Ultrasensitive Fluorescence Lateral Flow Assay for Simultaneous Detection of Pseudomonas aeruginosa and Salmonella typhimurium via Wheat Germ Agglutinin-Functionalized Magnetic Quantum Dot Nanoprobe
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials, Chemicals, and Instruments
2.2. Synthesis of Mag@QDs Nanobeads
2.3. Synthesis of WGA-Modified Mag@QDs
2.4. Fabrication of Mag@QDs-WGA-Based LFA for Bacterial Detection
2.5. Preparation of Bacterial Sample
2.6. Bacterial Detection with Mag@QDs-WGA-Based LFA
3. Results
3.1. Principle of Mag@QDs-WGA-LFA for Bacteria Detection
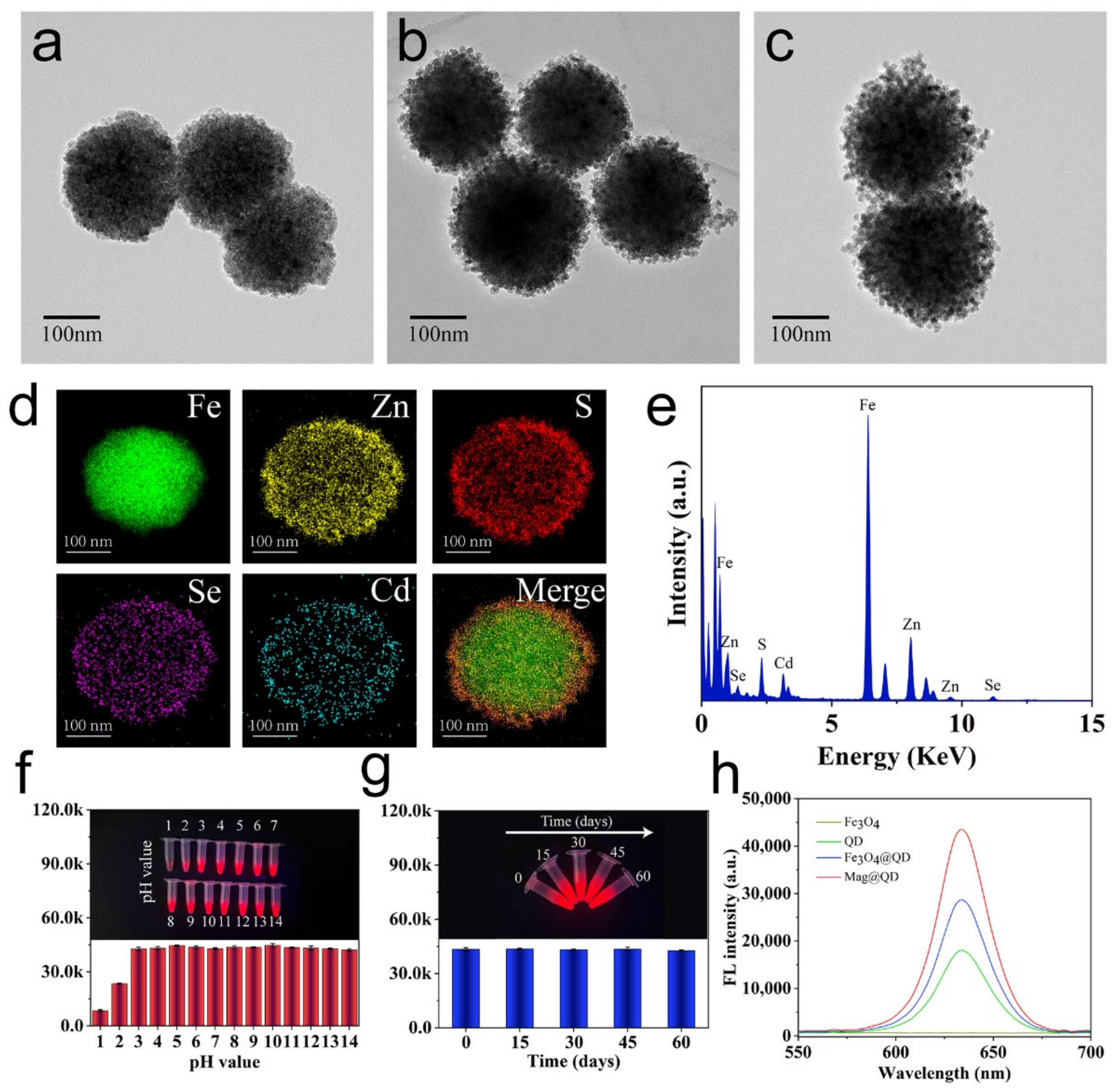
3.2. Characterization of Mag@QDs-WGA
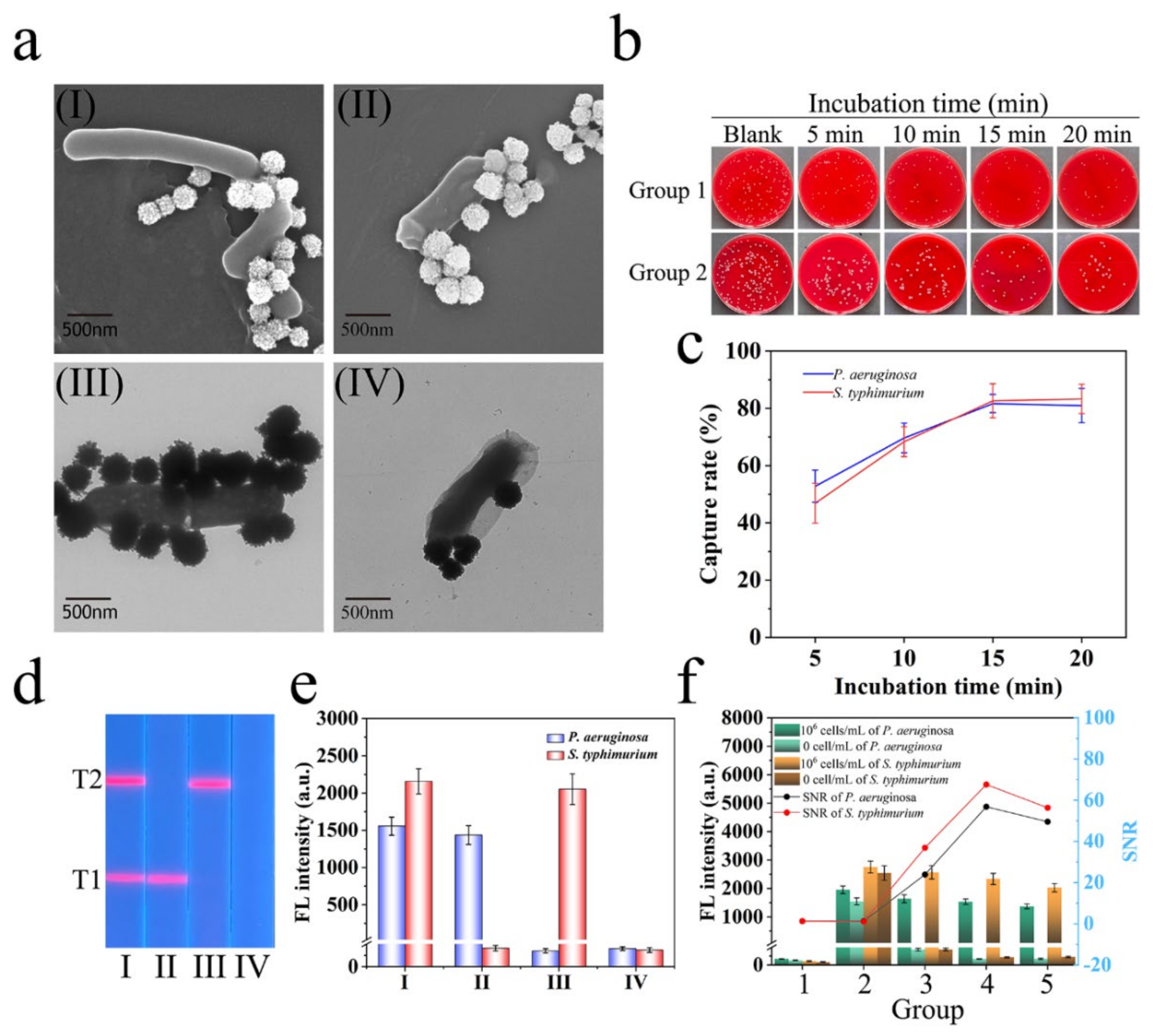
3.3. Verification of the Capture Ability of Mag@QDs-WGA for Bacteria
3.4. Construction of Mag@QDs-WGA-Based LFA Biosensor
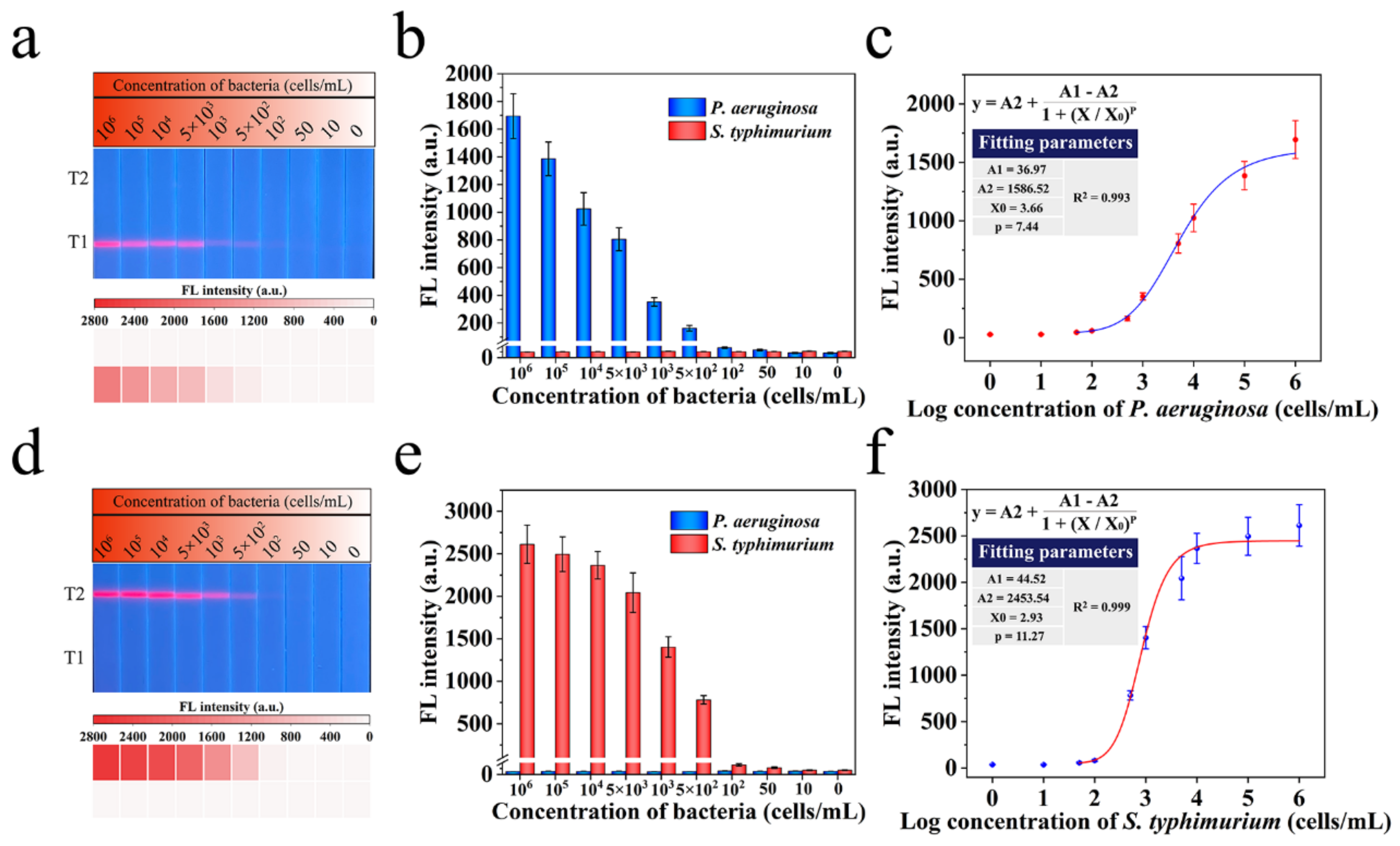
3.5. Evaluation of Mag@QDs-WGA-LFA System for Bacteria Detection
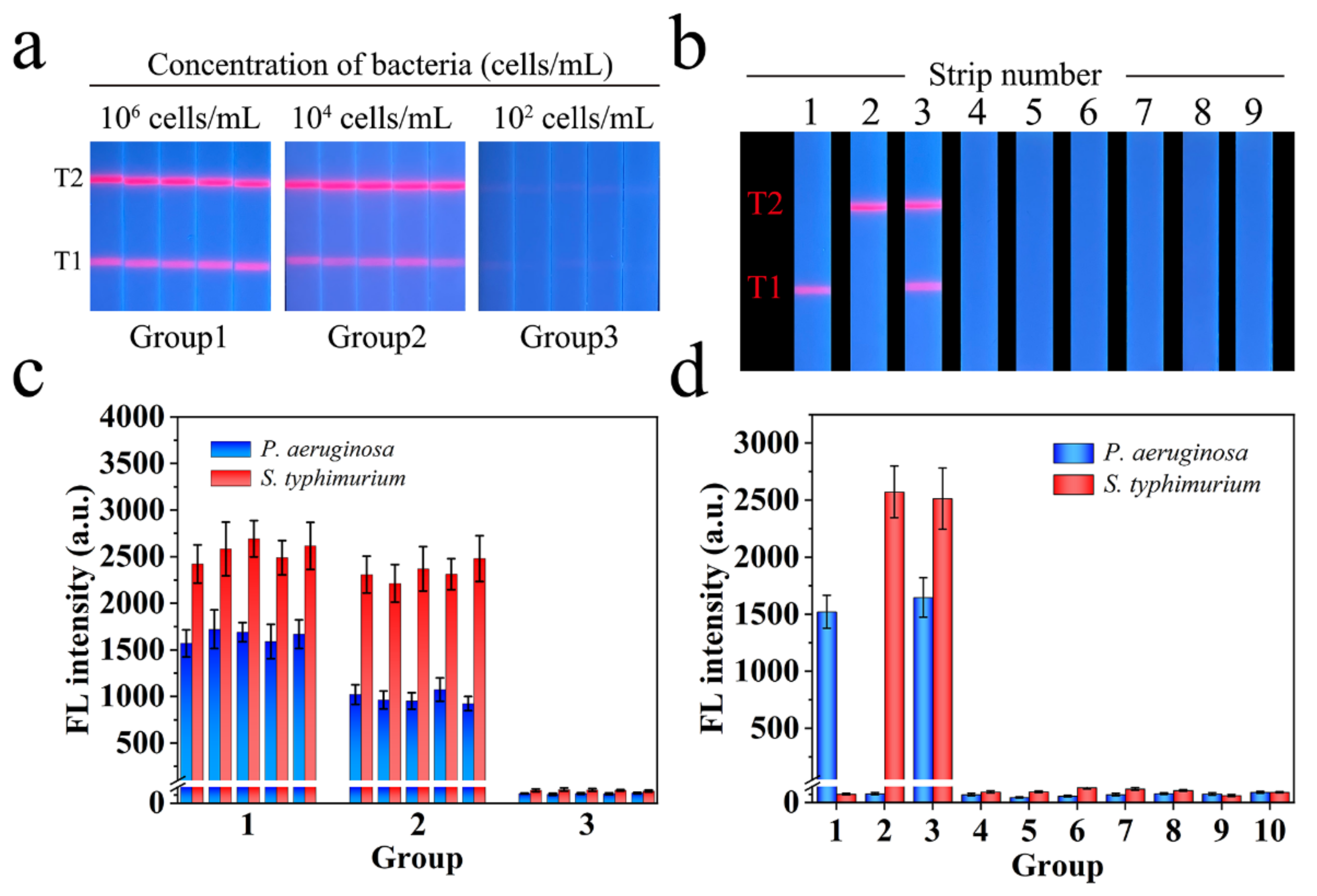
3.6. Specificity and Repeatability of Mag@QDs-WGA-LFA
3.7. Detection of Real Food and Environment Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, H.; Lee, S.; Seo, H.W.; Kang, B.; Moon, J.; Lee, K.G.; Yong, D.; Kang, H.; Jung, J.; Lim, E.-K.; et al. Clustered regularly interspaced short palindromic repeats-mediated surface-enhanced Raman scattering assay for multidrug-resistant bacteria. ACS Nano 2020, 14, 17241–17253. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Andler, S.M.; Goddard, J.M.; Nugen, S.R.; Rotello, V.M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2017, 46, 1272–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Zhou, X.; Shan, Y.; Yue, H.; Huang, R.; Hu, J.; Xing, D. Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat. Commun. 2020, 11, 267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Huang, J.; Li, Y.; Lv, J.; Ding, X. A simple and rapid colorimetric bacteria detection method based on bacterial inhibition of glucose oxidase-catalyzed reaction. Talanta 2019, 197, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Xiao, R.; Cheng, S.; Wang, S.; Shi, L.; Wang, C.; Qi, K.; Wang, S. A universal SERS-label immunoassay for pathogen bacteria detection based on Fe3O4@Au-aptamer separation and antibody-protein A orientation recognition. Anal. Chim. Acta 2021, 1160, 338421. [Google Scholar] [CrossRef]
- Roth-Konforti, M.; Green, O.; Hupfeld, M.; Fieseler, L.; Heinrich, N.; Ihssen, J.; Vorberg, R.; Wick, L.; Spitz, U.; Shabat, D. Ultrasensitive detection of salmonella and listeria monocytogenes by small-molecule chemiluminescence probes. Angew. Chem. 2019, 58, 10361–10367. [Google Scholar] [CrossRef]
- Jia, F.; Bai, X.; Zhang, X.; Fu, Y.; Li, Y.; Li, X.; Kokini, J.L. A Low-field magnetic resonance imaging Aptasensor for the rapid and visual sensing of Pseudomonas aeruginosa in food, juice, and water. Anal. Chem. 2021, 93, 8631–8637. [Google Scholar] [CrossRef]
- Dietvorst, J.; Vilaplana, L.; Uria, N.; Marco, M.-P.; Muñoz-Berbel, X. Current and near-future technologies for antibiotic susceptibility testing and resistant bacteria detection. TrAC Trends Anal. Chem. 2020, 127, 115891. [Google Scholar] [CrossRef]
- Boardman, A.K.; Wong, W.S.; Premasiri, W.R.; Ziegler, L.D.; Lee, J.C.; Miljkovic, M.; Klapperich, C.M.; Sharon, A.; Sauer-Budge, A.F. Rapid detection of bacteria from blood with surface-enhanced Raman spectroscopy. Anal. Chem. 2016, 88, 8026–8035. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, R.; Wang, H.; Slavik, M.; Wei, H.; Li, Y. An aptamer-based PCR method coupled with magnetic immunoseparation for sensitive detection of Salmonella typhimurium in ground turkey. Anal. Biochem. 2017, 533, 34–40. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, C.; Jin, J.; Huang, L.; Yu, W.; Su, B.; Hu, J. Ratiometric fluorescent lateral flow immunoassay for point-of-care testing of acute myocardial infarction. Angew. Chem. 2021, 60, 13042–13049. [Google Scholar] [CrossRef]
- Cheng, X.; Zheng, S.; Wang, W.; Han, H.; Yang, X.; Shen, W.; Wang, C.; Wang, S. Synthesis of two-dimensional graphene oxide-fluorescent nanoprobe for ultrasensitive and multiplex immunochromatographic detection of respiratory bacteria. Chem. Eng. J. 2021, 426, 131836. [Google Scholar] [CrossRef]
- Wang, C.; Wang, C.; Li, J.; Tu, Z.; Gu, B.; Wang, S. Ultrasensitive and multiplex detection of four pathogenic bacteria on a bi-channel lateral flow immunoassay strip with three-dimensional membrane-like SERS nanostickers. Biosens. Bioelectron. 2022, 214, 114525. [Google Scholar] [CrossRef]
- Wang, C.; Shen, W.; Rong, Z.; Liu, X.; Gu, B.; Xiao, R.; Wang, S. Layer-by-layer assembly of magnetic-core dual quantum dot-shell nanocomposites for fluorescence lateral flow detection of bacteria. Nanoscale 2020, 12, 795–807. [Google Scholar] [CrossRef]
- Lai, X.; Zhang, G.; Zeng, L.; Xiao, X.; Peng, J.; Guo, P.; Zhang, W.; Lai, W. Synthesis of PDA-mediated magnetic bimetallic nanozyme and its application in immunochromatographic assay. ACS Appl. Mater. Interfaces 2020, 13, 1413–1423. [Google Scholar] [CrossRef]
- Chen, R.; Chen, X.; Zhou, Y.; Lin, T.; Leng, Y.; Huang, X.; Xiong, Y. “Three-in-One” Multifunctional nanohybrids with colorimetric magnetic catalytic activities to enhance immunochromatographic diagnosis. ACS Nano 2022, 16, 3351–3361. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, L.; Liu, B.; Ni, H.; Sun, L.; Su, E.; Chen, H.; Gu, Z.; Zhao, X. Quantitative and ultrasensitive detection of multiplex cardiac biomarkers in lateral flow assay with core-shell SERS nanotags. Biosens. Bioelectron. 2018, 106, 204–211. [Google Scholar] [CrossRef]
- Lee, S.H.; Hwang, J.; Kim, K.; Jeon, J.; Lee, S.; Ko, J.; Lee, J.; Kang, M.; Chung, D.R.; Choo, J. Quantitative serodiagnosis of scrub typhus using surface-enhanced Raman scattering-based lateral flow assay platforms. Anal. Chem. 2019, 91, 12275–12282. [Google Scholar] [CrossRef]
- Zheng, S.; Wang, C.; Li, J.; Wang, W.; Yu, Q.; Wang, C.; Wang, S. Graphene oxide-based three-dimensional Au nanofilm with high-density and controllable hotspots: A powerful film-type SERS tag for immunochromatographic analysis of multiple mycotoxins in complex samples. Chem. Eng. J. 2022, 448, 137760. [Google Scholar] [CrossRef]
- Shen, W.; Wang, C.; Zheng, S.; Jiang, B.; Li, J.; Pang, Y.; Wang, C.; Hao, R.; Xiao, R. Ultrasensitive multichannel immunochromatographic assay for rapid detection of foodborne bacteria based on two-dimensional film-like SERS labels. J. Hazard. Mater. 2022, 437, 129347. [Google Scholar] [CrossRef]
- Gao, F.; Lei, C.; Liu, Y.; Song, H.; Kong, Y.; Wan, J.; Yu, C. Rational design of dendritic mesoporous silica nanoparticles’ surface chemistry for quantum dot enrichment and an ultrasensitive lateral flow immunoassay. ACS Appl. Mater. Interfaces 2021, 13, 21507–21515. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, X.; Zheng, S.; Cheng, X.; Xiao, R.; Li, Q.; Wang, W.; Liu, X.; Wang, S. Development of an ultrasensitive fluorescent immunochromatographic assay based on multilayer quantum dot nanobead for simultaneous detection of SARS-CoV-2 antigen and influenza A virus. Sensor Actuat. B Chem. 2021, 345, 130372. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Yang, X.; Zhang, B.; Cheng, S.; Han, H.; Jin, Q.; Wang, C.; Xiao, R. Sensitive detection of Escherichia coli O157:H7 and Salmonella typhimurium in food samples using two-channel fluorescence lateral flow assay with liquid Si@quantum dot. Food Chem. 2021, 363, 130400. [Google Scholar] [CrossRef] [PubMed]
- Mi, F.; Guan, M.; Hu, C.; Peng, F.; Sun, S.; Wang, X. Application of lectin-based biosensor technology in the detection of foodborne pathogenic bacteria: A review. Analyst 2021, 146, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Meng, X.; Wang, Y.; Yan, M.; Aguilar, Z.P.; Xu, H. 2-Step lectin-magnetic separation (LMS) strategy combined with AuNPs-based colorimetric system for S. aureus detection in blood. Sensor Actuat B Chem. 2019, 279, 87–94. [Google Scholar] [CrossRef]
- Kearns, H.; Goodacre, R.; Jamieson, L.E.; Graham, D.; Faulds, K. SERS detection of multiple antimicrobial-resistant pathogens using Nanosensors. Anal. Chem. 2017, 89, 12666–12673. [Google Scholar] [CrossRef] [Green Version]
- Masigol, M.; Fattahi, N.; Barua, N.; Lokitz, B.S.; Retterer, S.T.; Platt, T.G.; Hansen, R.R. Identification of critical surface parameters driving lectin-mediated capture of bacteria from solution. Biomacromolecules 2019, 20, 2852–2863. [Google Scholar] [CrossRef]
- Cheng, S.; Tu, Z.; Zheng, S.; Cheng, X.; Han, H.; Wang, C.; Xiao, R.; Gu, B. An efficient SERS platform for the ultrasensitive detection of Staphylococcus aureus and Listeria monocytogenes via wheat germ agglutinin-modified magnetic SERS substrate and streptavidin/aptamer co-functionalized SERS tags. Anal. Chim. Acta 2021, 1187, 339155. [Google Scholar] [CrossRef]
- Shen, W.; Wang, C.; Yang, X.; Wang, C.; Zhou, Z.; Liu, X.; Xiao, R.; Gu, B.; Wang, S. Synthesis of raspberry-like nanogapped Fe3O4@Au nanocomposites for SERS-based lateral flow detection of multiple tumor biomarkers. J. Mater. Chem. C 2020, 8, 12854–12864. [Google Scholar] [CrossRef]
- Wu, T.; Li, J.; Zheng, S.; Yu, Q.; Qi, K.; Shao, Y.; Wang, C.; Tu, J.; Xiao, R. Magnetic nanotag-based colorimetric/SERS dual-readout immunochromatography for ultrasensitive detection of clenbuterol hydrochloride and ractopamine in food samples. Biosensors 2022, 12, 709. [Google Scholar] [CrossRef]
- Jia, X.; Liu, Z.; Peng, Y.; Hou, G.; Chen, W.; Xiao, R. Automatic and sensitive detection of West Nile virus non-structural protein 1 with a portable SERS-LFIA detector. Microchim. Acta 2021, 188, 206. [Google Scholar] [CrossRef]
- Rajendran, V.K.; Bakthavathsalam, P.; Jaffar Ali, B.M. Smartphone based bacterial detection using biofunctionalized fluorescent nanoparticles. Microchim. Acta 2014, 181, 1815–1821. [Google Scholar] [CrossRef]
- Song, C.; Liu, C.; Wu, S.; Li, H.; Guo, H.; Yang, B.; Qiu, S.; Li, J.; Liu, L.; Zeng, J.; et al. Development of a lateral flow colloidal gold immunoassay strip for the simultaneous detection of Shigella boydii and Escherichia coli O157:H7 in bread, milk and jelly samples. Food Control 2016, 59, 345–351. [Google Scholar] [CrossRef]
- Tominaga, T. Rapid detection of Klebsiella pneumoniae, Klebsiella oxytoca, Raoultella ornithinolytica and other related bacteria in food by lateral-flow test strip immunoassays. J. Microbiol. Methods 2018, 147, 43–49. [Google Scholar] [CrossRef]
- Hu, J.; Jiang, Y.Z.; Tang, M.; Wu, L.L.; Xie, H.Y.; Zhang, Z.L.; Pang, D.W. Colorimetric-fluorescent-magnetic nanosphere-based multimodal assay platform for salmonella detection. Anal. Chem. 2019, 91, 1178–1184. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, X.; Liu, X.; Li, J.; Wang, C.; Wang, S. Polyethyleneimine-interlayered silica-core quantum dot-shell nanocomposites for sensitive detection of Salmonella typhimurium via a lateral flow immunoassay. RSC Adv. 2020, 10, 2483–2489. [Google Scholar] [CrossRef] [Green Version]
- Tang, R.; Yang, H.; Gong, Y.; You, M.; Liu, Z.; Choi, J.R.; Wen, T.; Qu, Z.; Mei, Q.; Xu, F. A fully disposable and integrated paper-based device for nucleic acid extraction, amplification and detection. Lab. Chip. 2017, 17, 1270–1279. [Google Scholar] [CrossRef]
- Gao, P.; Wang, L.; He, Y.; Wang, Y.; Yang, X.; Fu, S.; Qin, X.; Chen, Q.; Man, C.; Jiang, Y. An enhanced lateral flow assay based on aptamer-magnetic separation and multifold AuNPs for ultrasensitive detection of Salmonella Typhimurium in milk. Foods 2021, 10, 1605. [Google Scholar] [CrossRef]



| Detection Method | Bacteria | LOD (Cells mL−1) | Reference |
|---|---|---|---|
| Magnetic-Fluorescent LFA | S. typhimurium | 3.75 × 103 | [35] |
| Fluorescent LFA | S. typhimurium | 5 × 102 | [36] |
| Colorimetric LFA | S. typhimurium | 102 | [37] |
| Fluorescent LFA | S. typhimurium | 8.6 × 102 | [38] |
| Fluorescent LFA | S. typhimurium | 50 | [23] |
| Colorimetric LFA | P. aeruginosa | 3.3 × 102 | [36] |
| Magnetic-Fluorescent LFA | P. aeruginosa S. typhimurium | 25 28 | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, Z.; Yang, X.; Dong, H.; Yu, Q.; Zheng, S.; Cheng, X.; Wang, C.; Rong, Z.; Wang, S. Ultrasensitive Fluorescence Lateral Flow Assay for Simultaneous Detection of Pseudomonas aeruginosa and Salmonella typhimurium via Wheat Germ Agglutinin-Functionalized Magnetic Quantum Dot Nanoprobe. Biosensors 2022, 12, 942. https://doi.org/10.3390/bios12110942
Tu Z, Yang X, Dong H, Yu Q, Zheng S, Cheng X, Wang C, Rong Z, Wang S. Ultrasensitive Fluorescence Lateral Flow Assay for Simultaneous Detection of Pseudomonas aeruginosa and Salmonella typhimurium via Wheat Germ Agglutinin-Functionalized Magnetic Quantum Dot Nanoprobe. Biosensors. 2022; 12(11):942. https://doi.org/10.3390/bios12110942
Chicago/Turabian StyleTu, Zhijie, Xingsheng Yang, Hao Dong, Qing Yu, Shuai Zheng, Xiaodan Cheng, Chongwen Wang, Zhen Rong, and Shengqi Wang. 2022. "Ultrasensitive Fluorescence Lateral Flow Assay for Simultaneous Detection of Pseudomonas aeruginosa and Salmonella typhimurium via Wheat Germ Agglutinin-Functionalized Magnetic Quantum Dot Nanoprobe" Biosensors 12, no. 11: 942. https://doi.org/10.3390/bios12110942





