, Osmolytes, and Macromolecular Crowding. Protein J. 2009, 28, 305–325. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Hughes, M.P.; Rodriguez, J.A.; Riek, R.; Eisenberg, D.S. The Expanding Amyloid Family: Structure, Stability, Function, and Pathogenesis. Cell 2021, 184, 4857–4873. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P. Intrinsically Unstructured Proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Babu, M.M.; Barbar, E.; Blackledge, M.; Bondos, S.E.; Dosztányi, Z.; Dyson, H.J.; Forman-Kay, J.; Fuxreiter, M.; Gsponer, J.; et al. What’s in a Name? Why These Proteins Are Intrinsically Disordered: Why These Proteins Are Intrinsically Disordered. Intrinsically Disord. Proteins 2013, 1, e24157. [Google Scholar] [CrossRef] [PubMed]
- Quaglia, F.; Mészáros, B.; Salladini, E.; Hatos, A.; Pancsa, R.; Chemes, L.B.; Pajkos, M.; Lazar, T.; Peña-Díaz, S.; Santos, J.; et al. DisProt in 2022: Improved Quality and Accessibility of Protein Intrinsic Disorder Annotation. Nucleic Acids Res. 2022, 50, D480–D487. [Google Scholar] [CrossRef]
- Otzen, D.; Riek, R. Functional Amyloids. Cold Spring Harb. Perspect. Biol. 2019, 11, a033860. [Google Scholar] [CrossRef]
- Bhatia, S.; Udgaonkar, J.B. Heterogeneity in Protein Folding and Unfolding Reactions. Chem. Rev. 2022, 122, 8911–8935. [Google Scholar] [CrossRef]
- Dill, K.A.; MacCallum, J.L. The Protein-Folding Problem, 50 Years On. Science 2012, 338, 1042–1046. [Google Scholar] [CrossRef]
- Gianni, S.; Dogan, J.; Jemth, P. Coupled Binding and Folding of Intrinsically Disordered Proteins: What Can We Learn from Kinetics? Curr. Opin. Struct. Biol. 2016, 36, 18–24. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Lazar, T.; Martínez-Pérez, E.; Quaglia, F.; Hatos, A.; Chemes, L.B.; Iserte, J.A.; Méndez, N.A.; Garrone, N.A.; Saldaño, T.E.; Marchetti, J.; et al. PED in 2021: A Major Update of the Protein Ensemble Database for Intrinsically Disordered Proteins. Nucleic Acids Res. 2021, 49, D404–D411. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhang, M.; Wang, Y.; Ma, R.; Guo, C.; Feng, L.; Wu, J.; Yao, H.; Lin, D. Structural Basis for the Complete Resistance of the Human Prion Protein Mutant G127V to Prion Disease. Sci. Rep. 2018, 8, 13211. [Google Scholar] [CrossRef] [PubMed]
- Makhatadze, G.I.; Privalov, P.L. Energetics of Protein Structure. In Advances in Protein Chemistry; Elsevier: Amsterdam, The Netherlands, 1995; Volume 47, pp. 307–425. ISBN 978-0-12-034247-1. [Google Scholar]
- Robertson, A.D.; Murphy, K.P. Protein Structure and the Energetics of Protein Stability. Chem. Rev. 1997, 97, 1251–1268. [Google Scholar] [CrossRef] [PubMed]
- Rees, D.C.; Robertson, A.D. Some Thermodynamic Implications for the Thermostability of Proteins. Protein Sci. 2001, 10, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Sawle, L.; Ghosh, K. How Do Thermophilic Proteins and Proteomes Withstand High Temperature? Biophys. J. 2011, 101, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Pauling, L. The Nature of the Chemical Bond, 3rd ed.; The Cornell University Press: Ithaca, NY, USA, 1960; pp. 10–13. [Google Scholar]
- Shortle, D. The Denatured State (the Other Half of the Folding Equation) and Its Role in Protein Stability. FASEB J. 1996, 10, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, K.; Tanford, C. Viscosity and Density of Aqueous Solutions of Urea and Guanidine Hydrochloride. J. Biol. Chem. 1966, 241, 3228–3232. [Google Scholar] [CrossRef]
- Kumar, A. Aqueous Guanidinium Salts: Part I. Densities, Ultrasonic Velocities, and Apparent Molar Properties. J. Solut. Chem. 2001, 30, 281–290. [Google Scholar] [CrossRef]
- Halonen, S.; Kangas, T.; Haataja, M.; Lassi, U. Urea-Water-Solution Properties: Density, Viscosity, and Surface Tension in an Under-Saturated Solution. Emiss. Control Sci. Technol. 2017, 3, 161–170. [Google Scholar] [CrossRef]
- Breslow, R.; Guo, T. Surface Tension Measurements Show That Chaotropic Salting-in Denaturants Are Not Just Water-Structure Breakers. Proc. Natl. Acad. Sci. USA 1990, 87, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Soper, A.K.; Castner, E.W.; Luzar, A. Impact of Urea on Water Structure: A Clue to Its Properties as a Denaturant? Biophys. Chem. 2003, 105, 649–666. [Google Scholar] [CrossRef] [PubMed]
- Mason, P.E.; Neilson, G.W.; Enderby, J.E.; Saboungi, M.-L.; Dempsey, C.E.; MacKerell, A.D.; Brady, J.W. The Structure of Aqueous Guanidinium Chloride Solutions. J. Am. Chem. Soc. 2004, 126, 11462–11470. [Google Scholar] [CrossRef]
- Graziano, G. Contrasting the Denaturing Effect of Guanidinium Chloride with the Stabilizing Effect of Guanidinium Sulfate. Phys. Chem. Chem. Phys. 2011, 13, 12008. [Google Scholar] [CrossRef] [PubMed]
- Graziano, G. How Does Trimethylamine N-Oxide Counteract the Denaturing Activity of Urea? Phys. Chem. Chem. Phys. 2011, 13, 17689. [Google Scholar] [CrossRef] [PubMed]
- Graziano, G. On the Solubility of Aliphatic Hydrocarbons in 7 M Aqueous Urea. J. Phys. Chem. B 2001, 105, 2632–2637. [Google Scholar] [CrossRef]
- Trzesniak, D.; van der Vegt, N.F.A.; van Gunsteren, W.F. Computer Simulation Studies on the Solvation of Aliphatic Hydrocarbons in 6.9 M Aqueous Urea Solution. Phys. Chem. Chem. Phys. 2004, 6, 697. [Google Scholar] [CrossRef]
- Weerasinghe, S.; Smith, P.E. A Kirkwood−Buff Derived Force Field for Mixtures of Urea and Water. J. Phys. Chem. B 2003, 107, 3891–3898. [Google Scholar] [CrossRef]
- Weerasinghe, S.; Smith, P.E. A Kirkwood-Buff Derived Force Field for the Simulation of Aqueous Guanidinium Chloride Solutions. J. Chem. Phys. 2004, 121, 2180–2186. [Google Scholar] [CrossRef]
- Zangi, R.; Zhou, R.; Berne, B.J. Urea’s Action on Hydrophobic Interactions. J. Am. Chem. Soc. 2009, 131, 1535–1541. [Google Scholar] [CrossRef]
- Zheng, W.; Borgia, A.; Borgia, M.B.; Schuler, B.; Best, R.B. Empirical Optimization of Interactions between Proteins and Chemical Denaturants in Molecular Simulations. J. Chem. Theory Comput. 2015, 11, 5543–5553. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, S.; Balasco, N.; Vigorita, M.; Ruggiero, A.; Smaldone, G.; Del Vecchio, P.; Vitagliano, L.; Graziano, G. Guanidinium Binding to Proteins: The Intriguing Effects on the D1 and D2 Domains of Thermotoga Maritima Arginine Binding Protein and a Comprehensive Analysis of the Protein Data Bank. Int. J. Biol. Macromol. 2020, 163, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Paladino, A.; Balasco, N.; Graziano, G.; Vitagliano, L. A Protein Data Bank Survey of Multimodal Binding of Thiocyanate to Proteins: Evidence for Thiocyanate Promiscuity. Int. J. Biol. Macromol. 2022, 208, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Paladino, A.; Balasco, N.; Vitagliano, L.; Graziano, G. A Structure-Based Mechanism for the Denaturing Action of Urea, Guanidinium Ion and Thiocyanate Ion. Biology 2022, 11, 1764. [Google Scholar] [CrossRef]
- Makhatadze, G.I.; Privalov, P.L. Protein Interactions with Urea and Guanidinium Chloride. J. Mol. Biol. 1992, 226, 491–505. [Google Scholar] [CrossRef]
- Schellman, J.A. The Thermodynamics of Solvent Exchange. Biopolymers 1994, 34, 1015–1026. [Google Scholar] [CrossRef]
- Schellman, J.A. Protein Stability in Mixed Solvents: A Balance of Contact Interaction and Excluded Volume. Biophys. J. 2003, 85, 108–125. [Google Scholar] [CrossRef]
- Rembert, K.B.; Paterová, J.; Heyda, J.; Hilty, C.; Jungwirth, P.; Cremer, P.S. Molecular Mechanisms of Ion-Specific Effects on Proteins. J. Am. Chem. Soc. 2012, 134, 10039–10046. [Google Scholar] [CrossRef]
- Courtenay, E.S.; Capp, M.W.; Record, M.T. Thermodynamics of Interactions of Urea and Guanidinium Salts with Protein Surface: Relationship between Solute Effects on Protein Processes and Changes in Water-Accessible Surface Area. Protein Sci. 2009, 10, 2485–2497. [Google Scholar] [CrossRef]
- Record, M.T.; Guinn, E.; Pegram, L.; Capp, M. Introductory Lecture: Interpreting and Predicting Hofmeister Salt Ion and Solute Effects on Biopolymer and Model Processes Using the Solute Partitioning Model. Faraday Discuss 2013, 160, 9–44. [Google Scholar] [CrossRef]
- Pegram, L.M.; Record, M.T. Thermodynamic Origin of Hofmeister Ion Effects. J. Phys. Chem. B 2008, 112, 9428–9436. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved Side-Chain Torsion Potentials for the Amber Ff99SB Protein Force Field: Improved Protein Side-Chain Potentials. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L. Monte Carlo Simulation of n-butane in Water. Conformational Evidence for the Hydrophobic Effect. J. Chem. Phys. 1982, 77, 5757–5765. [Google Scholar] [CrossRef]
- O’Brien, E.P.; Dima, R.I.; Brooks, B.; Thirumalai, D. Interactions between Hydrophobic and Ionic Solutes in Aqueous Guanidinium Chloride and Urea Solutions: Lessons for Protein Denaturation Mechanism. J. Am. Chem. Soc. 2007, 129, 7346–7353. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.P.; Ziv, G.; Haran, G.; Brooks, B.R.; Thirumalai, D. Effects of Denaturants and Osmolytes on Proteins Are Accurately Predicted by the Molecular Transfer Model. Proc. Natl. Acad. Sci. USA 2008, 105, 13403–13408. [Google Scholar] [CrossRef]
- Liu, Z.; Reddy, G.; Thirumalai, D. Theory of the Molecular Transfer Model for Proteins with Applications to the Folding of the Src-SH3 Domain. J. Phys. Chem. B 2012, 116, 6707–6716. [Google Scholar] [CrossRef]
- Liu, Z.; Reddy, G.; Thirumalai, D. Folding PDZ2 Domain Using the Molecular Transfer Model. J. Phys. Chem. B 2016, 120, 8090–8101. [Google Scholar] [CrossRef]
- Klimov, D.K.; Thirumalai, D. Mechanisms and Kinetics of β-Hairpin Formation. Proc. Natl. Acad. Sci. USA 2000, 97, 2544–2549. [Google Scholar] [CrossRef]
- Tanford, C. Protein Denaturation. In Advances in Protein Chemistry; Elsevier: Amsterdam, The Netherlands, 1968; Volume 23, pp. 121–282. ISBN 978-0-12-034223-5. [Google Scholar]
- Flory, P.J. Principles of Polymer Chemistry; The Cornell University Press: Ithaca, NY, USA, 1953. [Google Scholar]
- Chan, H.S.; Dill, K.A. Polymer Principles in Protein Structure and Stability. Annu. Rev. Biophys. Biophys. Chem. 1991, 20, 447–490. [Google Scholar] [CrossRef]
- Clark, P.L.; Plaxco, K.W.; Sosnick, T.R. Water as a Good Solvent for Unfolded Proteins: Folding and Collapse Are Fundamentally Different. J. Mol. Biol. 2020, 432, 2882–2889. [Google Scholar] [CrossRef] [PubMed]
- Graziano, G. On the Mechanism of Cold Denaturation. Phys. Chem. Chem. Phys. 2014, 16, 21755–21767. [Google Scholar] [CrossRef] [PubMed]
- Merlino, A.; Pontillo, N.; Graziano, G. A Driving Force for Polypeptide and Protein Collapse. Phys. Chem. Chem. Phys. 2017, 19, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Graziano, G. Is Water a Good Solvent for the Denatured State of Globular Proteins? Chem. Phys. Lett. 2020, 759, 137949. [Google Scholar] [CrossRef]
- Dima, R.I.; Thirumalai, D. Asymmetry in the Shapes of Folded and Denatured States of Proteins. J. Phys. Chem. B 2004, 108, 6564–6570. [Google Scholar] [CrossRef]
- Holehouse, A.S.; Pappu, R.V. Collapse Transitions of Proteins and the Interplay Among Backbone, Sidechain, and Solvent Interactions. Annu. Rev. Biophys. 2018, 47, 19–39. [Google Scholar] [CrossRef]
- Best, R.B. Emerging Consensus on the Collapse of Unfolded and Intrinsically Disordered Proteins in Water. Curr. Opin. Struct. Biol. 2020, 60, 27–38. [Google Scholar] [CrossRef]
- Borgia, A.; Zheng, W.; Buholzer, K.; Borgia, M.B.; Schüler, A.; Hofmann, H.; Soranno, A.; Nettels, D.; Gast, K.; Grishaev, A.; et al. Consistent View of Polypeptide Chain Expansion in Chemical Denaturants from Multiple Experimental Methods. J. Am. Chem. Soc. 2016, 138, 11714–11726. [Google Scholar] [CrossRef]
- Zheng, W.; Borgia, A.; Buholzer, K.; Grishaev, A.; Schuler, B.; Best, R.B. Probing the Action of Chemical Denaturant on an Intrinsically Disordered Protein by Simulation and Experiment. J. Am. Chem. Soc. 2016, 138, 11702–11713. [Google Scholar] [CrossRef]
- Yu, L.; Brüschweiler, R. Quantitative Prediction of Ensemble Dynamics, Shapes and Contact Propensities of Intrinsically Disordered Proteins. PLoS Comput. Biol. 2022, 18, e1010036. [Google Scholar] [CrossRef]
- Merchant, K.A.; Best, R.B.; Louis, J.M.; Gopich, I.V.; Eaton, W.A. Characterizing the Unfolded States of Proteins Using Single-Molecule FRET Spectroscopy and Molecular Simulations. Proc. Natl. Acad. Sci. USA 2007, 104, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zerze, G.H.; Borgia, A.; Mittal, J.; Schuler, B.; Best, R.B. Inferring Properties of Disordered Chains from FRET Transfer Efficiencies. J. Chem. Phys. 2018, 148, 123329. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, I.C. Phase Transition Behavior of the Isolated Polymer Chain. Macromolecules 1979, 12, 980–988. [Google Scholar] [CrossRef]
- Hofmann, H.; Soranno, A.; Borgia, A.; Gast, K.; Nettels, D.; Schuler, B. Polymer Scaling Laws of Unfolded and Intrinsically Disordered Proteins Quantified with Single-Molecule Spectroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 16155–16160. [Google Scholar] [CrossRef]
- Aznauryan, M.; Delgado, L.; Soranno, A.; Nettels, D.; Huang, J.; Labhardt, A.M.; Grzesiek, S.; Schuler, B. Comprehensive Structural and Dynamical View of an Unfolded Protein from the Combination of Single-Molecule FRET, NMR, and SAXS. Proc. Natl. Acad. Sci. USA 2004, 108, 6564–6570. [Google Scholar] [CrossRef]
- O’Brien, E.P.; Morrison, G.; Brooks, B.R.; Thirumalai, D. How Accurate Are Polymer Models in the Analysis of Förster Resonance Energy Transfer Experiments on Proteins? J. Chem. Phys. 2009, 130, 124903. [Google Scholar] [CrossRef]
- Kohn, J.E.; Millett, I.S.; Jacob, J.; Zagrovic, B.; Dillon, T.M.; Cingel, N.; Dothager, R.S.; Seifert, S.; Thiyagarajan, P.; Sosnick, T.R.; et al. Random-Coil Behavior and the Dimensions of Chemically Unfolded Proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 12491–12496. [Google Scholar] [CrossRef]
- Narayan, A.; Bhattacharjee, K.; Naganathan, A.N. Thermally versus Chemically Denatured Protein States. Biochemistry 2019, 58, 2519–2523. [Google Scholar] [CrossRef]
- Nettels, D.; Müller-Späth, S.; Küster, F.; Hofmann, H.; Haenni, D.; Rüegger, S.; Reymond, L.; Hoffmann, A.; Kubelka, J.; Heinz, B.; et al. Single-Molecule Spectroscopy of the Temperature-Induced Collapse of Unfolded Proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 20740–20745. [Google Scholar] [CrossRef]
- Tompa, P. Intrinsically Disordered Proteins: A 10-Year Recap. Trends Biochem. Sci. 2012, 37, 509–516. [Google Scholar] [CrossRef]
- Turoverov, K.K.; Kuznetsova, I.M.; Fonin, A.V.; Darling, A.L.; Zaslavsky, B.Y.; Uversky, V.N. Stochasticity of Biological Soft Matter: Emerging Concepts in Intrinsically Disordered Proteins and Biological Phase Separation. Trends Biochem. Sci. 2019, 44, 716–728. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically Disordered Proteins and Their “Mysterious” (Meta)Physics. Front. Phys. 2019, 7, 10. [Google Scholar] [CrossRef]
- Forman-Kay, J.D.; Mittag, T. From Sequence and Forces to Structure, Function, and Evolution of Intrinsically Disordered Proteins. Structure 2013, 21, 1492–1499. [Google Scholar] [CrossRef] [PubMed]
- Das, R.K.; Ruff, K.M.; Pappu, R.V. Relating Sequence Encoded Information to Form and Function of Intrinsically Disordered Proteins. Curr. Opin. Struct. Biol. 2015, 32, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P.; Bhattacharya, S.; Achuthan, S.; Behal, A.; Jolly, M.K.; Kotnala, S.; Mohanty, A.; Rangarajan, G.; Salgia, R.; Uversky, V. Intrinsically Disordered Proteins: Critical Components of the Wetware. Chem. Rev. 2022, 122, 6614–6633. [Google Scholar] [CrossRef] [PubMed]
- Gomes, G.-N.W.; Krzeminski, M.; Namini, A.; Martin, E.W.; Mittag, T.; Head-Gordon, T.; Forman-Kay, J.D.; Gradinaru, C.C. Conformational Ensembles of an Intrinsically Disordered Protein Consistent with NMR, SAXS, and Single-Molecule FRET. J. Am. Chem. Soc. 2020, 142, 15697–15710. [Google Scholar] [CrossRef]
- Shrestha, U.R.; Smith, J.C.; Petridis, L. Full Structural Ensembles of Intrinsically Disordered Proteins from Unbiased Molecular Dynamics Simulations. Commun. Biol. 2021, 4, 243. [Google Scholar] [CrossRef]
- Milles, S.; Salvi, N.; Blackledge, M.; Jensen, M.R. Characterization of Intrinsically Disordered Proteins and Their Dynamic Complexes: From in Vitro to Cell-like Environments. Prog. Nucl. Magn. Reson. Spectrosc. 2018, 109, 79–100. [Google Scholar] [CrossRef]
- Ozenne, V.; Bauer, F.; Salmon, L.; Huang, J.; Jensen, M.R.; Segard, S.; Bernadó, P.; Charavay, C.; Blackledge, M. Flexible-Meccano: A Tool for the Generation of Explicit Ensemble Descriptions of Intrinsically Disordered Proteins and Their Associated Experimental Observables. Bioinformatics 2012, 28, 1463–1470. [Google Scholar] [CrossRef]
- Allison, J.R.; Varnai, P.; Dobson, C.M.; Vendruscolo, M. Determination of the Free Energy Landscape of α-Synuclein Using Spin Label Nuclear Magnetic Resonance Measurements. J. Am. Chem. Soc. 2009, 131, 18314–18326. [Google Scholar] [CrossRef]
- Ganguly, D.; Chen, J. Structural Interpretation of Paramagnetic Relaxation Enhancement-Derived Distances for Disordered Protein States. J. Mol. Biol. 2009, 390, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Silvestre-Ryan, J.; Bertoncini, C.W.; Fenwick, R.B.; Esteban-Martin, S.; Salvatella, X. Average Conformations Determined from PRE Data Provide High-Resolution Maps of Transient Tertiary Interactions in Disordered Proteins. Biophys. J. 2013, 104, 1740–1751. [Google Scholar] [CrossRef] [PubMed]
- Schwalbe, M.; Ozenne, V.; Bibow, S.; Jaremko, M.; Jaremko, L.; Gajda, M.; Jensen, M.R.; Biernat, J.; Becker, S.; Mandelkow, E.; et al. Predictive Atomic Resolution Descriptions of Intrinsically Disordered HTau40 and α-Synuclein in Solution from NMR and Small Angle Scattering. Structure 2014, 22, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.-K.; Nodet, G.; Kim, H.-Y.; Jensen, M.R.; Bernado, P.; Fernandez, C.O.; Becker, S.; Blackledge, M.; Zweckstetter, M. Structural Characterization of α-Synuclein in an Aggregation Prone State: α-Synuclein in an Aggregation-Prone State. Protein Sci. 2009, 18, 1840–1846. [Google Scholar] [CrossRef]
- Bibow, S.; Ozenne, V.; Biernat, J.; Blackledge, M.; Mandelkow, E.; Zweckstetter, M. Structural Impact of Proline-Directed Pseudophosphorylation at AT8, AT100, and PHF1 Epitopes on 441-Residue Tau. J. Am. Chem. Soc. 2011, 133, 15842–15845. [Google Scholar] [CrossRef]
- Sibille, N.; Bernadó, P. Structural Characterization of Intrinsically Disordered Proteins by the Combined Use of NMR and SAXS. Biochem. Soc. Trans. 2012, 40, 955–962. [Google Scholar] [CrossRef]
- Cordeiro, T.N.; Herranz-Trillo, F.; Urbanek, A.; Estaña, A.; Cortés, J.; Sibille, N.; Bernadó, P. Small-Angle Scattering Studies of Intrinsically Disordered Proteins and Their Complexes. Curr. Opin. Struct. Biol. 2017, 42, 15–23. [Google Scholar] [CrossRef]
- Naudi-Fabra, S.; Tengo, M.; Jensen, M.R.; Blackledge, M.; Milles, S. Quantitative Description of Intrinsically Disordered Proteins Using Single-Molecule FRET, NMR, and SAXS. J. Am. Chem. Soc. 2021, 143, 20109–20121. [Google Scholar] [CrossRef]
- Shea, J.-E.; Best, R.B.; Mittal, J. Physics-Based Computational and Theoretical Approaches to Intrinsically Disordered Proteins. Curr. Opin. Struct. Biol. 2021, 67, 219–225. [Google Scholar] [CrossRef]
- Mercadante, D.; Wagner, J.A.; Aramburu, I.V.; Lemke, E.A.; Gräter, F. Sampling Long- versus Short-Range Interactions Defines the Ability of Force Fields to Reproduce the Dynamics of Intrinsically Disordered Proteins. J. Chem. Theory Comput. 2017, 13, 3964–3974. [Google Scholar] [CrossRef]
- Robustelli, P.; Piana, S.; Shaw, D.E. Developing a Molecular Dynamics Force Field for Both Folded and Disordered Protein States. Proc. Natl. Acad. Sci. USA 2018, 115, E4758–E4766. [Google Scholar] [CrossRef] [PubMed]
- Pesce, F.; Lindorff-Larsen, K. Refining Conformational Ensembles of Flexible Proteins against Small-Angle X-ray Scattering Data. Biophys. J. 2022, 121, 1576–1579. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Benton, L.A.; Singh, V.; Pielak, G.J. Disordered Protein Diffusion under Crowded Conditions. J. Phys. Chem. Lett. 2012, 3, 2703–2706. [Google Scholar] [CrossRef] [PubMed]
- Szasz, C.; Alexa, A.; Toth, K.; Rakacs, M.; Langowski, J.; Tompa, P. Protein Disorder Prevails under Crowded Conditions. Biochemistry 2011, 50, 5834–5844. [Google Scholar] [CrossRef]
- Soranno, A.; Koenig, I.; Borgia, M.B.; Hofmann, H.; Zosel, F.; Nettels, D.; Schuler, B. Single-Molecule Spectroscopy Reveals Polymer Effects of Disordered Proteins in Crowded Environments. Proc. Natl. Acad. Sci. USA 2014, 111, 4874–4879. [Google Scholar] [CrossRef]
- Zosel, F.; Soranno, A.; Buholzer, K.J.; Nettels, D.; Schuler, B. Depletion Interactions Modulate the Binding between Disordered Proteins in Crowded Environments. Proc. Natl. Acad. Sci. USA 2020, 117, 13480–13489. [Google Scholar] [CrossRef]
- Shillcock, J.C.; Thomas, D.B.; Ipsen, J.H.; Brown, A.D. Macromolecular Crowding Is Surprisingly Unable to Deform the Structure of a Model Biomolecular Condensate. Biology 2023, 12, 181. [Google Scholar] [CrossRef]
- Speer, S.L.; Stewart, C.J.; Sapir, L.; Harries, D.; Pielak, G.J. Macromolecular Crowding Is More than Hard-Core Repulsions. Annu. Rev. Biophys. 2022, 51, 267–300. [Google Scholar] [CrossRef]
- Fagerberg, E.; Lenton, S.; Nylander, T.; Seydel, T.; Skepö, M. Self-Diffusive Properties of the Intrinsically Disordered Protein Histatin 5 and the Impact of Crowding Thereon: A Combined Neutron Spectroscopy and Molecular Dynamics Simulation Study. J. Phys. Chem. B 2022, 126, 789–801. [Google Scholar] [CrossRef]
- Moses, D.; Yu, F.; Ginell, G.M.; Shamoon, N.M.; Koenig, P.S.; Holehouse, A.S.; Sukenik, S. Revealing the Hidden Sensitivity of Intrinsically Disordered Proteins to Their Chemical Environment. J. Phys. Chem. Lett. 2020, 11, 10131–10136. [Google Scholar] [CrossRef]
- Vancraenenbroeck, R.; Harel, Y.S.; Zheng, W.; Hofmann, H. Polymer Effects Modulate Binding Affinities in Disordered Proteins. Proc. Natl. Acad. Sci. USA 2019, 116, 19506–19512. [Google Scholar] [CrossRef] [PubMed]
- Longo, L.M.; Blaber, M. Prebiotic Protein Design Supports a Halophile Origin of Foldable Proteins. Front. Microbiol. 2014, 4, 418. [Google Scholar] [CrossRef] [PubMed]
- Reddy, G.; Thirumalai, D. Collapse Precedes Folding in Denaturant-Dependent Assembly of Ubiquitin. J. Phys. Chem. B 2017, 121, 995–1009. [Google Scholar] [CrossRef] [PubMed]
- Riback, J.A.; Bowman, M.A.; Zmyslowski, A.M.; Knoverek, C.R.; Jumper, J.M.; Hinshaw, J.R.; Kaye, E.B.; Freed, K.F.; Clark, P.L.; Sosnick, T.R. Innovative Scattering Analysis Shows That Hydrophobic Disordered Proteins Are Expanded in Water. Science 2017, 358, 238–241. [Google Scholar] [CrossRef]
- Yoo, T.Y.; Meisburger, S.P.; Hinshaw, J.; Pollack, L.; Haran, G.; Sosnick, T.R.; Plaxco, K. Small-Angle X-Ray Scattering and Single-Molecule FRET Spectroscopy Produce Highly Divergent Views of the Low-Denaturant Unfolded State. J. Mol. Biol. 2012, 418, 226–236. [Google Scholar] [CrossRef]
- Watkins, H.M.; Simon, A.J.; Sosnick, T.R.; Lipman, E.A.; Hjelm, R.P.; Plaxco, K.W. Random Coil Negative Control Reproduces the Discrepancy between Scattering and FRET Measurements of Denatured Protein Dimensions. Proc. Natl. Acad. Sci. USA 2015, 112, 6631–6636. [Google Scholar] [CrossRef]
- Abascal, J.L.F.; Vega, C. A General Purpose Model for the Condensed Phases of Water: TIP4P/2005. J. Chem. Phys. 2005, 123, 234505. [Google Scholar] [CrossRef]
- Best, R.B. Computational and Theoretical Advances in Studies of Intrinsically Disordered Proteins. Curr. Opin. Struct. Biol. 2017, 42, 147–154. [Google Scholar] [CrossRef]
- Fuertes, G.; Banterle, N.; Ruff, K.M.; Chowdhury, A.; Mercadante, D.; Koehler, C.; Kachala, M.; Estrada Girona, G.; Milles, S.; Mishra, A.; et al. Decoupling of Size and Shape Fluctuations in Heteropolymeric Sequences Reconciles Discrepancies in SAXS vs. FRET Measurements. Proc. Natl. Acad. Sci. USA 2017, 114, E6342–E6351. [Google Scholar] [CrossRef]
- Song, J.; Gomes, G.-N.; Gradinaru, C.C.; Chan, H.S. An Adequate Account of Excluded Volume Is Necessary To Infer Compactness and Asphericity of Disordered Proteins by Förster Resonance Energy Transfer. J. Phys. Chem. B 2015, 119, 15191–15202. [Google Scholar] [CrossRef]
- Ke, P.C.; Zhou, R.; Serpell, L.C.; Riek, R.; Knowles, T.P.J.; Lashuel, H.A.; Gazit, E.; Hamley, I.W.; Davis, T.P.; Fändrich, M.; et al. Half a Century of Amyloids: Past, Present and Future. Chem. Soc. Rev. 2020, 49, 5473–5509. [Google Scholar] [CrossRef] [PubMed]
- Jahn, T.R.; Makin, O.S.; Morris, K.L.; Marshall, K.E.; Tian, P.; Sikorski, P.; Serpell, L.C. The Common Architecture of Cross-β Amyloid. J. Mol. Biol. 2010, 395, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.I.P.; Staniforth, R.A. General Principles Underpinning Amyloid Structure. Front. Neurosci. 2022, 16, 878869. [Google Scholar] [CrossRef] [PubMed]
- Reches, M.; Gazit, E. Casting Metal Nanowires Within Discrete Self-Assembled Peptide Nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef] [PubMed]
- Balasco, N.; Diaferia, C.; Morelli, G.; Vitagliano, L.; Accardo, A. Amyloid-Like Aggregation in Diseases and Biomaterials: Osmosis of Structural Information. Front. Bioeng. Biotechnol. 2021, 9, 641372. [Google Scholar] [CrossRef]
- Amit, M.; Yuran, S.; Gazit, E.; Reches, M.; Ashkenasy, N. Tailor-Made Functional Peptide Self-Assembling Nanostructures. Adv. Mater. 2018, 30, 1707083. [Google Scholar] [CrossRef]
- Buell, A.K.; Dobson, C.M.; Knowles, T.P.J. The Physical Chemistry of the Amyloid Phenomenon: Thermodynamics and Kinetics of Filamentous Protein Aggregation. Essays Biochem. 2014, 56, 11–39. [Google Scholar] [CrossRef]
- Baldwin, A.J.; Knowles, T.P.J.; Tartaglia, G.G.; Fitzpatrick, A.W.; Devlin, G.L.; Shammas, S.L.; Waudby, C.A.; Mossuto, M.F.; Meehan, S.; Gras, S.L.; et al. Metastability of Native Proteins and the Phenomenon of Amyloid Formation. J. Am. Chem. Soc. 2011, 133, 14160–14163. [Google Scholar] [CrossRef]
- Michaels, T.C.T.; Šarić, A.; Habchi, J.; Chia, S.; Meisl, G.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P.J. Chemical Kinetics for Bridging Molecular Mechanisms and Macroscopic Measurements of Amyloid Fibril Formation. Annu. Rev. Phys. Chem. 2018, 69, 273–298. [Google Scholar] [CrossRef]
- Arosio, P.; Knowles, T.P.J.; Linse, S. On the Lag Phase in Amyloid Fibril Formation. Phys. Chem. Chem. Phys. 2015, 17, 7606–7618. [Google Scholar] [CrossRef]
- Weiffert, T.; Meisl, G.; Curk, S.; Cukalevski, R.; Šarić, A.; Knowles, T.P.J.; Linse, S. Influence of Denaturants on Amyloid Β42 Aggregation Kinetics. Front. Neurosci. 2022, 16, 943355. [Google Scholar] [CrossRef]
- Khan, M.A.I.; Respondek, M.; Kjellström, S.; Deep, S.; Linse, S.; Akke, M. Cu/Zn Superoxide Dismutase Forms Amyloid Fibrils under Near-Physiological Quiescent Conditions: The Roles of Disulfide Bonds and Effects of Denaturant. ACS Chem. Neurosci. 2017, 8, 2019–2026. [Google Scholar] [CrossRef] [PubMed]
- Hamada, D.; Dobson, C.M. A Kinetic Study of β-Lactoglobulin Amyloid Fibril Formation Promoted by Urea. Protein Sci. 2009, 11, 2417–2426. [Google Scholar] [CrossRef] [PubMed]
- Vernaglia, B.A.; Huang, J.; Clark, E.D. Guanidine Hydrochloride Can Induce Amyloid Fibril Formation from Hen Egg-White Lysozyme. Biomacromolecules 2004, 5, 1362–1370. [Google Scholar] [CrossRef] [PubMed]
- Dueholm, M.S.; Petersen, S.V.; Sønderkaer, M.; Larsen, P.; Christiansen, G.; Hein, K.L.; Enghild, J.J.; Nielsen, J.L.; Nielsen, K.L.; Nielsen, P.H.; et al. Functional Amyloid in Pseudomonas: Functional Amyloid in Pseudomonas. Mol. Microbiol. 2010, 77, 1009–1020. [Google Scholar] [CrossRef]
- Buell, A.K. Stability Matters, Too–the Thermodynamics of Amyloid Fibril Formation. Chem. Sci. 2022, 13, 10177–10192. [Google Scholar] [CrossRef]
- Sulatsky, M.I.; Sulatskaya, A.I.; Stepanenko, O.V.; Povarova, O.I.; Kuznetsova, I.M.; Turoverov, K.K. Denaturant Effect on Amyloid Fibrils: Declasterization, Depolymerization, Denaturation and Reassembly. Int. J. Biol. Macromol. 2020, 150, 681–694. [Google Scholar] [CrossRef]
- Vettore, N.; Buell, A.K. Thermodynamics of Amyloid Fibril Formation from Chemical Depolymerization. Phys. Chem. Chem. Phys. 2019, 21, 26184–26194. [Google Scholar] [CrossRef]
- Chen, S.; Berthelier, V.; Hamilton, J.B.; O’Nuallai, B.; Wetzel, R. Amyloid-like Features of Polyglutamine Aggregates and Their Assembly Kinetics. Biochemistry 2002, 41, 7391–7399. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Sambashivan, S.; Nelson, R.; Ivanova, M.I.; Sievers, S.A.; Apostol, M.I.; Thompson, M.J.; Balbirnie, M.; Wiltzius, J.J.W.; McFarlane, H.T.; et al. Atomic Structures of Amyloid Cross-β Spines Reveal Varied Steric Zippers. Nature 2007, 447, 453–457. [Google Scholar] [CrossRef]
- Pesce, F.; Newcombe, E.A.; Seiffert, P.; Tranchant, E.E.; Olsen, J.G.; Grace, C.R.; Kragelund, B.B.; Lindorff-Larsen, K. Assessment of Models for Calculating the Hydrodynamic Radius of Intrinsically Disordered Proteins. Biophys. J. 2023, 122, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Thomasen, F.E.; Pesce, F.; Roesgaard, M.A.; Tesei, G.; Lindorff-Larsen, K. Improving Martini 3 for Disordered and Multidomain Proteins. J. Chem. Theory Comput. 2022, 18, 2033–2041. [Google Scholar] [CrossRef] [PubMed]




| ρ g L−1 | [H2O] M | ξ3 | γ Dyne cm−1 | |
|---|---|---|---|---|
| H2O | 997 | 55.3 | 0.383 | 72.8 |
| 2 M urea | 1029 | 50.4 | 0.413 | 72.9 |
| 4 M urea | 1059 | 45.4 | 0.442 | 73.3 |
| 6 M urea | 1088 | 40.4 | 0.471 | 74.4 |
| 2 M GdmCl | 1047 | 47.5 | 0.424 | 74.0 |
| 4 M GdmCl | 1095 | 39.6 | 0.465 | 74.8 |
| 6 M GdmCl | 1140 | 31.5 | 0.504 | 75.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paladino, A.; Vitagliano, L.; Graziano, G. The Action of Chemical Denaturants: From Globular to Intrinsically Disordered Proteins. Biology 2023, 12, 754. https://doi.org/10.3390/biology12050754
Paladino A, Vitagliano L, Graziano G. The Action of Chemical Denaturants: From Globular to Intrinsically Disordered Proteins. Biology. 2023; 12(5):754. https://doi.org/10.3390/biology12050754
Chicago/Turabian StylePaladino, Antonella, Luigi Vitagliano, and Giuseppe Graziano. 2023. "The Action of Chemical Denaturants: From Globular to Intrinsically Disordered Proteins" Biology 12, no. 5: 754. https://doi.org/10.3390/biology12050754