Risk for Seasonal Affective Disorder (SAD) Linked to Circadian Clock Gene Variants
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Self-Report Surveys
2.3. Genotyping
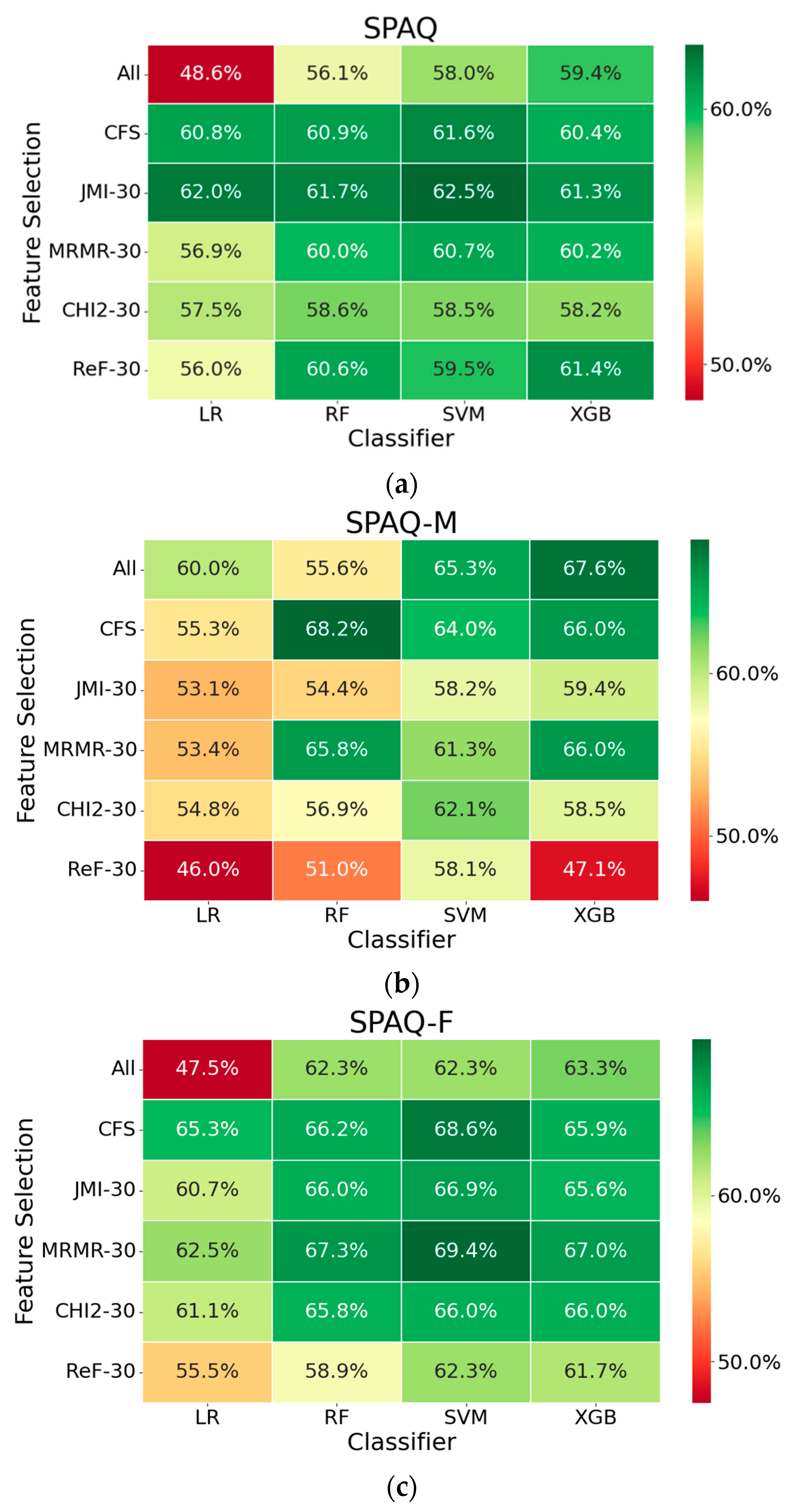
2.4. Feature Generation, Selection and Classification
2.5. Statistical Analyses
3. Results
3.1. Seasonality Prediction
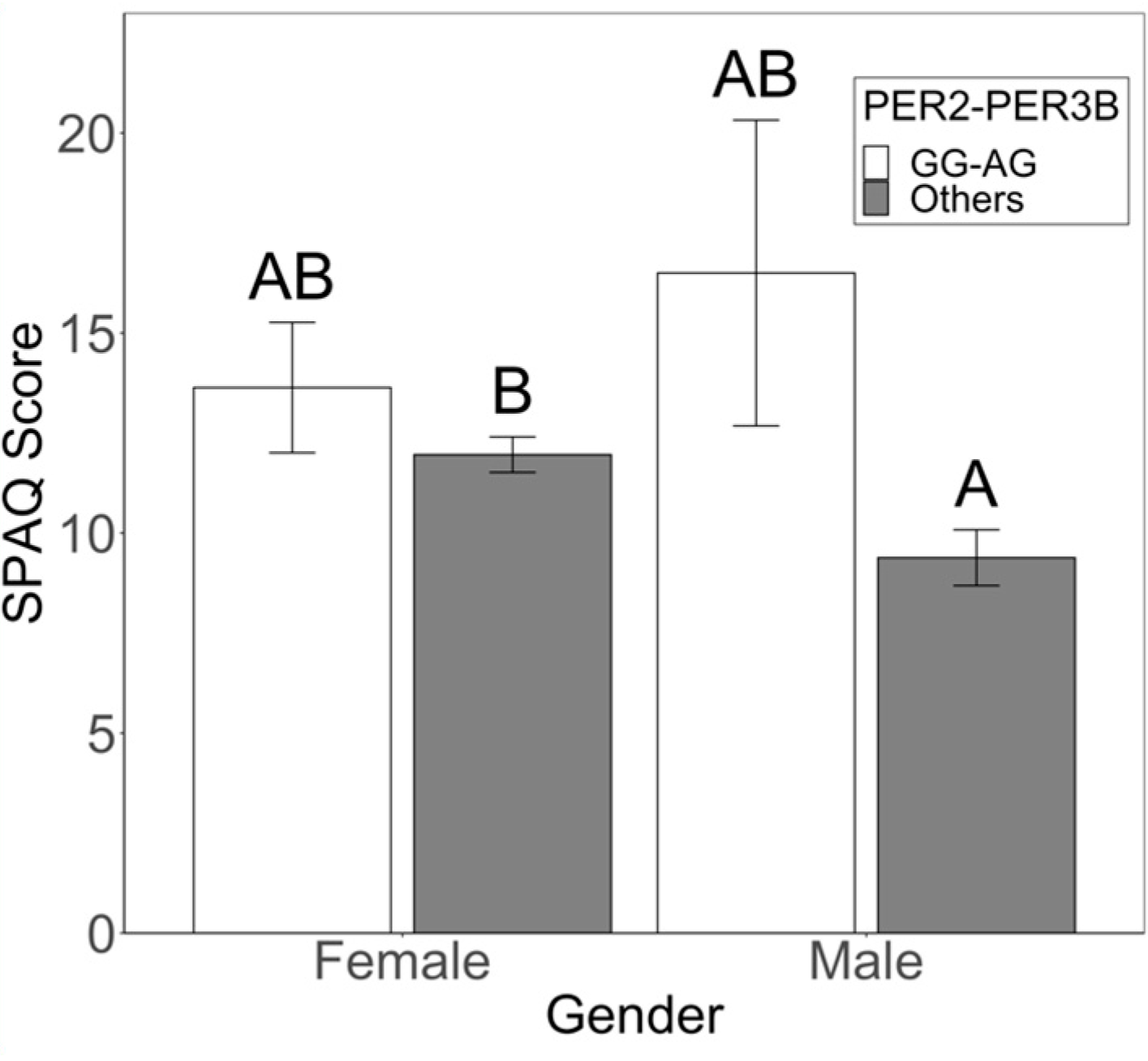
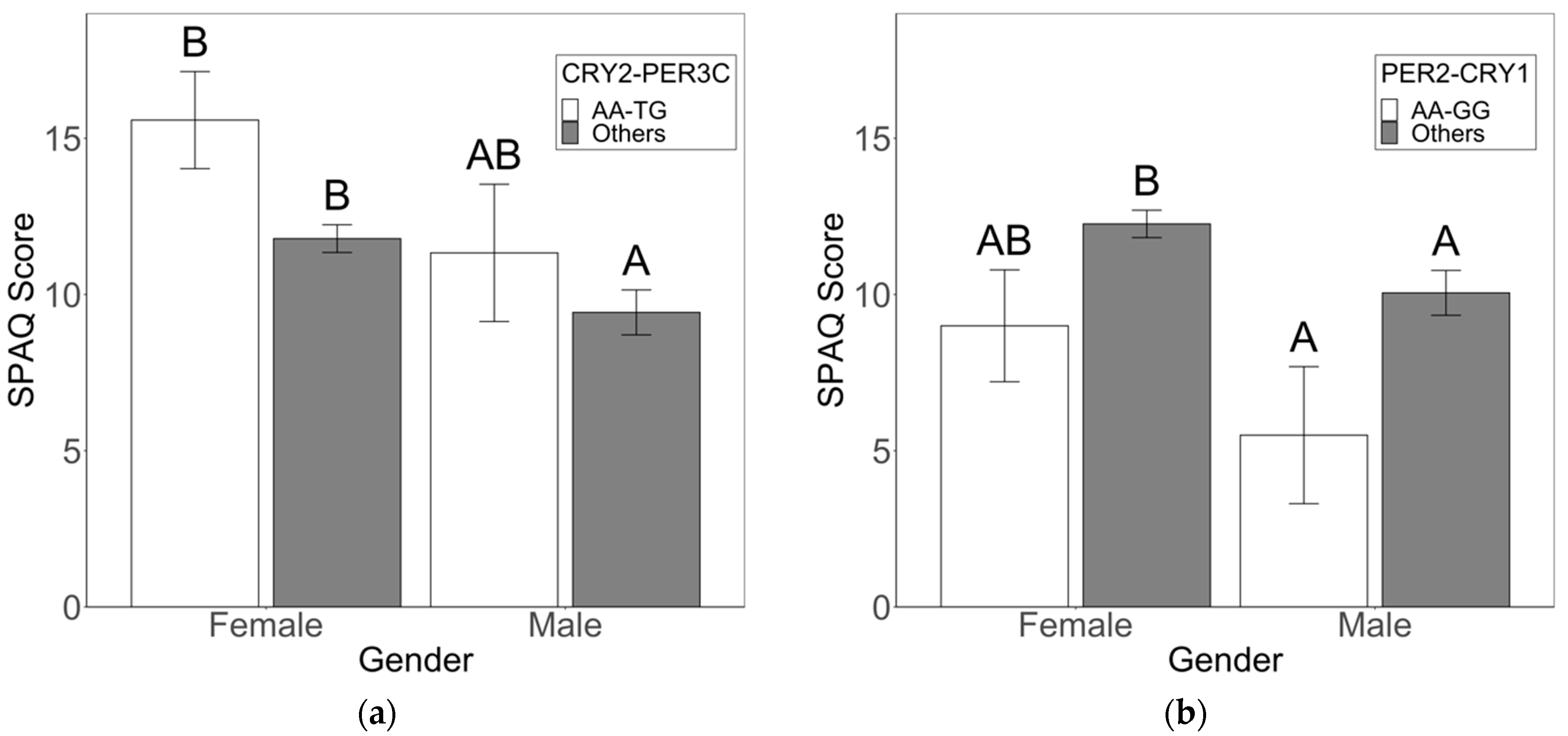
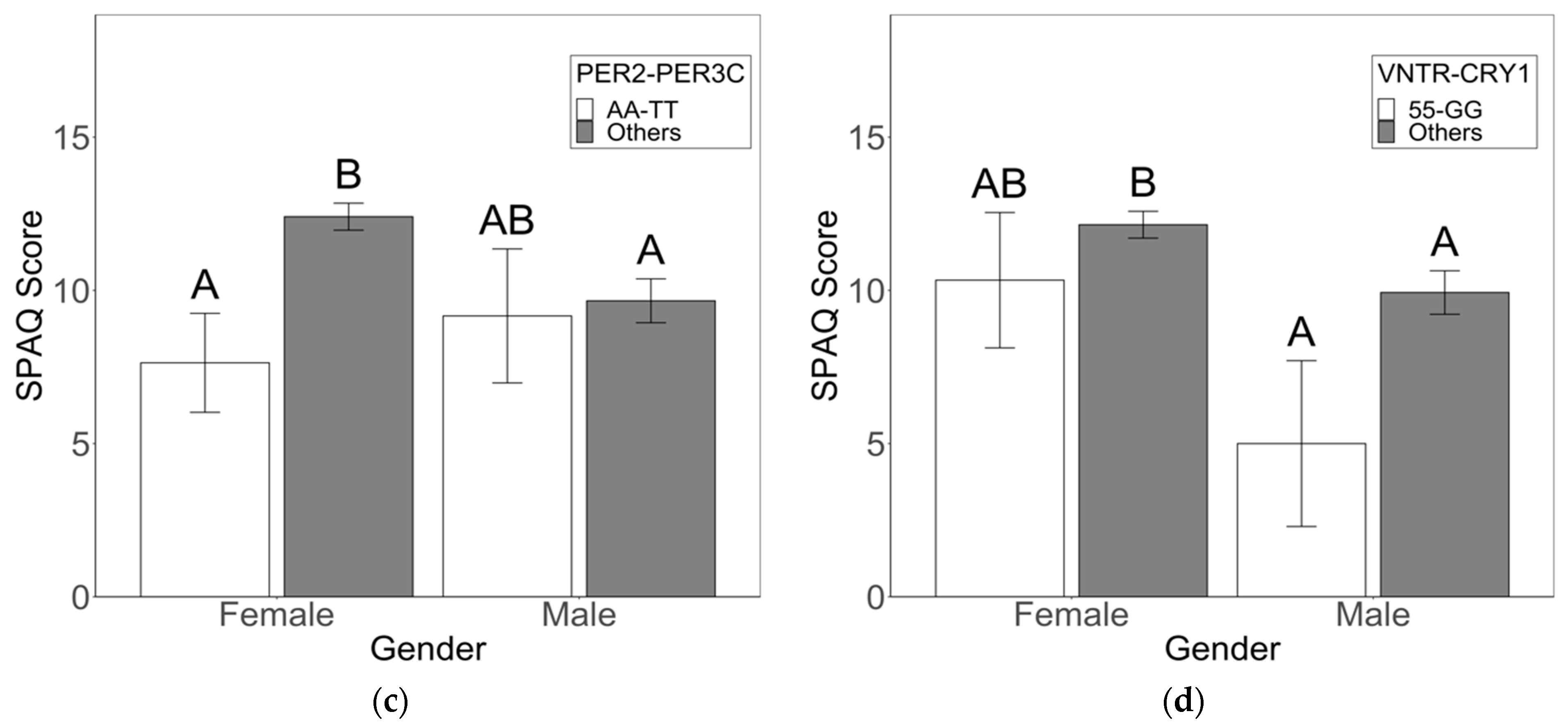
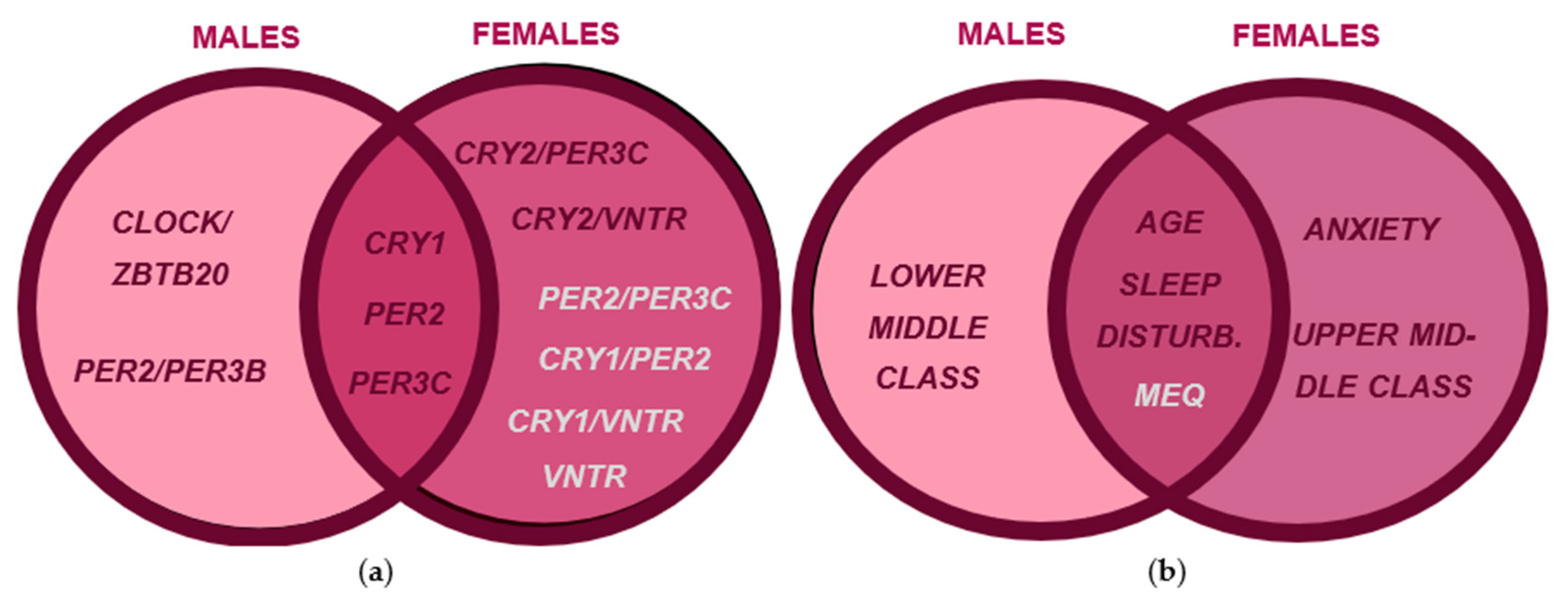
3.2. Sex-Specific Risk and Protective Factors for Seasonality and SAD Symptoms
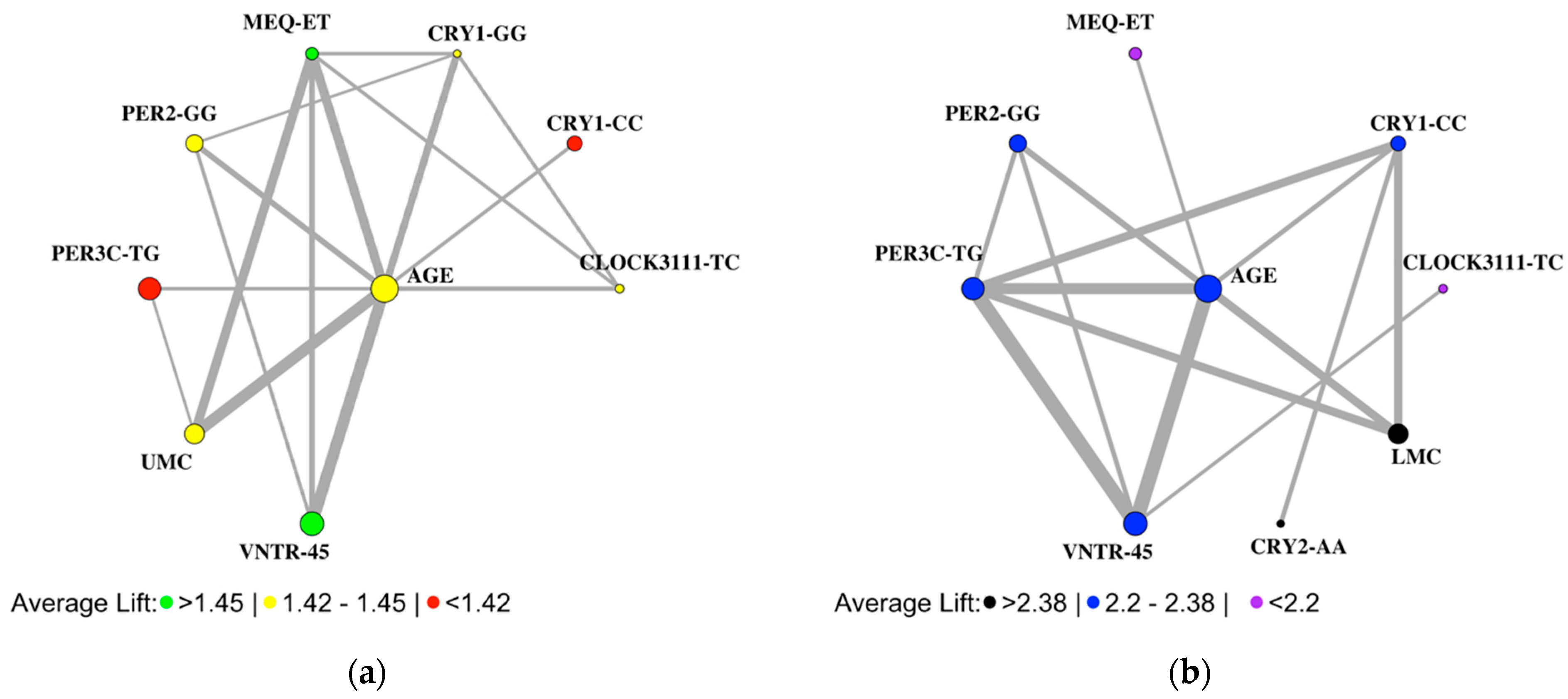
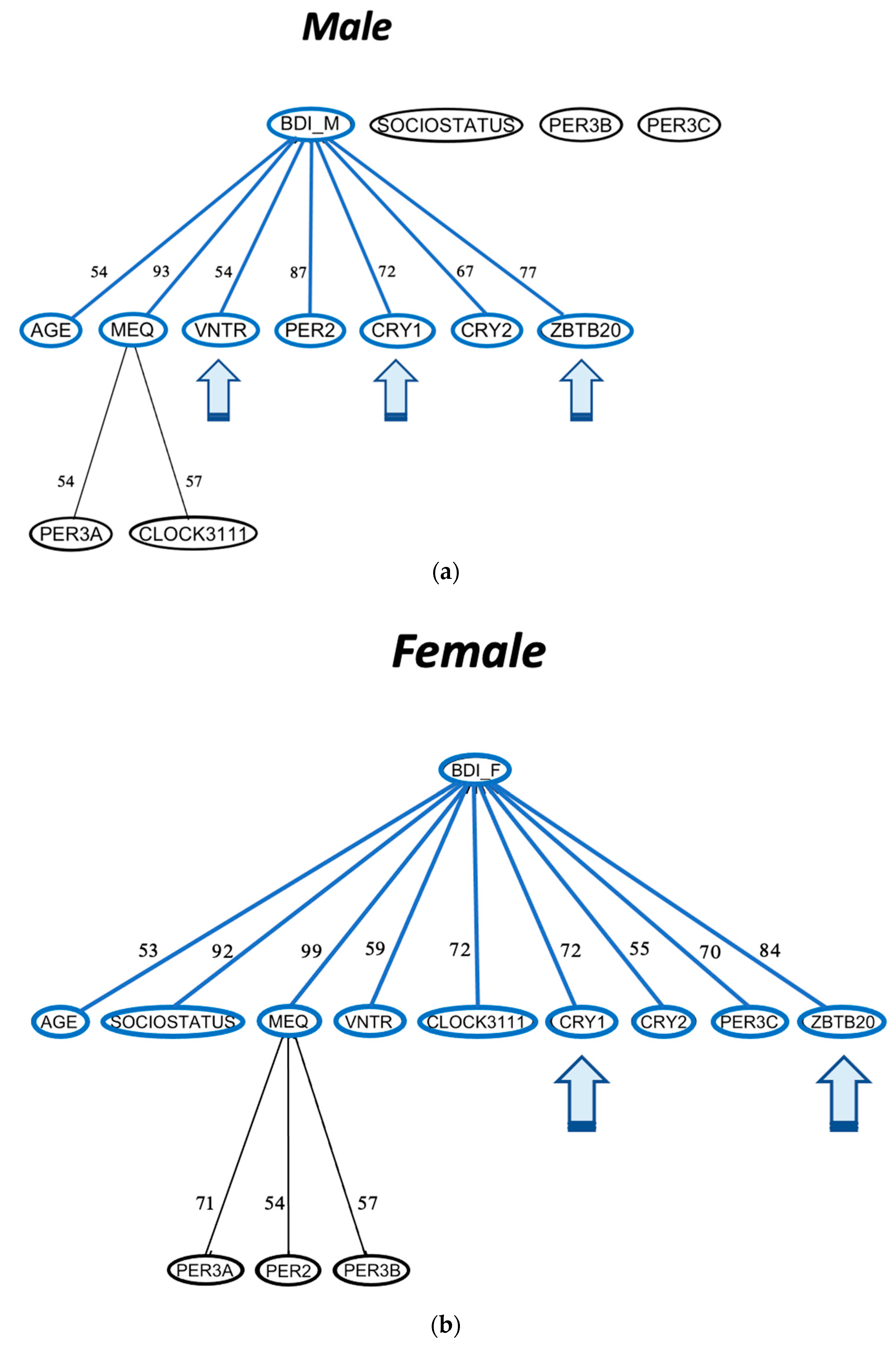
3.3. Association Rule Learning Networks Also Reveal Sex-Specific Rules for Seasonality
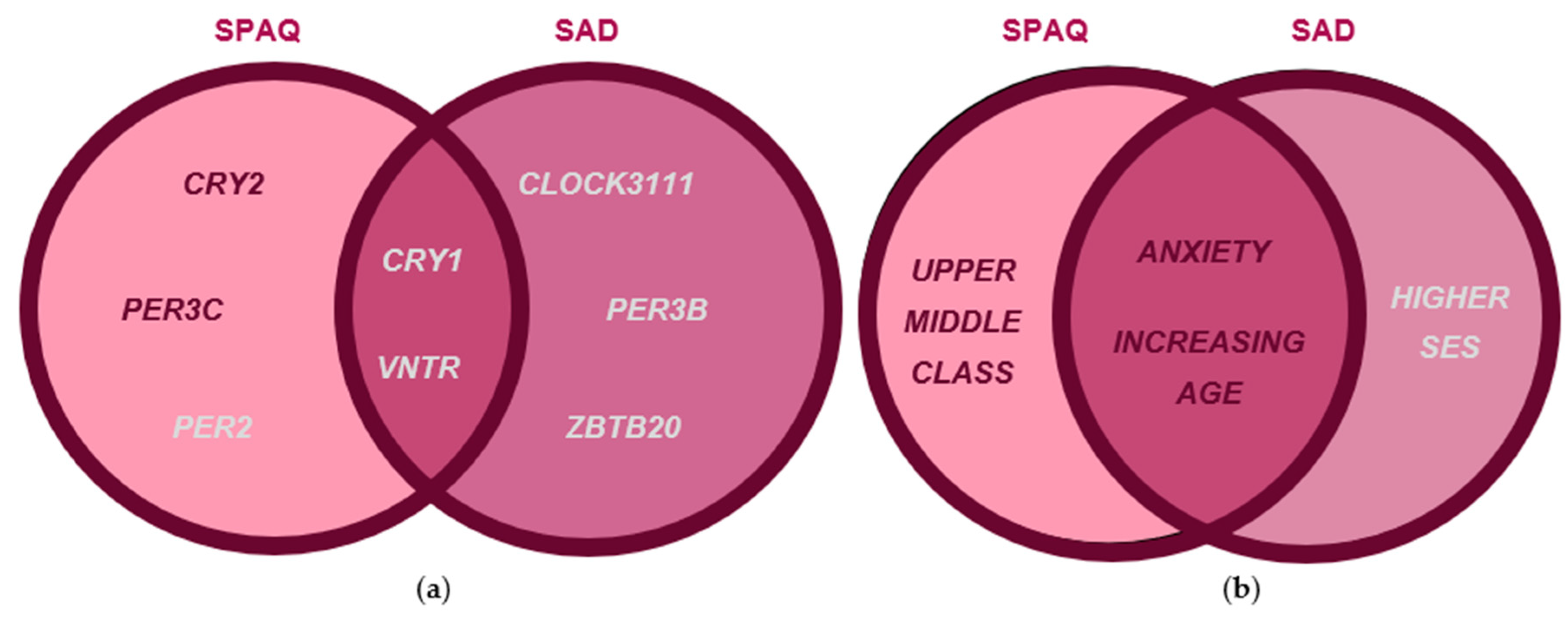
3.4. Comparisons of Genotypic and Clinical Feature Associations with Seasonality and SAD
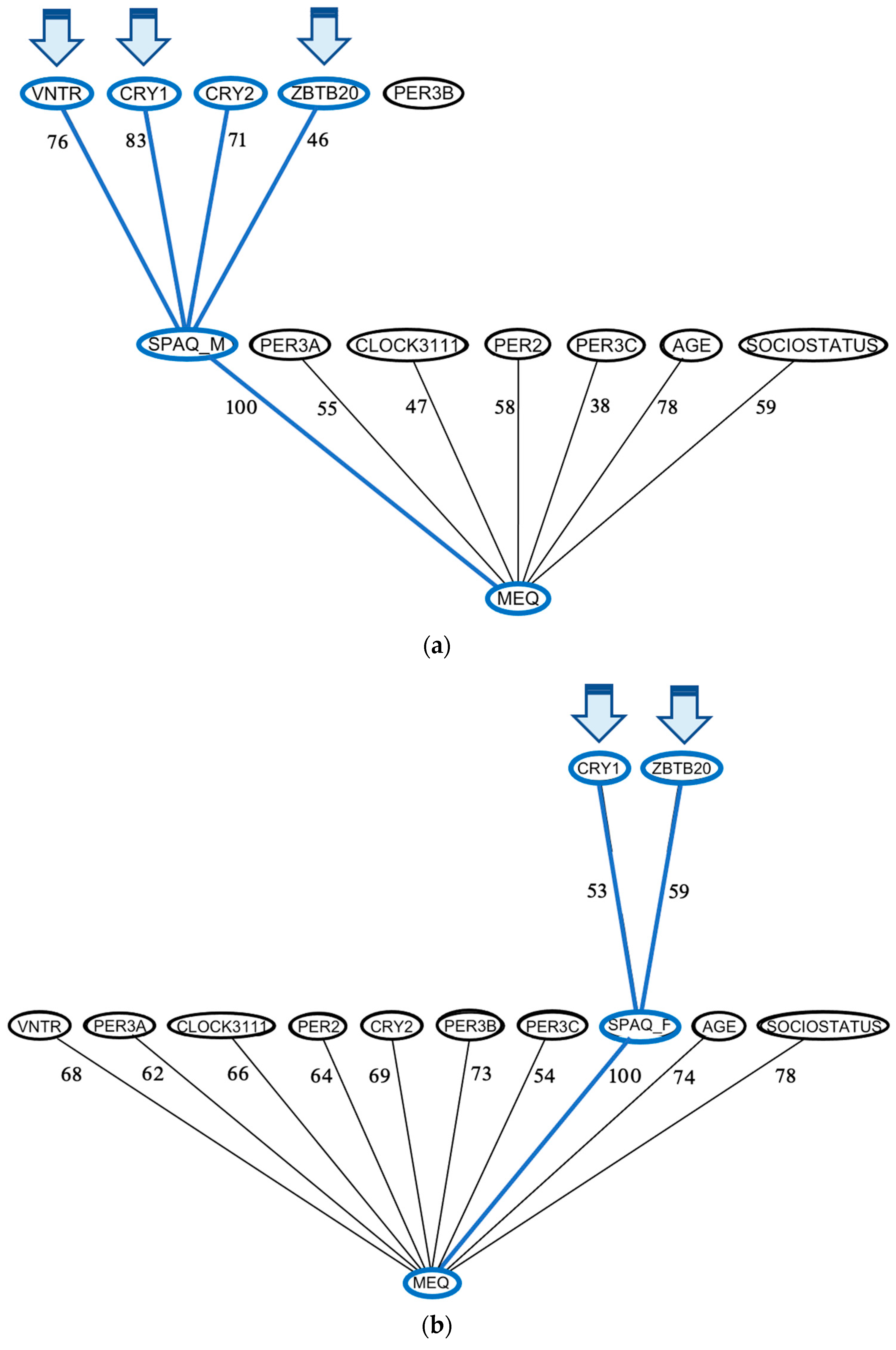
3.5. Mediation Analyses Reveal Robust Mediation of Seasonality Relative to SAD Symptoms
3.6. Mutual Information Analyses
4. Discussion
4.1. Circadian Genotypes Exhibit Sex-Specific Effects on Seasonality and SAD Symptoms
4.2. Clock Gene Effects on Seasonality and SAD May Be Direct or Mediated by Diurnal Preference
4.3. Clock Variants with Direct Effects on Seasonality Symptoms Also Have Direct Effects on Depression Symptoms
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fineberg, N.A.; Haddad, P.M.; Carpenter, L.; Gannon, B.; Sharpe, R.; Young, A.H.; Joyce, E.; Rowe, J.; Wellsted, D.; Nutt, D.J.; et al. The size, burden and cost of disorders of the brain in the UK. J. Psychopharmacol. 2013, 27, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Stepanek, M.; Taylor, J.; Troxel, W.M.; Van Stolk, C. Why Sleep Matters-The Economic Costs of Insufficient Sleep: A Cross-Country Comparative Analysis. Rand Health Q. 2017, 6, 11. [Google Scholar]
- McCarthy, M.J.; Welsh, D.K. Cellular Circadian Clocks in Mood Disorders. J. Biol. Rhythm. 2012, 27, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Soria, V.; Martinez-Amoros, E.; Escaramís, G.; Valero, J.; Pérez-Egea, R.; Garcia, C.; Gutiérrez-Zotes, A.; Puigdemont, D.; Bayés, M.; Crespo, J.M.; et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology 2010, 35, 1279–1289. [Google Scholar] [CrossRef]
- Zhang, L.; Hirano, A.; Hsu, P.-K.; Jones, C.R.; Sakai, N.; Okuro, M.; McMahon, T.; Yamazaki, M.; Xu, Y.; Saigoh, N.; et al. A PERIOD3 variant causes a circadian phenotype and is associated with a seasonal mood trait. Proc. Natl. Acad. Sci. USA 2016, 113, 1536–1544. [Google Scholar] [CrossRef]
- Lee, H.-J.; Rex, K.M.; Nievergelt, C.M.; Kelsoe, J.R.; Kripke, D.F. Delayed sleep phase syndrome is related to seasonal affective disorder. J. Affect. Disord. 2011, 133, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Henderson, S.E.M.; Brady, E.M.; Robertson, N. Associations between social jetlag and mental health in young people: A systematic review. Chronobiol. Int. 2019, 36, 1316–1333. [Google Scholar] [CrossRef]
- Islam, Z.; Hu, H.; Akter, S.; Kuwahara, K.; Kochi, T.; Eguchi, M.; Kurotani, K.; Nanri, A.; Kabe, I.; Mizoue, T. Social jetlag is associated with an increased likelihood of having depressive symptoms among the Japanese working popu-lation: The Furukawa Nutrition and Health Study. Sleep 2020, 43, zsz204. [Google Scholar] [CrossRef]
- Levandovski, R.; Dantas, G.; Fernandes, L.C.; Caumo, W.; Torres, I.; Roenneberg, T.; Hidalgo, M.P.L.; Allebrandt, K.V. Depression Scores Associate with Chronotype and Social Jetlag in a Rural Population. Chronobiol. Int. 2011, 28, 771–778. [Google Scholar] [CrossRef]
- Murray, J.M.; Sletten, T.L.; Magee, M.; Gordon, C.; Lovato, N.; Bartlett, D.J.; Kennaway, G.J.; Lack, L.C.; Grunstein, R.R.; Lockley, S.W.; et al. Prevalence of Circadian Misalignment and Its Association with Depressive Symptoms in Delayed Sleep Phase Dis-order. Sleep 2017, 40, zsw002. [Google Scholar]
- Nguyen, C.; Murray, G.; Anderson, S.; Filipowicz, A.; Ingram, K.K. In vivo molecular chronotyping, circadian misalignment, and high rates of depression in young adults. J. Affect. Disord. 2019, 250, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Archer, S.N.; Robilliard, D.L.; Skene, D.J.; Smits, M.; Williams, A.; Arendt, J.; von Schantz, M. A Length Polymorphism in the Circadian Clock Gene Per3 is Linked to Delayed Sleep Phase Syndrome and Extreme Diurnal Preference. Sleep 2003, 26, 413–415. [Google Scholar] [CrossRef] [PubMed]
- Hida, A.; Kitamura, S.; Katayose, Y.; Kato, M.; Ono, H.; Kadotani, H.; Uchiyama, M.; Ebisawa, T.; Inoue, Y.; Kamei, Y.; et al. Screening of Clock Gene Polymorphisms Demonstrates Association of a PER3 Polymorphism with Morningness–Eveningness Preference and Circadian Rhythm Sleep Disorder. Sci. Rep. 2014, 4, srep06309. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.I.; Lee, H.J.; Cho, C.H.; Kang, S.G.; Yoon, H.K.; Park, Y.M.; Lee, S.-H.; Moon, J.-H.; Song, H.-M.; Lee, E.; et al. Association of CLOCK, ARNTL, and NPAS2 gene polymorphisms and seasonal variations in mood and behavior. Chronobiol. Int. 2015, 32, 785–791. [Google Scholar] [CrossRef]
- Lavebratt, C.; Sjöholm, L.K.; Partonen, T.; Schalling, M.; Forsell, Y. PER2 variantion is associated with depression vulnerability. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2010, 153, 570–581. [Google Scholar] [CrossRef]
- Lavebratt, C.; Sjöholm, L.K.; Soronen, P.; Paunio, T.; Vawter, M.P.; Bunney, W.E.; Adolfsson, R.; Forsell, Y.; Wu, J.C.; Kelsoe, J.R.; et al. CRY2 Is Associated with Depression. PLoS ONE 2010, 5, e9407. [Google Scholar] [CrossRef]
- Partonen, T.; Treutlein, J.; Alpman, A.; Frank, J.; Johansson, C.; Depner, M.; Aron, L.; Rietschel, M.; Wellek, S.; Soronen, P.; et al. Three circadian clock genes Per2, Arntl, and Npas2 contribute to winter depression. Ann. Med. 2007, 39, 229–238. [Google Scholar] [CrossRef]
- Shi, S.-Q.; White, M.J.; Borsetti, H.M.; Pendergast, J.S.; Hida, A.; Ciarleglio, C.M.; de Verteuil, P.A.; Cadar, A.G.; Cala, C.; McMahon, D.G.; et al. Molecular analyses of circadian gene variants reveal sex-dependent links between depression and clocks. Transl. Psychiatry 2016, 6, e748. [Google Scholar] [CrossRef]
- Courtet, P.; Olie, E. Circadian dimension and severity of depression. Eur. Neuropsychopharmacol. 2012, 22, S476–S481. [Google Scholar] [CrossRef]
- McCarthy, M.J.; Nievergelt, C.M.; Kelsoe, J.R.; Welsh, D.K. A Survey of Genomic Studies Supports Association of Circadian Clock Genes with Bipolar Disorder Spectrum Illnesses and Lithium Response. PLoS ONE 2012, 7, e32091. [Google Scholar] [CrossRef]
- Terracciano, A.; Tanaka, T.; Sutin, A.R.; Sanna, S.; Deiana, B.; Lai, S.; Uda, M.; Schlessinger, D.; Abecasis, G.R.; Ferrucci, L.; et al. Genome-Wide Association Scan of Trait Depression. Biol. Psychiatry 2010, 68, 811–817. [Google Scholar] [CrossRef]
- Mendlewicz, J. Disruption of the circadian timing systems: Molecular mechanisms in mood disorders. CNS Drugs 2009, 23 (Suppl. 2), 15–26. [Google Scholar] [CrossRef]
- Robillard, R.; Naismith, S.L.; Rogers, N.L.; Ip, T.K.; Hermens, D.F.; Scott, E.M.; Hickie, I.B. Delayed sleep phase in young people with unipolar or bipolar affective disorders. J. Affect. Disord. 2013, 145, 260–263. [Google Scholar] [CrossRef]
- Liberman, A.R.; Halitjaha, L.; Ay, A.; Ingram, K.K. Modeling strengthens molecular link between circadian polymorphisms and major mood disorders. J. Biol. Rhythm. 2018, 33, 318–336. [Google Scholar] [CrossRef]
- Meesters, Y.; Gordijn, M.C. Seasonal affective disorder, winter type: Current insights and treatment options. Psychol. Res. Behav. Manag. 2016, 9, 317–327. [Google Scholar] [CrossRef]
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An expanded view of complex traits: From polygenic to omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef]
- Ciarleglio, C.M.; Ryckman, K.K.; Servick, S.V.; Hida, A.; Robbins, S.; Wells, N.; Hicks, J.; Larson, S.A.; Wiedermann, J.P.; Johnson, C.H. Genetic differences in human circadian clock genes among worldwide populations. J. Biol. Rhythm. 2008, 23, 330–340. [Google Scholar] [CrossRef]
- Robillard, R.; Hermens, D.F.; Naismith, S.L.; White, D.; Rogers, N.L.; Ip, T.K.; Mullin, S.J.; Alvares, G.A.; Guastella, A.J.; Smith, K.L.; et al. Ambulatory sleep-wake patterns and variability in young people with emerging mental disorders. J. Psychiatry Neurosci. 2015, 40, 28–37. [Google Scholar] [CrossRef]
- Partonen, T. Clock gene variants in mood and anxiety disorders. J. Neural Transm. 2012, 119, 1133–1145. [Google Scholar] [CrossRef]
- Partonen, T. Circadian Clock Proteins in Mood Regulation. Front. Psychiatry 2014, 5, 195. [Google Scholar] [CrossRef]
- Zafar, A.; Overton, R.; Attia, Z.; Ay, A.; Ingram, K. Machine learning and expression analyses reveal circadian clock features predictive of anxiety. Sci. Rep. 2022, 12, 5508. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, N.; Bradt, G.; Wehr, T. Seasonal Pattern Assessment Questionnaire (SPAQ); National Institute of Mental Health: Bethesda, MD, USA, 1984.
- Beck, A.T.; Beamesderfer, A. Assessment of depression: The depression inventory. Mod. Probl. Pharmacopsychiatry 1974, 7, 151–169. [Google Scholar] [PubMed]
- Spielberger, C.D.; Gorsuch, R.L.; Lushene, R.; Vagg, P.R.; Jacobs, G.A. Manual for the State-Trait Anxiety Inventory; Consulting Psychologists Press: Washington, DC, USA, 1983. [Google Scholar]
- Yu, L.; Buysse, D.J.; Germain, A.; Moul, D.E.; Stover, A.; Dodds, N.E.; Johnston, K.L.; Pilkonis, P.A. Development of Short Forms From the PROMIS™ Sleep Disturbance and Sleep-Related Impairment Item Banks. Behav. Sleep Med. 2011, 10, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Roenneberg, T.; Kuehnle, T.; Juda, M.; Kantermann, T.; Allebrandt, K.; Gordijn, M.; Merrow, M. Epidemiology of the human circadian clock. Sleep Med. Rev. 2007, 11, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Horne, J.A.; Ostberg, O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int. J. Chronobiol. 1976, 4, 97–110. [Google Scholar]
- Short, R.; Fukunaga, K. The optimal distance measure for nearest neighbor classification. IEEE Trans. Inf. Theory 1981, 27, 622–627. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Huan, L.; Setiono, R. Chi2: Feature selection and discretization of numeric attributes. In Proceedings of the 7th IEEE International Conference on Tools with Artificial Intelligence, Herndon, VA, USA, 5–8 November 1995. [Google Scholar]
- Robnik-Sikonja, M.; Kononenko, I. An adaptation of Relief for attribute estimation in regression. In International Conference on Machine Learning; Citeseer: Princeton, NJ, USA, 1997; Volume 5, pp. 296–304. [Google Scholar]
- Ding, C.; Peng, H. Minimum redundancy feature selection from microarray gene expression data. J. Bioinform. Comput. Biol. 2005, 3, 185–205. [Google Scholar] [CrossRef]
- Hall, M. Correlation-Based Feature Selection for Machine Learning. Ph.D. Thesis, The University of Waikato, Department of Computer Science, Hamilton, New Zealand, 2000; p. 19. [Google Scholar]
- Yang, H.H.; Moody, J.E. Feature Selection Based on Joint Mutual Information. In Proceedings of the International ICSC Symposium on Advances in Intelligent Data Analysis, Rochester, NY, USA, 15 October 1999. [Google Scholar]
- Boser, B.; Guyon, I.; Vapnik, V. A Training Algorithm for Optimal Margin Classifier. In Proceedings of the Fifth Annual ACM Workshop on Computational Learning Theory; Association for Computing Machinery: New York, NY, USA, 1992. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Washington, DC, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 10 December 2023).
- Bozdogan, H. Model selection and Akaike’s Information Criterion (AIC): The general theory and its analytical extensions. Psychometrika 1987, 52, 345–370. [Google Scholar] [CrossRef]
- Imai, K.; Keele, L.; Tingley, D.; Yamamoto, T. Causal Mediation Analysis Using R. In Advances in Social Science Research Using R; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression; Sage: New York, NY, USA, 2019. [Google Scholar]
- Hahsler, M.; Chelluboina, S.; Hornik, K.; Buchta, C. The arules R-package ecosystem: Analyzing interesting patterns from large transaction datasets. J. Mach. Learn. Res. 2011, 12, 1977–1981. [Google Scholar]
- Csardi, G.; Nepusz, T. The Igraph Software Package for Complex Network Research. InterJournal Complex Syst. 2005, 1695, 1–9. [Google Scholar]
- Margolin, A.A.; Nemenman, I.; Basso, K.; Wiggins, C.; Stolovitzky, G.; Favera, R.D.; Califano, A. ARACNE: An Algorithm for the Reconstruction of Gene Regulatory Networks in a Mammalian Cellular Context. BMC Bioinform. 2006, 7, S7. [Google Scholar] [CrossRef]
- Hansen, K.D.; Gentry, J.; Long, L.; Gentleman, R.; Falcon, S.; Hahne, F.; Sarkar, D. Rgraphviz: Provides Plotting Capabilities for R Graph Objects, R package version 2.42.0. 2022. [Google Scholar]
- Meyer, P.E.; Lafitte, F.; Bontempi, G. minet: A R/Bioconductor Package for Inferring Large Transcriptional Networks Using Mutual Information. BMC Bioinform. 2008, 9, 461. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, F.; Dallaspezia, S.; Fulgosi, M.C.; Lorenzi, C.; Serretti, A.; Barbini, B.; Colombo, C.; Smeraldi, E. Actimetric evidence that CLOCK 3111 T/C SNP influences sleep and activity patterns in patients affected by bipolar depression. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2007, 144, 631–635. [Google Scholar] [CrossRef] [PubMed]
- Katzenberg, D.; Young, T.; Finn, L.; Lin, L.; King, D.P.; Takahashi, J.S.; Mignot, E. A CLOCK Polymorphism Associated with Human Diurnal Preference. Sleep 1998, 21, 569–576. [Google Scholar] [CrossRef]
- Daut, R.A.; Fonken, L.K. Circadian regulation of depression: A role for serotonin. Front. Neuroendocr. 2019, 54, 100746. [Google Scholar] [CrossRef] [PubMed]
- Pjrek, E.; Konstantinidis, A.; Assem-Hilger, E.; Praschak-Rieder, N.; Willeit, M.; Kasper, S.; Winkler, D. Therapeutic effects of escitalopram and reboxetine in seasonal affective disorder: A pooled analysis. J. Psychiatr. Res. 2009, 43, 792–797. [Google Scholar] [CrossRef]
- Natale, V.; Adan, A.; Scapellato, P. Are seasonality of mood and eveningness closely associated? Psychiatry Res. 2005, 136, 51–60. [Google Scholar] [CrossRef]
- Tonetti, L.; Fabbri, M.; Martoni, M.; Natale, V. Circadian type and mood seasonality in adolescents. Psychiatry Clin. Neurosci. 2012, 66, 157–159. [Google Scholar] [CrossRef]
- Zhang, L.; Evans, D.S.; Raheja, U.K.; Stephens, S.H.; Stiller, J.W.; Reeves, G.M.; Johnson, M.; Ryan, K.A.; Weizel, N.; Vaswani, D.; et al. Chronotype and seasonality: Morningness is associated with lower seasonal mood and behavior changes in the Old Order Amish. J. Affect. Disord. 2015, 174, 209–214. [Google Scholar] [CrossRef]
- Merikanto, I.; Suvisaari, J.; Lahti, T.; Partonen, T. Eveningness relates to burnout and seasonal sleep and mood problems among young adults. Nord. J. Psychiatry 2016, 70, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Groeger, J.A.; Viola, A.U.; Lo, J.C.; von Schantz, M.; Archer, S.N.; Dijk, D.-J. Early Morning Executive Functioning During Sleep Deprivation Is Compromised by a PERIOD3 Polymorphism. Sleep 2008, 31, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Hirano, A.; Shi, G.; Jones, C.R.; Lipzen, A.; Pennacchio, L.A.; Xu, Y.; Hallows, W.C.; McMahon, T.; Yamazaki, M.; Ptáček, L.J.; et al. A Cryptochrome 2 mutation yields advanced sleep phase in humans. Elife 2016, 5, e16695. [Google Scholar] [CrossRef] [PubMed]
- Liberman, A.R.; Bin Kwon, S.; Vu, H.T.; Filipowicz, A.; Ay, A.; Ingram, K.K. Circadian Clock Model Supports Molecular Link Between PER3 and Human Anxiety. Sci. Rep. 2017, 7, 9893. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.C.P.E.; Dos Santos, M.J.; Gitaí, D.L.G.; de Miranda Coelho, J.A.P.; de Andrade, T.G. Depression and anxiety symptoms correlate with diurnal preference, sleep habits, and Per3 VNTR polymorphism (rs57875989) in a non-clinical sample. J. Affect. Disord. 2020, 277, 260–270. [Google Scholar] [CrossRef]
- Viena, T.; Gobin, C.; Fins, A.; Craddock, T.; Tartar, A.; Tartar, J. A PER3 Polymorphism Interacts with Sleep Duration to Influence Transient Mood States in Women. J. Circadian Rhythm. 2016, 14, 3. [Google Scholar] [CrossRef]
- Jones, S.E.; Lane, J.M.; Wood, A.R.; van Hees, V.T.; Tyrrell, J.; Beaumont, R.N.; Jeffries, A.R.; Dashti, H.S.; Hillsdon, M.; Ruth, K.S.; et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat. Commun. 2019, 10, 343. [Google Scholar] [CrossRef]
- Johansson, C.; Willeit, M.; Smedh, C.; Ekholm, J.; Paunio, T.; Kieseppä, T.; Lichtermann, D.; Praschak-Rieder, N.; Neumeister, A.; Nilsson, L.-G.; et al. Circadian Clock-Related Polymorphisms in Seasonal Affective Disorder and their Relevance to Diurnal Preference. Neuropsychopharmacology 2003, 28, 734–739. [Google Scholar] [CrossRef]
- Kovanen, L.; Saarikoski, S.T.; Aromaa, A.; Lönnqvist, J.; Partonen, T. ARNTL (BMAL1) and NPAS2 Gene Variants Contribute to Fertility and Seasonality. PLoS ONE 2010, 5, e10007. [Google Scholar] [CrossRef]
- Weiss, C.; Woods, K.; Filipowicz, A.; Ingram, K.K. Sleep Quality, Sleep Structure, and PER3 Genotype Mediate Chronotype Effects on Depressive Symptoms in Young Adults. Front. Psychol. 2020, 11, 2028. [Google Scholar] [CrossRef] [PubMed]
- Lamia, K.A.; Papp, S.J.; Yu, R.T.; Barish, G.D.; Uhlenhaut, N.H.; Jonker, J.W.; Downes, M.; Evans, R.M. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 2011, 480, 552–556. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.A. How might circadian rhythms control mood? Let me count the ways. Biol. Psychiatry 2013, 74, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.W.D.; Han, S.; Nielsen, J.V.; Jancic, D.; Hing, B.; Fiedorowicz, J.; Weissman, M.M.; Levinson, D.F.; Potash, J.B. Genome-wide association study of seasonal affective disorder. Transl. Psychiatry 2018, 8, 190. [Google Scholar] [CrossRef]
- Davies, M.N.; Krause, L.; Bell, J.T.; Gao, F.; Ward, K.J.; Wu, H.; Lu, H.; Liu, Y.; Tsai, P.-C.; Collier, D.A.; et al. Hypermethylation in the ZBTB20 gene is associated with major depressive disorder. Genome Biol. 2014, 15, R56. [Google Scholar] [CrossRef]
- Qu, Z.; Zhang, H.; Huang, M.; Shi, G.; Liu, Z.; Xie, P.; Li, H.; Wang, W.; Xu, G.; Zhang, Y.; et al. Loss of ZBTB20 impairs circadian output and leads to unimodal behavioral rhythms. Elife 2016, 5, e17171. [Google Scholar] [CrossRef] [PubMed]
| Feature | Male | Female | |
|---|---|---|---|
| Risk | SPAQ M OR [estimate] | SPAQ F OR [estimate] | SAD F OR [estimate] |
| CLOCK3111-TC/ZBTB20-AT | 3.975 | ||
| CRY2-AA/PER3C-TG | [4.279] | ||
| CRY2-AA/VNTR-4,5 | *8.919* | ||
| PER2-GG/PER3B-AG | 7.262 | ||
| Anxiety | 1.061 | ||
| Evening type | *1.783* | ||
| Increasing Age | *6.406* | ||
| Sleep Disturbance | [0.320] | [0.173] | |
| Protective | |||
| VNTR-4,5 | 0.258 | ||
| CLOCK3111-TC/PER3B-GG | 0.357 | ||
| CLOCK3111-TC/PER3C-TT | 0.383 | ||
| CRY1-GG | 0.491 | ||
| CRY1-GG/PER2-AA | [−3.595] | ||
| CRY1-GG/VNTR-5,5 | *0.112* | ||
| CRY1-GG/ZBTB20-AA | 0.454 | ||
| PER2-AA/PER3C-TT | *0.203* | ||
| PER3B-GG/VNTR-4,5 | 0.304 | ||
| PER3C-TT/VNTR-4,5 | 0.361 | ||
| VNTR-4,5/ZBTB20-AA | 0.321 | ||
| Morning type | 0.937 | ||
| Genotypic Feature | SPAQ F | SPAQ M |
|---|---|---|
| PER2-GG | Full MEQ, partial MSF | |
| CRY1-CC/PER2-AG | Full MSF | |
| CRY1-CG/VNTR-55 | Partial MEQ | |
| PER2-GG/PER3B-GG | Partial MEQ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dang, T.; Russel, W.A.; Saad, T.; Dhawka, L.; Ay, A.; Ingram, K.K. Risk for Seasonal Affective Disorder (SAD) Linked to Circadian Clock Gene Variants. Biology 2023, 12, 1532. https://doi.org/10.3390/biology12121532
Dang T, Russel WA, Saad T, Dhawka L, Ay A, Ingram KK. Risk for Seasonal Affective Disorder (SAD) Linked to Circadian Clock Gene Variants. Biology. 2023; 12(12):1532. https://doi.org/10.3390/biology12121532
Chicago/Turabian StyleDang, Thanh, William A. Russel, Tazmilur Saad, Luvna Dhawka, Ahmet Ay, and Krista K. Ingram. 2023. "Risk for Seasonal Affective Disorder (SAD) Linked to Circadian Clock Gene Variants" Biology 12, no. 12: 1532. https://doi.org/10.3390/biology12121532





