Generation of High-Value Genomic Resource in Rice: A “Subgenomic Library” of Low-Light Tolerant Rice Cultivar Swarnaprabha
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth of Plants
2.2. Isolation of Total Genomic DNA of Rice
2.3. Subgenomic Library Preparation
2.4. Screening of Recombinant Clones by α-Complementation
2.5. Estimation of Efficiency of Library by Colony PCR
2.6. Long-Term Storage of Subgenomic Clones
3. Results
3.1. Plant Growth
3.2. Isolation of Genomic DNA from Rice Tissue
3.3. Subgenomic Library Construction
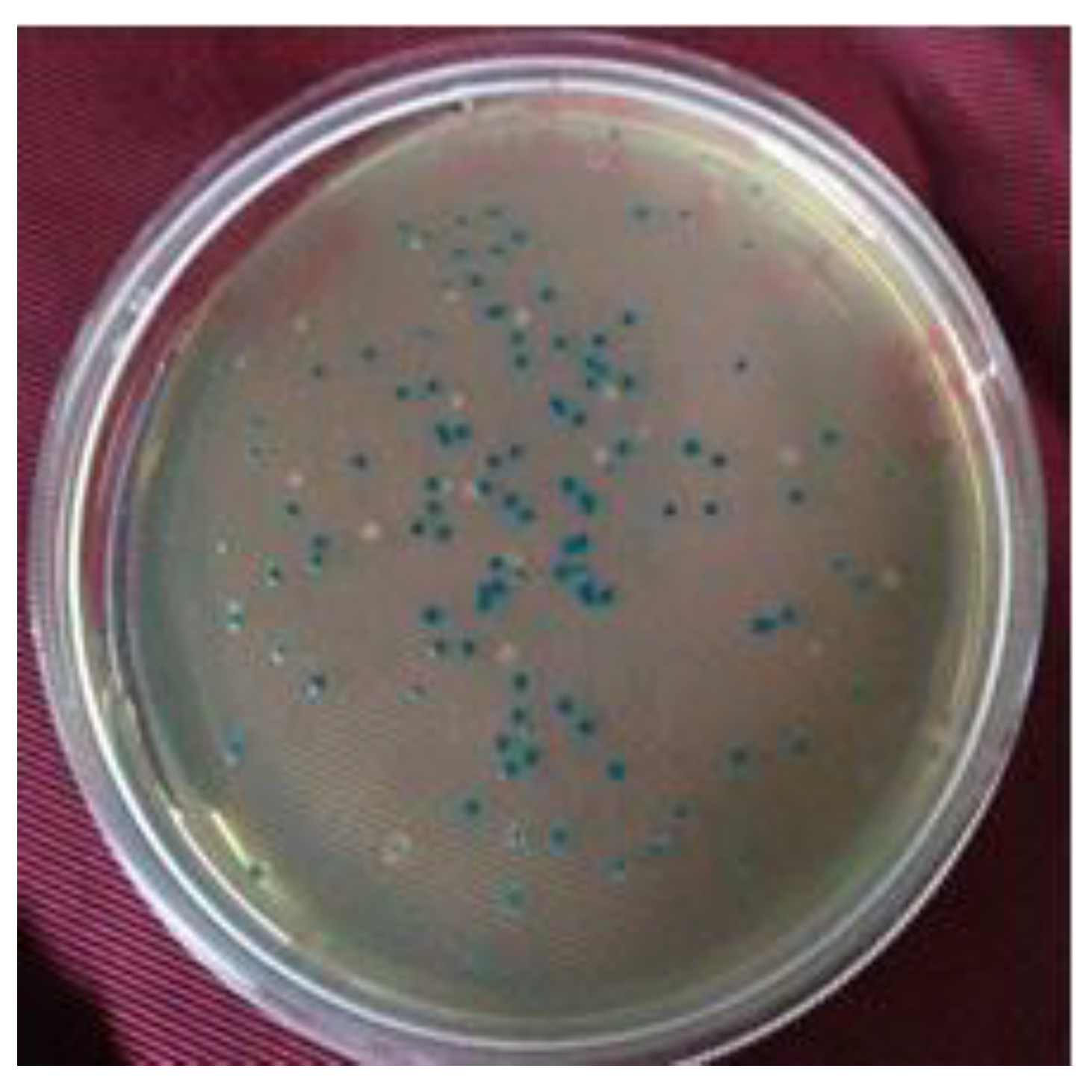
3.4. Authentication of Library by Colony PCR
3.5. Long-Term Storage of Subgenomic Clones
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Bharati, B.; Chatopadhyay, K.; Singh, S.; Dash, P.; Reddy, J.N. Association Mapping Reveals Multiple QTLs for Grain Protein Content in Rice Useful for Biofortification. Mol. Genet. Genom. 2019, 294, 963–983. [Google Scholar] [CrossRef] [PubMed]
- Bastia, R.; Pandit, E.; Sanghamitra, P.; Barik, S.R.; Nayak, D.K.; Sahoo, A.; Moharana, A.; Meher, J.; Dash, P.K.; Raj, R.; et al. Association Mapping for Quantitative Trait Loci Controlling Superoxide Dismutase, Flavonoids, Anthocyanins, Carotenoids, γ-Oryzanol and Antioxidant Activity in Rice. Agronomy 2022, 12, 3036. [Google Scholar] [CrossRef]
- Laskowski, W.; Górska-Warsewicz, H.; Rejman, K.; Czeczotko, M.; Zwolińska, J. How Important Are Cereals and Cereal Products in the Average Polish Diet? Nutrients 2019, 11, 679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dash, P.K.; Gupta, P.; Pradhan, S.K.; Shasany, A.K.; Rai, R. Analysis of Homologous Regions of Small RNAs MIR397 and MIR408 Reveals the Conservation of Microsynteny among Rice Crop-Wild Relatives. Cells 2022, 11, 3461. [Google Scholar] [CrossRef] [PubMed]
- Maloof, J.N.; Borevitz, J.O.; Dabi, T.; Lutes, J.; Nehring, R.B.; Redfern, J.L.; Trainer, G.T.; Wilson, J.M.; Asami, T.; Berry, C.C.; et al. Natural Variation in Light Sensitivity of Arabidopsis. Nat. Genet. 2001, 29, 441–446. [Google Scholar] [CrossRef]
- Arsovski, A.A.; Zemke, J.E.; Haagen, B.D.; Kim, S.-H.; Nemhauser, J.L. Phytochrome B Regulates Resource Allocation in Brassica Rapa. J. Exp. Bot. 2018, 69, 2837–2846. [Google Scholar] [CrossRef] [Green Version]
- Bharali, B.; Barmudoi, B. Effects of Light Intensity and Quality on Physiological Changes in Winter Rice (Oryza Sativa L.). Int. J. Environ. Agric. Res. 2016, 2, 65–76. [Google Scholar]
- Fraser, D.P.; Hayes, S.; Franklin, K.A. Photoreceptor Crosstalk in Shade Avoidance. Curr. Opin. Plant Biol. 2016, 33, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, J.; Dudley, S.A.; Pigliucci, M. Manipulative Approaches to Testing Adaptive Plasticity: Phytochrome-Mediated Shade-Avoidance Responses in Plants. Am. Nat. 1999, 154, S43–S54. [Google Scholar] [CrossRef]
- Huang, C.-L.; Hung, C.-Y.; Chiang, Y.-C.; Hwang, C.-C.; Hsu, T.-W.; Huang, C.-C.; Hung, K.-H.; Tsai, K.-C.; Wang, K.-H.; Osada, N.; et al. Footprints of Natural and Artificial Selection for Photoperiod Pathway Genes in Oryza. Plant J. 2012, 70, 769–782. [Google Scholar] [CrossRef]
- Han, X.; Tohge, T.; Lalor, P.; Dockery, P.; Devaney, N.; Esteves-Ferreira, A.A.; Fernie, A.R.; Sulpice, R. Phytochrome A and B Regulate Primary Metabolism in Arabidopsis Leaves in Response to Light. Front. Plant Sci. 2017, 8, 1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sineshchekov, V.A. Phytochrome A: Functional Diversity and Polymorphism. Photochem. Photobiol. Sci. 2004, 3, 596. [Google Scholar] [CrossRef] [PubMed]
- Borthwick, H.A.; Hendricks, S.B.; Parker, M.W.; Toole, E.H.; Toole, V.K. A Reversible Photoreaction Controlling Seed Germination. Proc. Natl. Acad. Sci. USA 1952, 38, 662–666. [Google Scholar] [CrossRef] [Green Version]
- Su, L.; Hou, P.; Song, M.; Zheng, X.; Guo, L.; Xiao, Y.; Yan, L.; Li, W.; Yang, J. Synergistic and Antagonistic Action of Phytochrome (Phy) A and PhyB during Seedling De-Etiolation in Arabidopsis Thaliana. Int. J. Mol. Sci. 2015, 16, 12199–12212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Xu, X.X.; Lu, X.; Xie, L.; Bai, B.; Zheng, C.; Sun, H.; He, Y.; Xie, X. The Rice Phytochrome Genes, PHYA and PHYB, Have Synergistic Effects on Anther Development and Pollen Viability. Sci. Rep. 2017, 7, 6439. [Google Scholar] [CrossRef] [Green Version]
- Oide, M.; Nakasako, M. Red Light-Induced Structure Changes in Phytochrome A from Pisum Sativum. Sci. Rep. 2021, 11, 2827. [Google Scholar] [CrossRef]
- Liu, P.; Sharrock, R.A. Biological Activity and Dimerization State of Modified Phytochrome A Proteins. PLoS ONE 2017, 12, e0186468. [Google Scholar] [CrossRef] [Green Version]
- Bae, G.; Choi, G. Decoding of Light Signals by Plant Phytochromes and Their Interacting Proteins. Annu. Rev. Plant Biol. 2008, 59, 281–311. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.-C.; Kathare, P.K.; Paik, I.; Huq, E. Phytochrome Signaling Networks. Annu. Rev. Plant Biol. 2021, 72, 217–244. [Google Scholar] [CrossRef]
- Nagatani, A. Phytochrome: Structural Basis for Its Functions. Curr. Opin. Plant Biol. 2010, 13, 565–570. [Google Scholar] [CrossRef]
- Wu, S.-H.; Lagarias, J.C. Defining the Bilin Lyase Domain: Lessons from the Extended Phytochrome Superfamily. Biochemistry 2000, 39, 13487–13495. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Su, Y.-S.; Lagarias, J.C. Phytochrome Structure and Signaling Mechanisms. Annu. Rev. Plant Biol. 2006, 57, 837–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montgomery, B.L.; Lagarias, J.C. Phytochrome Ancestry: Sensors of Bilins and Light. Trends Plant Sci. 2002, 7, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Halliday, K.J.; Salter, M.G.; Thingnaes, E.; Whitelam, G.C. Phytochrome Control of Flowering Is Temperature Sensitive and Correlates with Expression of the Floral Integrator FT. Plant J. 2003, 33, 875–885. [Google Scholar] [CrossRef]
- Strasser, B.; Sánchez-Lamas, M.; Yanovsky, M.J.; Casal, J.J.; Cerdán, P.D. Arabidopsis Thaliana Life without Phytochromes. Proc. Natl. Acad. Sci. USA 2010, 107, 4776–4781. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Franklin, K.A.; Sharrock, R.A.; Jones, M.A.; Harmer, S.L.; Lagarias, J.C. Unanticipated Regulatory Roles for Arabidopsis Phytochromes Revealed by Null Mutant Analysis. Proc. Natl. Acad. Sci. USA 2013, 110, 1542–1547. [Google Scholar] [CrossRef] [Green Version]
- Oh, S.; Montgomery, B.L. Phytochrome-Dependent Coordinate Control of Distinct Aspects of Nuclear and Plastid Gene Expression during Anterograde Signaling and Photomorphogenesis. Front. Plant Sci. 2014, 5, 171. [Google Scholar] [CrossRef] [Green Version]
- Casson, S.A.; Hetherington, A.M. Phytochrome B Is Required for Light-Mediated Systemic Control of Stomatal Development. Curr. Biol. 2014, 24, 1216–1221. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Seaton, D.D.; Krahmer, J.; Halliday, K.J. Photoreceptor Effects on Plant Biomass, Resource Allocation, and Metabolic State. Proc. Natl. Acad. Sci. USA 2016, 113, 7667–7672. [Google Scholar] [CrossRef] [Green Version]
- Gupta, P.; Dash, P.K.; Sanjay, T.D.; Pradhan, S.K.; Sreevathsa, R.; Rai, R. Cloning and Molecular Characterization of the PhlD Gene Involved in the Biosynthesis of “Phloroglucinol”, a Compound with Antibiotic Properties from Plant Growth Promoting Bacteria Pseudomonas Spp. Antibiotics 2023, 12, 260. [Google Scholar] [CrossRef]
- Kim, H.; Yang, T.; Kudrna, D.; Wing, R. Construction and Application of Genomic DNA Libraries. In Handbook of Plant Biotechnology; Wiley: Hoboken, NJ, USA, 2004. [Google Scholar]
- Gupta, P.; Duong, H.; Repalli, S.; Sreevathsa, R.; Yadava, D.; Dash, P. Cloning and Structural Elucidation of a Brassinosteroids Biosynthetic Gene (Atdwarf4) and Genetic Transformation of Indian Mustard (Brassica juncea L.). Indian J. Biochem. Biophys. 2022, 59, 320–330. [Google Scholar]
- Dash, P.; Gupta, P.; Panwar, B.S.; Rai, R. Isolation, Cloning and Characterization of PhlB Gene from an Indian Strain of Gram Negative Soil Bacteria Pseudomonas Fluorescens. Indian J. Exp. Biol. 2020, 58, 412–419. [Google Scholar]
- Ruberti, I.; Sessa, G.; Ciolfi, A.; Possenti, M.; Carabelli, M.; Morelli, G. Plant Adaptation to Dynamically Changing Environment: The Shade Avoidance Response. Biotechnol. Adv. 2012, 30, 1047–1058. [Google Scholar] [CrossRef]
- Ballaré, C.L.; Pierik, R. The Shade-Avoidance Syndrome: Multiple Signals and Ecological Consequences. Plant Cell Environ. 2017, 40, 2530–2543. [Google Scholar] [CrossRef]
- Smith, H. Light Quality, Photoperception, and Plant Strategy. Annu. Rev. Plant Physiol. 1982, 33, 481–518. [Google Scholar] [CrossRef]
- Clay, S.A.; Clay, D.E.; Horvath, D.P.; Pullis, J.; Carlson, C.G.; Hansen, S.; Reicks, G. Corn Response to Competition: Growth Alteration vs. Yield Limiting Factors. Agron. J. 2009, 101, 1522–1529. [Google Scholar] [CrossRef] [Green Version]
- Ciolfi, A.; Sessa, G.; Sassi, M.; Possenti, M.; Salvucci, S.; Carabelli, M.; Morelli, G.; Ruberti, I. Dynamics of the Shade-Avoidance Response in Arabidopsis. Plant Physiol. 2013, 163, 331–353. [Google Scholar] [CrossRef] [Green Version]
- Casal, J.J. Photoreceptor Signaling Networks in Plant Responses to Shade. Annu. Rev. Plant Biol. 2013, 64, 403–427. [Google Scholar] [CrossRef]
- Sheerin, D.J.; Menon, C.; zur Oven-Krockhaus, S.; Enderle, B.; Zhu, L.; Johnen, P.; Schleifenbaum, F.; Stierhof, Y.-D.; Huq, E.; Hiltbrunner, A. Light-Activated Phytochrome A and B Interact with Members of the SPA Family to Promote Photomorphogenesis in Arabidopsis by Reorganizing the COP1/SPA Complex. Plant Cell 2015, 27, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Sheerin, D.J.; Hiltbrunner, A. Molecular Mechanisms and Ecological Function of Far-Red Light Signalling. Plant Cell Environ. 2017, 40, 2509–2529. [Google Scholar] [CrossRef]
- Yang, C.; Li, L. Hormonal Regulation in Shade Avoidance. Front. Plant Sci. 2017, 8, 1527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legris, M.; Ince, Y.Ç.; Fankhauser, C. Molecular Mechanisms Underlying Phytochrome-Controlled Morphogenesis in Plants. Nat. Commun. 2019, 10, 5219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Kang, H.; Park, J.; Kim, W.; Yoo, J.; Lee, N.; Kim, J.; Yoon, T.; Choi, G. PIF1-Interacting Transcription Factors and Their Binding Sequence Elements Determine the in Vivo Targeting Sites of PIF1. Plant Cell 2016, 28, 1388–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, N.; Choi, G. Phytochrome-Interacting Factor from Arabidopsis to Liverwort. Curr. Opin. Plant Biol. 2017, 35, 54–60. [Google Scholar] [CrossRef]
- Chen, D.; Xu, G.; Tang, W.; Jing, Y.; Ji, Q.; Fei, Z.; Lin, R. Antagonistic Basic Helix-Loop-Helix/BZIP Transcription Factors Form Transcriptional Modules That Integrate Light and Reactive Oxygen Species Signaling in Arabidopsis. Plant Cell 2013, 25, 1657–1673. [Google Scholar] [CrossRef] [Green Version]
- Castillon, A.; Shen, H.; Huq, E. Phytochrome Interacting Factors: Central Players in Phytochrome-Mediated Light Signaling Networks. Trends Plant Sci. 2007, 12, 514–521. [Google Scholar] [CrossRef]
- Jiao, Y.; Lau, O.S.; Deng, X.W. Light-Regulated Transcriptional Networks in Higher Plants. Nat. Rev. Genet. 2007, 8, 217–230. [Google Scholar] [CrossRef]
- Shen, Y.; Khanna, R.; Carle, C.M.; Quail, P.H. Phytochrome Induces Rapid PIF5 Phosphorylation and Degradation in Response to Red-Light Activation. Plant Physiol. 2007, 145, 1043–1051. [Google Scholar] [CrossRef] [Green Version]
- Fiorucci, A.-S.; Fankhauser, C. Plant Strategies for Enhancing Access to Sunlight. Curr. Biol. 2017, 27, R931–R940. [Google Scholar] [CrossRef] [Green Version]
- Feng, L.; Raza, M.A.; Li, Z.; Chen, Y.; bin Khalid, M.H.; Du, J.; Liu, W.; Wu, X.; Song, C.; Yu, L.; et al. The Influence of Light Intensity and Leaf Movement on Photosynthesis Characteristics and Carbon Balance of Soybean. Front. Plant Sci. 2019, 9, 1952. [Google Scholar] [CrossRef]
- Dash, P.K.; Cao, Y.; Jailani, A.K.; Gupta, P.; Venglat, P.; Xiang, D.; Rai, R.; Sharma, R.; Thirunavukkarasu, N.; Abdin, M.Z.; et al. Genome-Wide Analysis of Drought Induced Gene Expression Changes in Flax (Linum usitatissimum). GM Crops Food 2014, 5, 106–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Hobson, N.; Galindo, L.; Zhu, S.; Shi, D.; McDill, J.; Yang, L.; Hawkins, S.; Neutelings, G.; Datla, R.; et al. The Genome of Flax (Linum usitatissimum) Assembled de Novo from Short Shotgun Sequence Reads. Plant J. 2012, 72, 461–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dash, P.K.; Rai, R.; Mahato, A.K.; Gaikwad, K.; Singh, N.K. Transcriptome Landscape at Different Developmental Stages of a Drought Tolerant Cultivar of Flax (Linum usitatissimum). Front. Chem. 2017, 5, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivakumara, T.N.; Sreevathsa, R.; Dash, P.K.; Sheshshayee, M.S.; Papolu, P.K.; Rao, U.; Tuteja, N.; UdayaKumar, M. Overexpression of Pea DNA Helicase 45 (PDH45) Imparts Tolerance to Multiple Abiotic Stresses in Chili (Capsicum annuum L.). Sci. Rep. 2017, 7, 2760. [Google Scholar] [CrossRef]
- Kesiraju, K.; Tyagi, S.; Mukherjee, S.; Rai, R.; Singh, N.K.; Sreevathsa, R.; Dash, P.K. An Apical Meristem-Targeted in Planta Transformation Method for the Development of Transgenics in Flax (Linum usitatissimum): Optimization and Validation. Front. Plant Sci. 2021, 11, 562056. [Google Scholar] [CrossRef]
- Gupta, P.; Dash, P.K. Precise Method of in Situ Drought Stress Induction in Flax (Linum usitatissimum) for RNA Isolation towards down-Stream Analysis. An. Agric. Res. 2015, 36, 10–17. [Google Scholar]
- Pradhan, K.C.; Pandit, E.; Mohanty, S.P.; Moharana, A.; Sanghamitra, P.; Meher, J.; Jena, B.K.; Dash, P.K.; Behera, L.; Mohapatra, P.M.; et al. Development of Broad Spectrum and Durable Bacterial Blight Resistant Variety through Pyramiding of Four Resistance Genes in Rice. Agronomy 2022, 12, 1903. [Google Scholar] [CrossRef]
- Sedeek, K.E.M.; Mahas, A.; Mahfouz, M. Plant Genome Engineering for Targeted Improvement of Crop Traits. Front. Plant Sci. 2019, 10, 114. [Google Scholar] [CrossRef] [Green Version]









| Vector DNA Preparation (EcoRI Digestion) | Vector (pBS) and Subgenomic DNA (Rice) Ligation | PCR Components | |||
|---|---|---|---|---|---|
| Vector DNA (pBS) | 20 μL | Vector (pBS) | 8 μL | Template DNA | colony |
| EcoRI buffer | 3 μL | Insert (4–9 kb) | 15 μL | Forward Primer (0.5 μM) | 0.5 μL |
| EcoRI (Enzyme) | 2 μL | Ligase buffer | 3 μL | Reverse Primer (0.5 μM) | 0.5 μL |
| Water | 5 μL | T4 DNA ligase | 2.5 μL | dNTPs (0.2 mM) | 0.5 μL |
| Total | 30 μL | Water | 1.5 μL | PCR Buffer (10X) | 2.0 μL |
| Total | 30 μL | Taq polymerase (2 units) | 0.5 μL | ||
| Water | 16 μL | ||||
| Total | 20 μL | ||||
| S. No. | Primer Name | Primer Sequence |
|---|---|---|
| 1 | M13 Forward Primer | AGCGGATAACAATTTCACACAGG |
| 2 | M13 Reverse Primer | CCCAGTCACGACGTTGTAAAACG |
| S. No. | Primer Name | Primer Sequence |
|---|---|---|
| 1 | phyA Forward Primer | CATCCATCGAGTTCTAGCAGG |
| 2 | phyA Reverse Primer | AAATCAATCAACCCACAACG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sahu, S.; Gupta, P.; Gowtham, T.P.; Yogesh, K.S.; Sanjay, T.D.; Singh, A.; Duong, H.V.; Pradhan, S.K.; Bisht, D.S.; Singh, N.K.; et al. Generation of High-Value Genomic Resource in Rice: A “Subgenomic Library” of Low-Light Tolerant Rice Cultivar Swarnaprabha. Biology 2023, 12, 428. https://doi.org/10.3390/biology12030428
Sahu S, Gupta P, Gowtham TP, Yogesh KS, Sanjay TD, Singh A, Duong HV, Pradhan SK, Bisht DS, Singh NK, et al. Generation of High-Value Genomic Resource in Rice: A “Subgenomic Library” of Low-Light Tolerant Rice Cultivar Swarnaprabha. Biology. 2023; 12(3):428. https://doi.org/10.3390/biology12030428
Chicago/Turabian StyleSahu, Sovanlal, Payal Gupta, Thirumalanahalli Prakash Gowtham, Kumar Shiva Yogesh, Tenkabailu Dharmanna Sanjay, Ayushi Singh, Hay Van Duong, Sharat Kumar Pradhan, Deepak Singh Bisht, Nagendra Kumar Singh, and et al. 2023. "Generation of High-Value Genomic Resource in Rice: A “Subgenomic Library” of Low-Light Tolerant Rice Cultivar Swarnaprabha" Biology 12, no. 3: 428. https://doi.org/10.3390/biology12030428






