A Self-Referencing Detection of Microorganisms Using Surface Enhanced Raman Scattering Nanoprobes in a Test-in-a-Tube Platform
Abstract
:1. Introduction
2. Experimental Section
2.1. Reagents and Antibodies
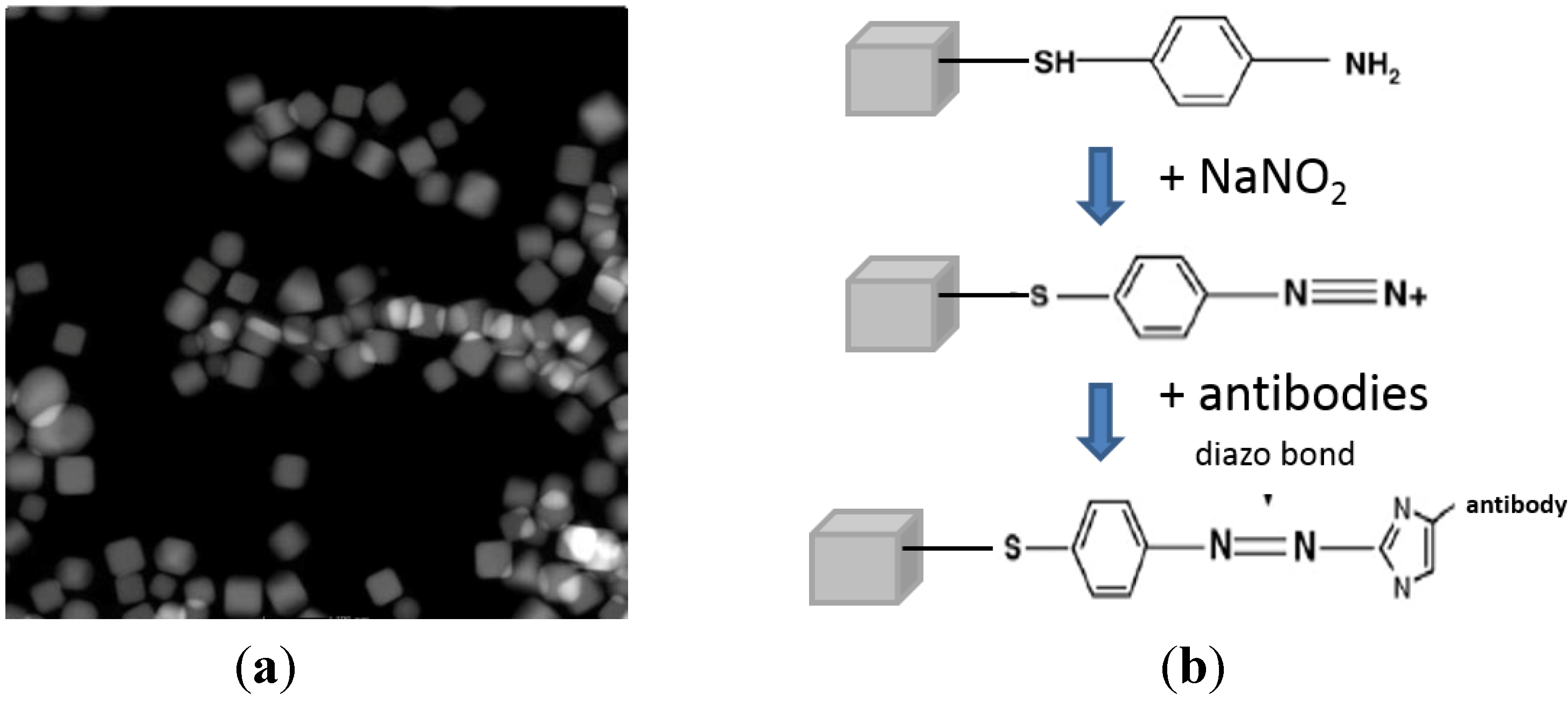
2.2. Nanoprobe Fabrication and Functionalization
2.3. Bacterial Cell Culture
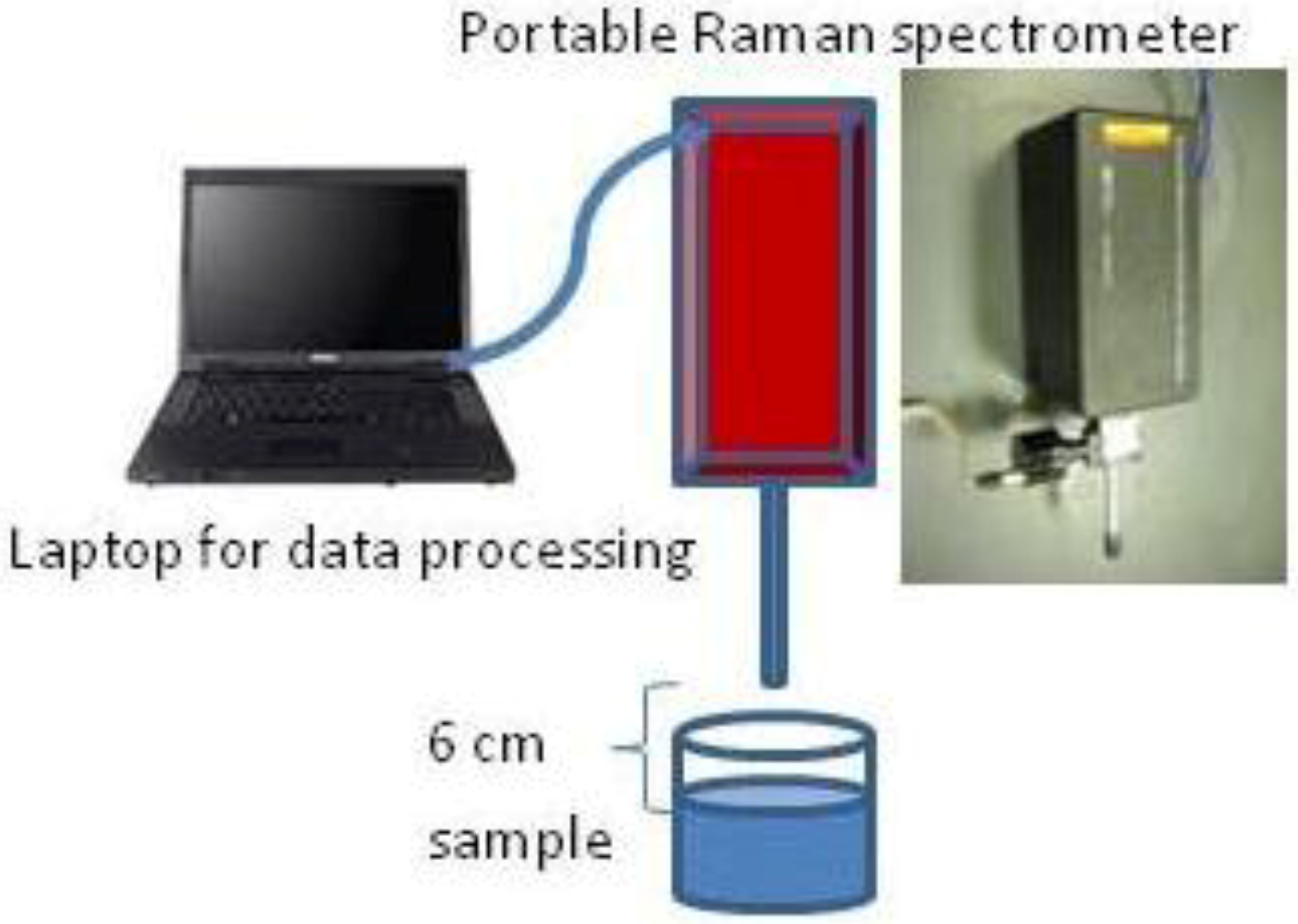
2.4. One-Step Raman Spectroscopic Measurement
2.5. Spectral Data Processing
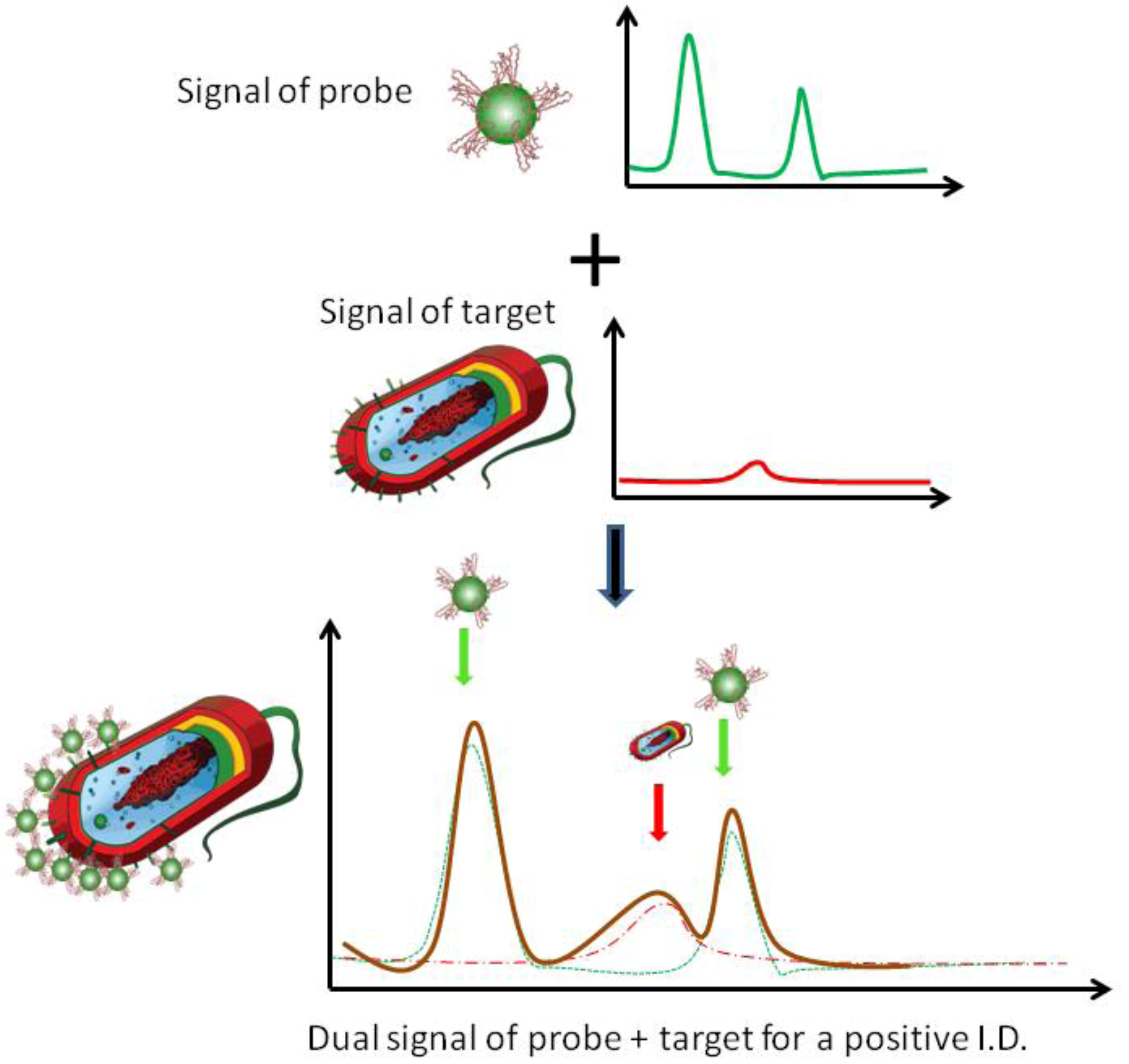
3. Results and Discussion
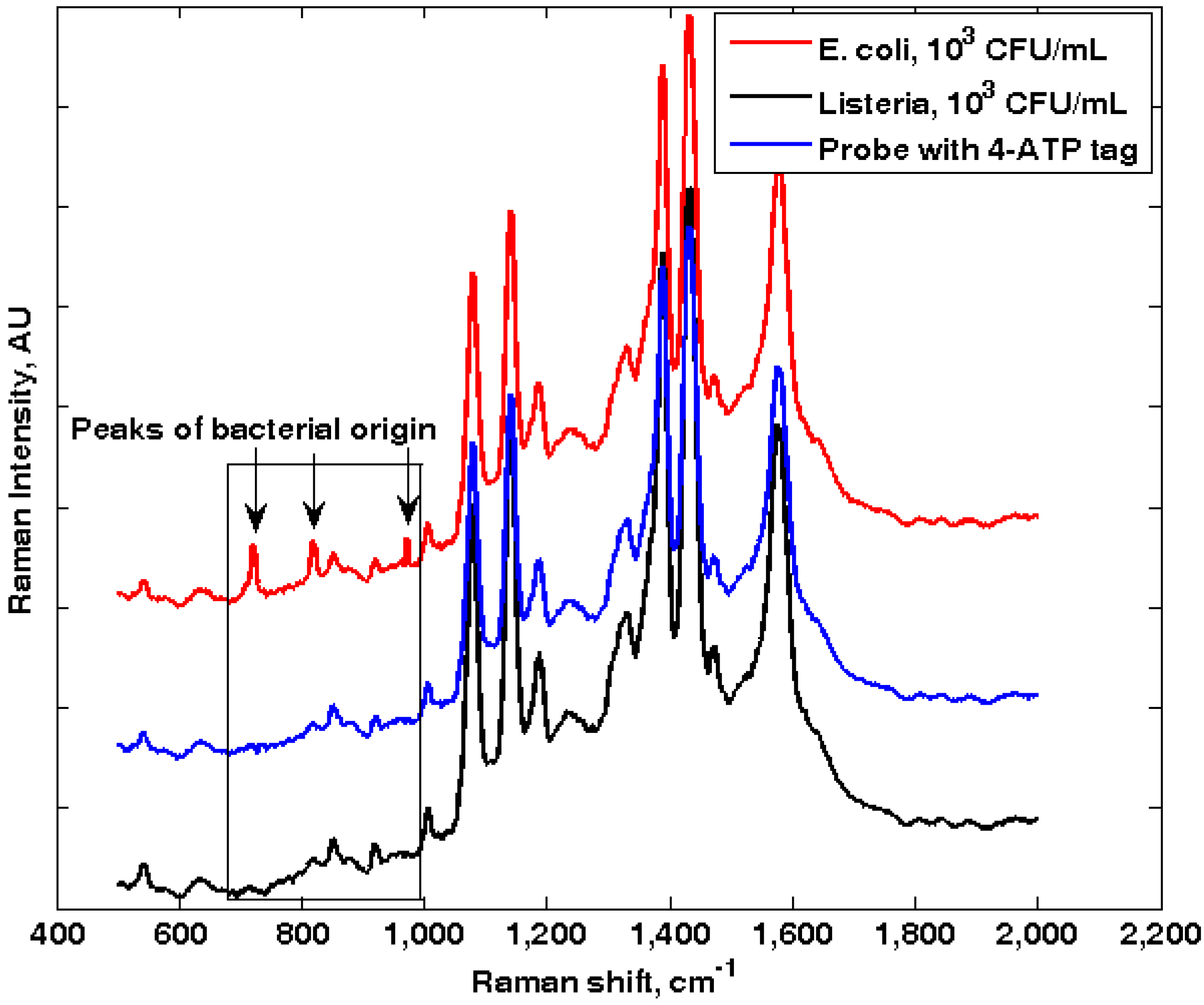
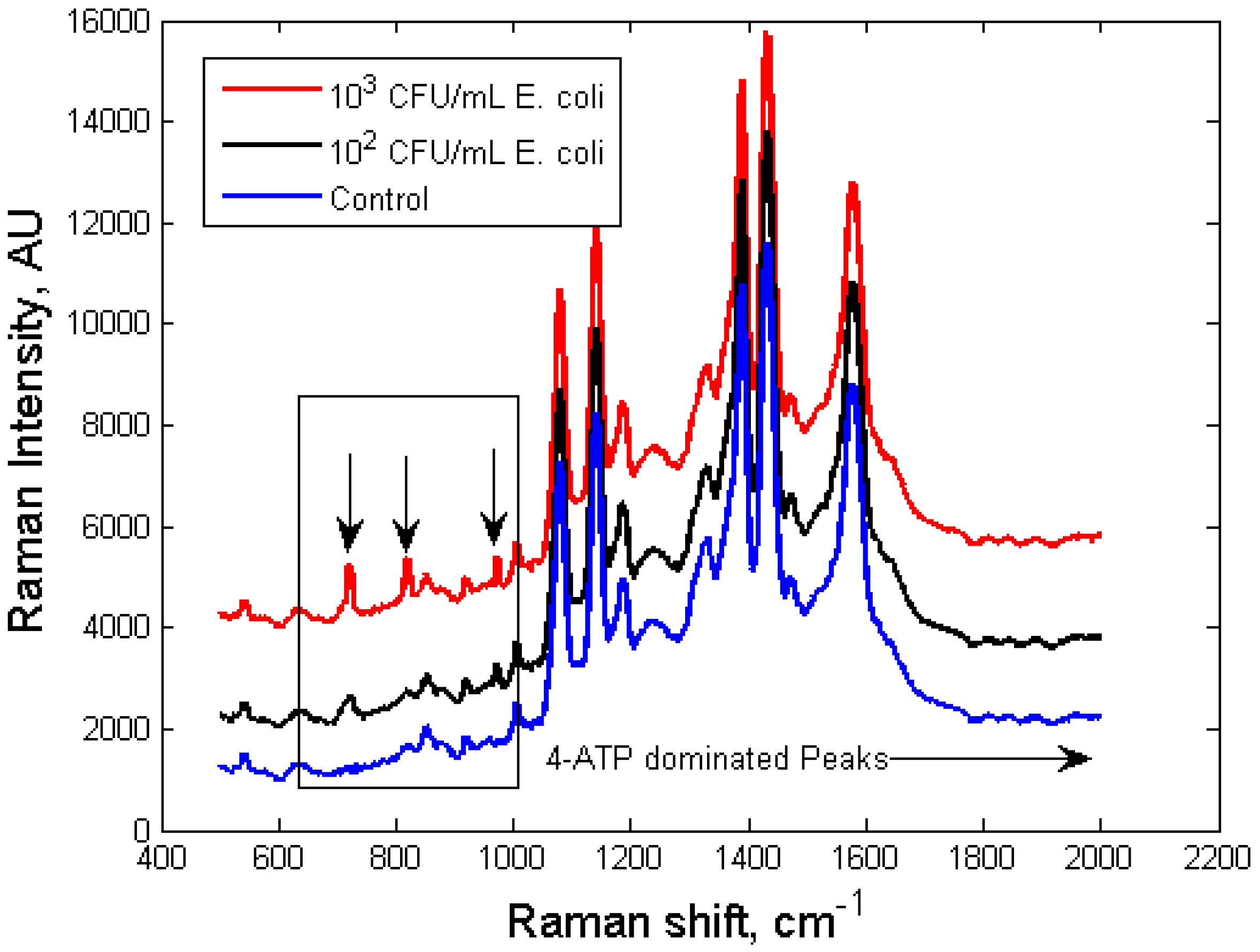
3.1. Self-Referencing Detection of Target Bacteria in a Test-in-a-Tube Platform
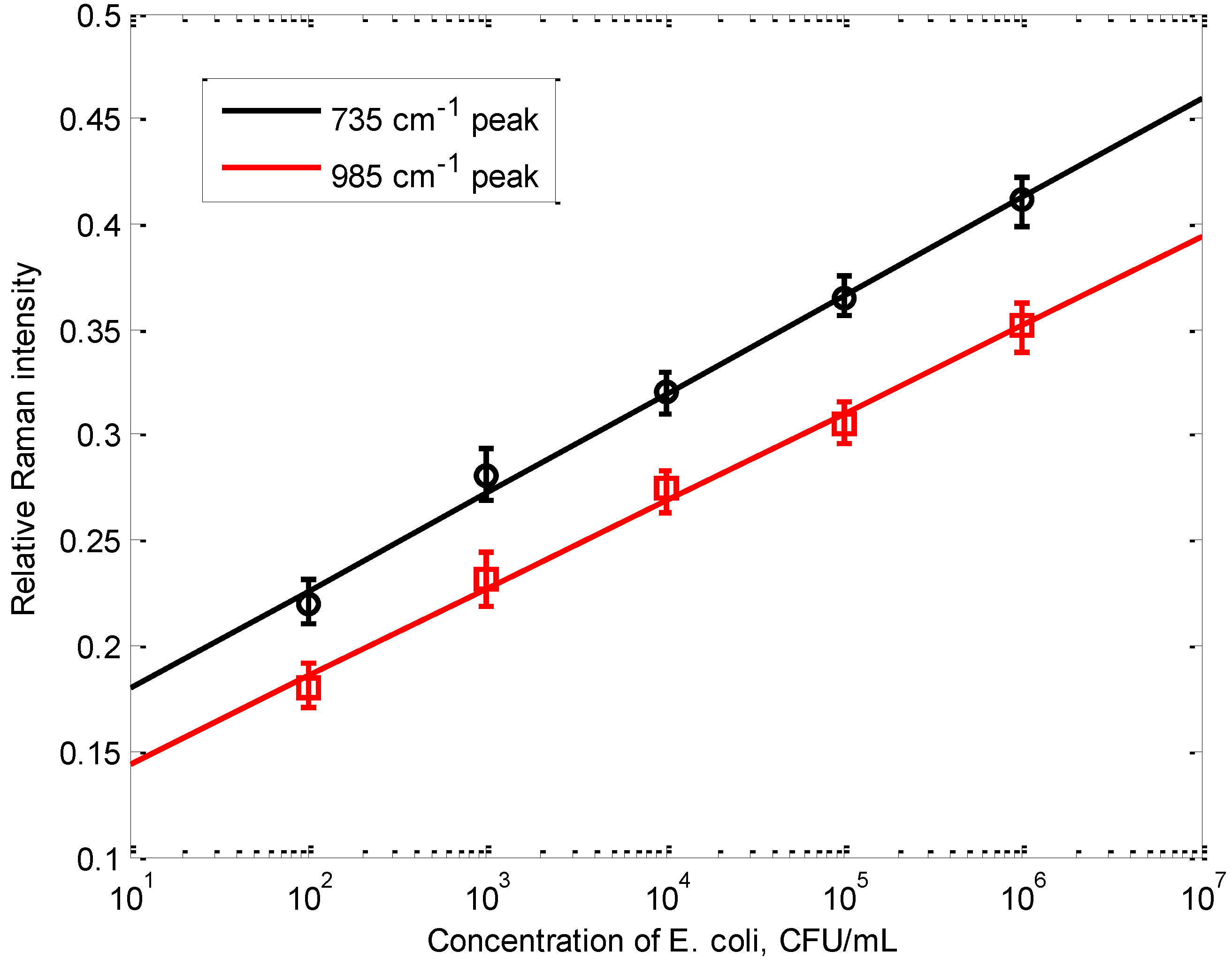
3.2. Limit of Detection of the Dual-Recognition Probing Scheme
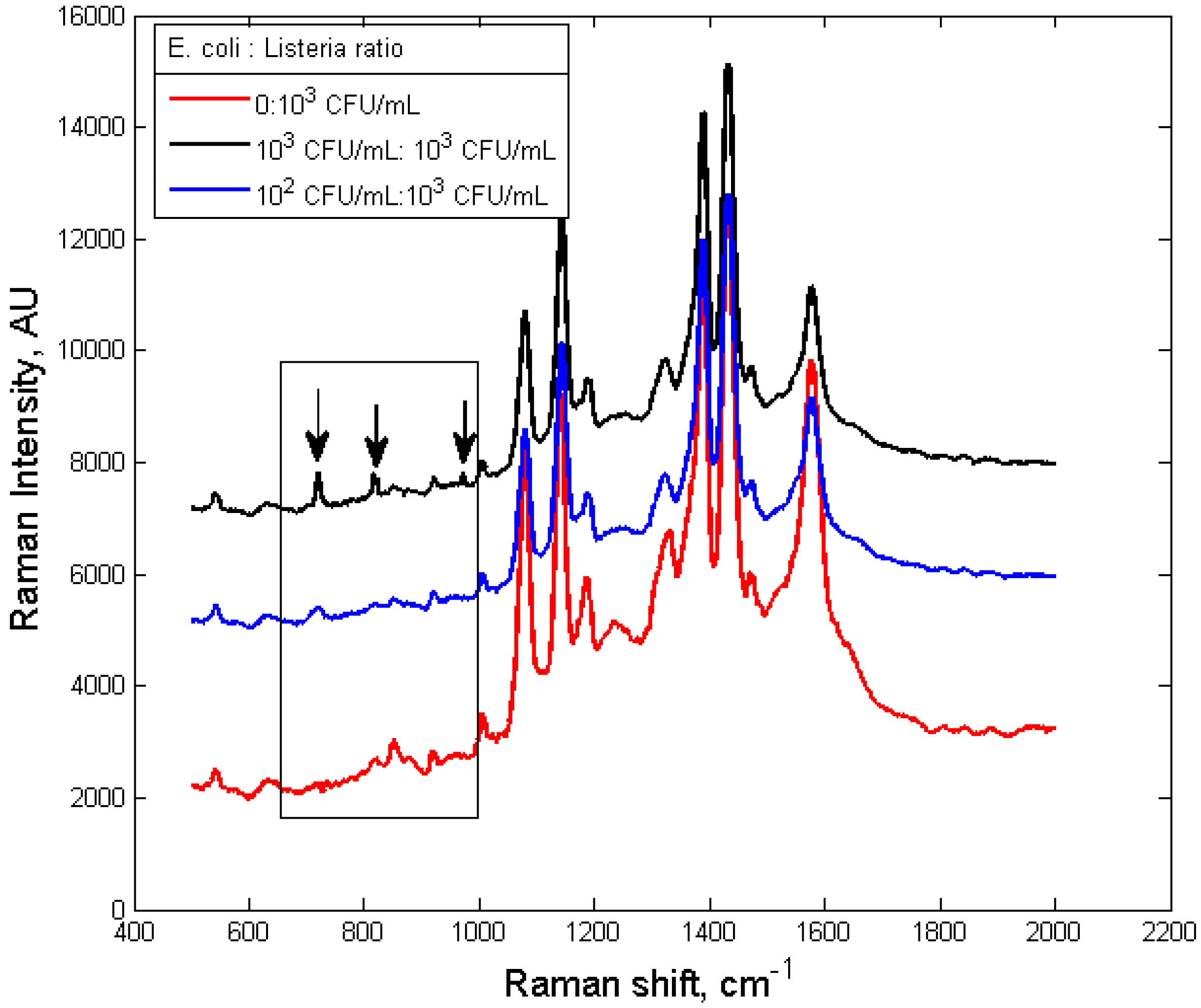
3.3. Specificity of the Dual-Recognition Probing Scheme
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Grifin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar]
- Jarvis, R.M.; Goodacre, R. Characterisation and identification of bacteria using SERS. Chem. Soc. Rev. 2008, 37, 931–936. [Google Scholar] [CrossRef]
- Lam, H.; Kostov, Y. Optical instrumentation for bioprocess monitoring. Adv. Biochem. Eng. Biotechnol. 2010, 116, 125–142. [Google Scholar]
- Sun, L.; Yu, C.; Irudayaraj, I. Surface-enhanced Raman scattering based nonfluorescent probe for multiplex DNA detection. Anal. Chem. 2007, 79, 3981–3988. [Google Scholar] [CrossRef]
- Sun, L.; Yu, C.; Irudayaraj, I. Raman multiplexers for alternative gene splicing. Anal. Chem. 2008, 80, 3342–3349. [Google Scholar] [CrossRef]
- Le Ru, E.C.; Blackie, E.; Meyer, M.; Etchegoin, P.G. Surface Enhanced Raman Scattering enhancement factors: A comprehensive study. J. Phys. Chem. 2007, 111, 13794–13803. [Google Scholar]
- Haran, G. Single-molecule Raman spectroscopy: A probe of surface dynamics and plasmonic fields. Acc. Chem. Res. 2010, 43, 1135–1143. [Google Scholar] [CrossRef]
- Neacsu, C.C.; Dreyer, J.; Behr, N.; Raschke, M.B. Scanning-probe Raman spectroscopy with single-molecule sensitivity. Phys. Rev. B 2006, 73, 193406. [Google Scholar] [CrossRef]
- Bizzarri, A.R.; Cannistraro, S. SERS detection of thrombin by protein recognition using functionalized gold nanoparticles. Nanomedicine NBM 2007, 3, 306–310. [Google Scholar] [CrossRef]
- Drachev, V.P.; Thoreson, M.D.; Khaliullin, E.N.; Davisson, J.; Shalaev, V.M. Surface-enhanced Raman difference between human insulin and insulin lispro detected with adaptive nanostructures. J. Phys. Chem. B. 2004, 108, 18046–18052. [Google Scholar]
- Tripp, R.; Dluhy, R.; Zhao, Y. Novel nanostructures for SERS biosensing. Nano Today 2008, 3, 31–37. [Google Scholar] [CrossRef]
- Wang, Y.; Lee, K.; Irudayaraj, J. Silver nanosphere SERS probes for sensitive identification of pathogens. J. Phys. Chem. C 2010, 114, 16122–16128. [Google Scholar] [CrossRef]
- Kahraman, M.; Zamaleeva, A.I.; Fakhrullin, R.F.; Culha, M. Layer-by-layer coating of bacteria with noble metal nanoparticles for surface-enhanced Raman scattering. Anal. Bioanal. Chem. 2009, 395, 2559–2567. [Google Scholar] [CrossRef]
- Patel, I.S.; Premasiri, W.R.; Moir, D.T.; Ziegler, L.D. Barcoding bacterial cells: A SERS based methodology for pathogen identification. J. Raman Spectrosc. 2008, 39, 1660–1672. [Google Scholar] [CrossRef]
- Jarvis, R.M.; Brooker, A.; Goodacre, R. Surface-enhanced Raman spectroscopy for bacterial discrimination utilizing a scanning electron microscope with a Raman spectroscopy interface. Anal. Chem. 2004, 76, 5198–5202. [Google Scholar] [CrossRef]
- Kahraman, M.; Yazici, M.M.; Sahin, F.; Culha, M. Convective assembly of bacteria for surface-enhanced Raman scattering. Langmuir 2008, 24, 894–901. [Google Scholar] [CrossRef]
- Premasiri, W.R.; Moir, D.T.; Klempner, M.S.; Krieger, N.; Jones, G., II.; Ziegler, L.D. Characterization of the surface enhanced Raman scattering (SERS) of bacteria. J. Phys. Chem. B 2005, 109, 312–320. [Google Scholar]
- Javis, R.M.; Goodacre, R. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal. Chem. 2004, 76, 40–47. [Google Scholar] [CrossRef]
- Efrima, S.; Bronk, B.V. Silver colloids impregnating or coating bacteria. J. Phys. Chem. B 1998, 102, 5947–5950. [Google Scholar] [CrossRef]
- Efrima, S.; Zeiri, L. Understanding SERS of bacteria. J. Raman Spectrosc. 2009, 40, 277–288. [Google Scholar] [CrossRef]
- Murphy, C.J.; Sau, T.K.; Gole, A.M.; Orendorff, C.J.; Gao, J.; Gou, L.; Hunyadi, S.E.; Li, T. Anisotropic metal nanoparticles: Synthesis, assembly, and optical applications. J. Phys. Chem. B 2005, 109, 13857–13870. [Google Scholar]
- Potara, M.; Baia, M.; Farcau, C.; Astilean, S. Chitosan-coated anisotropic silver nanoparticles as a SERS substrate for single-molecule detection. Nanotechnology 2012, 23. [Google Scholar] [CrossRef]
- Qian, X.M.; Nie, S.M. Single-molecule and single-nanoparticle SERS: From fundamental mechanisms to biomedical applications. Chem. Soc. Rev. 2008, 37, 912–920. [Google Scholar] [CrossRef]
- Li, Z.Y.; Xia, Y. Metal nanoparticles with gain toward single-molecule detection by surface-enhanced Raman scattering. Nano Lett. 2010, 10, 243–249. [Google Scholar] [CrossRef]
- Xiao, N.; Yu, C. Dual Recognition Mechanism for Bacteria Detection Using Surface Enhanced Raman Spectroscopic Nanoprobes. In Proceedings of ASABE Annual International Meeting, Pittsburgh, PA, USA, 20–23 June 2010.
- Skrabalak, S.E.; Au, L.; Li, X.D.; Xia, Y. Facile synthesis of Ag nanocubes and Au nanocages. Nat. Protocol. 2007, 2, 2182–2190. [Google Scholar] [CrossRef]
- Zhao, J.; Lui, H.; McLean, D.I.; Zeng, H. Automated autofluorescence background subtraction algorithm for biomedical Raman spectroscopy. Appl. Spectros. 2007, 61, 1225–1232. [Google Scholar] [CrossRef]
- Wang, Q.; Grozdanic, S.D.; Harper, M.M.; Hamouche, K.; Hamouche, N.; Kecova, H.; Lazic, T.; Yu, C. Detection and characterization of glaucoma-like canine retinal tissues using Raman spectroscopy. J. Biomed. Opt. 2013, 18. [Google Scholar] [CrossRef]
- Rycenga, M.; Kim, M.H.; Camargo, P.H.C.; Cobley, C.; Li, Z.; Xia, Y. Surface-enhanced Raman scattering: Comparison of three different molecules on single-crystal nanocubes and nanospheres of silver. J. Phys. Chem. A 2009, 113, 3932–3939. [Google Scholar]
- Frey, S.; Stadler, V.; Heister, K.; Eck, W.; Zharnikov, M.; Grunze, M.; Zeysing, B.; Tertort, A. Structure of thioaromatic self-assembled monolayers on gold and silver. Langmuir 2001, 17, 2408–2415. [Google Scholar] [CrossRef]
- Zheng, J.; Zhou, Y.; Li, X.; Ji, Y.; Lu, T.; Gu, R. Surface-enhanced Raman scattering of 4-aminothiophenol in assemblies of nanosized particles and the macroscopic surface of silver. Langmuir 2003, 19, 632–636. [Google Scholar] [CrossRef]
- Jiao, L.; Niu, L.; Shen, J.; You, T.; Dong, S.; Ivaska, A. Simple azo derivatization on 4-aminothiophenol/Au monolayer. Electrochem. Comm. 2005, 7, 219–222. [Google Scholar] [CrossRef]
- Xiao, N.; Yu, C. Rapid-response and highly sensitive non-crosslinking colorimetric nitrite sensor using 4-aminothiophenol modified gold nanorods. Anal. Chem. 2010, 82, 3659–3663. [Google Scholar] [CrossRef]
- Zheng, Y.; Carey, P.R.; Palfey, B.A. Raman spectrum of fully reduced flavin. J. Raman Spectrosc. 2004, 35, 521–524. [Google Scholar] [CrossRef]
Appendix
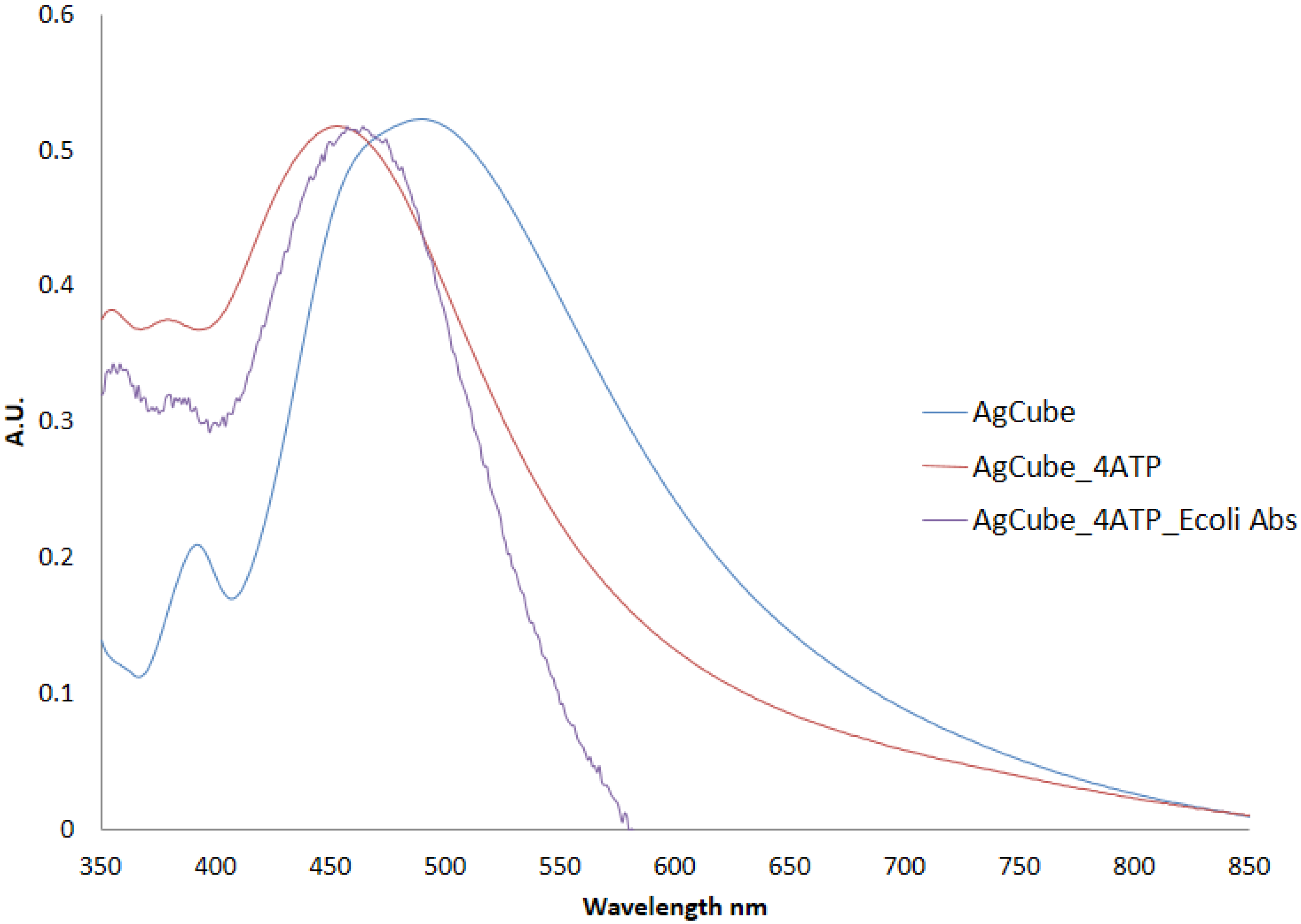
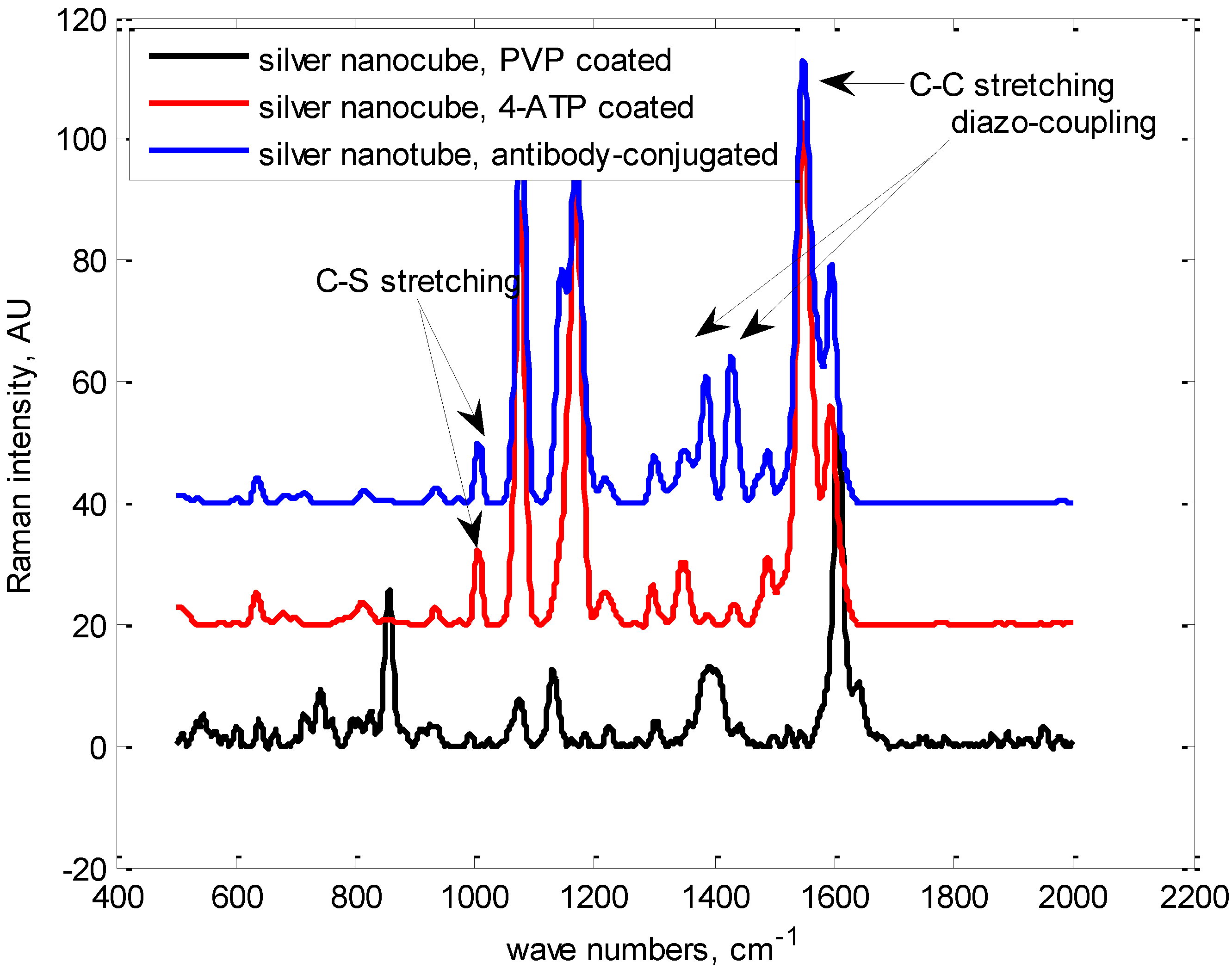
Synthesis and Bio-Functionalization of Silver Nanocubes
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Xiao, N.; Wang, C.; Yu, C. A Self-Referencing Detection of Microorganisms Using Surface Enhanced Raman Scattering Nanoprobes in a Test-in-a-Tube Platform. Biosensors 2013, 3, 312-326. https://doi.org/10.3390/bios3030312
Xiao N, Wang C, Yu C. A Self-Referencing Detection of Microorganisms Using Surface Enhanced Raman Scattering Nanoprobes in a Test-in-a-Tube Platform. Biosensors. 2013; 3(3):312-326. https://doi.org/10.3390/bios3030312
Chicago/Turabian StyleXiao, Nan, Chao Wang, and Chenxu Yu. 2013. "A Self-Referencing Detection of Microorganisms Using Surface Enhanced Raman Scattering Nanoprobes in a Test-in-a-Tube Platform" Biosensors 3, no. 3: 312-326. https://doi.org/10.3390/bios3030312
APA StyleXiao, N., Wang, C., & Yu, C. (2013). A Self-Referencing Detection of Microorganisms Using Surface Enhanced Raman Scattering Nanoprobes in a Test-in-a-Tube Platform. Biosensors, 3(3), 312-326. https://doi.org/10.3390/bios3030312




