Information as a Manifestation of Development
Abstract
: Information manifests a reduction in uncertainty or indeterminacy. As such it can emerge in two ways: by measurement, which involves the intentional choices of an observer; or more generally, by development, which involves systemically mutual (‘self-organizing’) processes that break symmetry. The developmental emergence of information is most obvious in ontogeny, but pertains as well to the evolution of ecosystems and abiotic dissipative structures. In this review, a seminal, well-characterized ontogenetic paradigm—the sea urchin embryo—is used to show how cybernetic causality engenders the developmental emergence of biological information at multiple hierarchical levels of organization. The relevance of information theory to developmental genomics is also discussed.1. Introduction: Indeterminacy, Information, Feedback and History
In colloquial usage information is widely understood as being that which imparts knowledge: The structural qualities by which things, events, phenomena, etc., can be distinguished and described (and from thence explained and predicted). Knowledge can be more or less certain however, indicating that information is also quantitative. It is therefore reasonable to define information in two ways: Qualitatively, as that which reduces uncertainty, and quantitatively, as the extent to which uncertainty is reduced. These definitions can be objectively generalized by substituting the term indeterminacy for uncertainty.
Indeterminacy is related to statistical randomness, and hence to the concept of entropy developed by Ludwig Boltzmann to explain the 2nd law of thermodynamics, and later by Claude Shannon to quantify the capacity of a noisy communication channel to transmit information. The higher a system's entropy, the more uncertain are the microstates or outcomes of the system—in other words entropy quantifies a system's indeterminacy. Thus, if a system's entropy can be measured, any information that emerges within the system can be quantified as a decrease in that entropy.
From the foregoing it should be clear that information is a constraint on degrees of freedom, randomness or the production of entropy. As such it can emerge through measurements that reduce the uncertainty associated with the state or outcome of an observed process; or more generally, through symmetry breaking processes that locally reduce the indeterminacy of a system. A symmetry-breaking trajectory of change producing an increase in determinacy is commonly referred to as development; hence, in general, information emerges developmentally.
Development occurs within localized systems of interacting processes comprising nested levels of inclusiveness, and while it is initiated from the bottom-up (by way of growth) it is ultimately controlled from the top-down (by way of selection); it can thus be modeled hierarchically whenever enough information exists to locate a system [1-4]. For living systems this is straightforward because life is based on cells, systems that are spatially bounded by lipid membranes. Moreover, cells contain complex polymers in which functionally significant, anticipatory information is both stored and faithfully reproduced during cellular reproduction. This allows for the heritable propagation of information during exponential growth, the basis for evolution by Darwinian selection. Within any complex system, the de novo emergence of information at a given focal level is governed both by lower-level internal processes and by higher level environmental context; thus, if the focal level is a tissue within a multicellular organism, developmental morphogenesis of that tissue is governed both by cell physiology and organismal constraints.
The key to the heritable reproducibility of biological information is the DNA molecule: A complex polymer that, by virtue of its deoxyribose sugar backbone and double helical structure involving specific Watson and Crick base-pairs, is ideally suited to both storing and reproducing information. It is probable that early in the evolution of life this job was carried out by RNA and later co-opted by DNA [5]. In any case the major challenge for understanding the origin of life—an emergence of information and hence (by the thesis of this review) a developmental phenomenon—is to identify how pre-biotic chemistry gave rise to a complex chemical polymer that allowed for Darwinian selection— which is strictly a biological phenomenon.
The thesis of this review is that the developmental emergence of information is caused by feedback (‘cybernetic causality’). This is obvious in organismal ontogeny, which will be the main focus of this review. However, it is also apparent in higher-level phenomena such as ecological succession, phylogenetic evolution, and perhaps even cosmology [3,4,6]. In all of these cases localized trajectories of systemic change can be identified that manifest increasing determinacy (i.e., increasing predictability of outcome owing to structural constraint) owing to autocatalytic feedback cycles. The case of ontogeny is particularly revealing however, because it shows not only how information emerges de novo via development, but also how this de novo developmental emergence is controlled by the inherited record of historical information contained within the system (including the deep historical information stored in DNA). It can thus be seen that development occurs only to the extent that the outcome of a process is cyclically constrained (or controlled) by the history of that process. By extension, history is implicit in the concept of information. Nevertheless, it is useful to distinguish between ‘pre-formed’ historical information that affords causal constraint or control, and emergent information that is produced de novo during development.
2. A Developmental Paradigm: Sea Urchin Embryogenesis
The sea urchin embryo has been a paradigm of developmental biology since the inception of experimental embryology as a research discipline in the late 19th century. The seminal experiment of Hans Driesch [7] demonstrating that each cell of the 4-cell embryo retains the ability to develop into a complete organism (‘totipotency’) reinvigorated the age-old debate on whether development is merely the unmasking of preformed structures (‘preformation’), or a self-organizing process (‘epigenesis’). Driesch's experiment proved that the inherited historical (i.e., genetically preformed) information that guides organismal reproduction does not do so by its differential partitioning among the parts of the developing embryo, as many thought to be the case at the time. Rather, as was subsequently proved by the experiments of Theodor Boveri on polyspermic sea urchin embryos [8], each cell of the developing embryo must retain the whole of the organism's chromosomal inheritance in order for development to reproduce a fully formed organism.
Sea urchin embryogenesis is illustrative of both the developmental origin of emergent information and the causal role of historical information in determining the outcome of development. If this sounds suspiciously circular, that's because it is: Development generally occurs via cybernetic or reflexive (i.e., circular) causality, wherein an historical event or process stimulates and/or regulates its own recurrence by interacting with other processes that provide positive and negative feedback [3]. The de novo emergence of information (symmetry breaking) by way of this cybernetic principle, which occurs ubiquitously in nature, is illustrated here by the development of axial polarity in the sea urchin embryo.
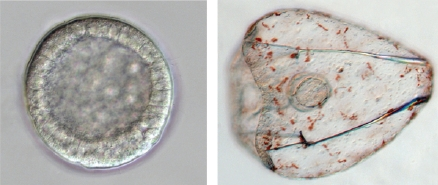
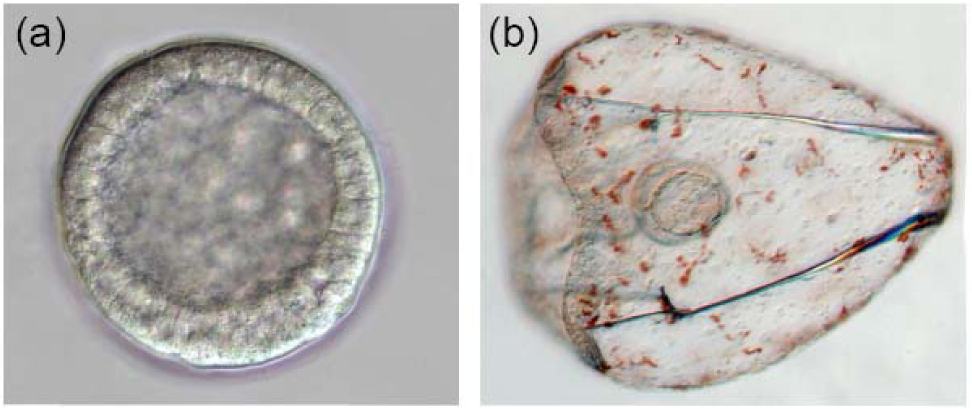
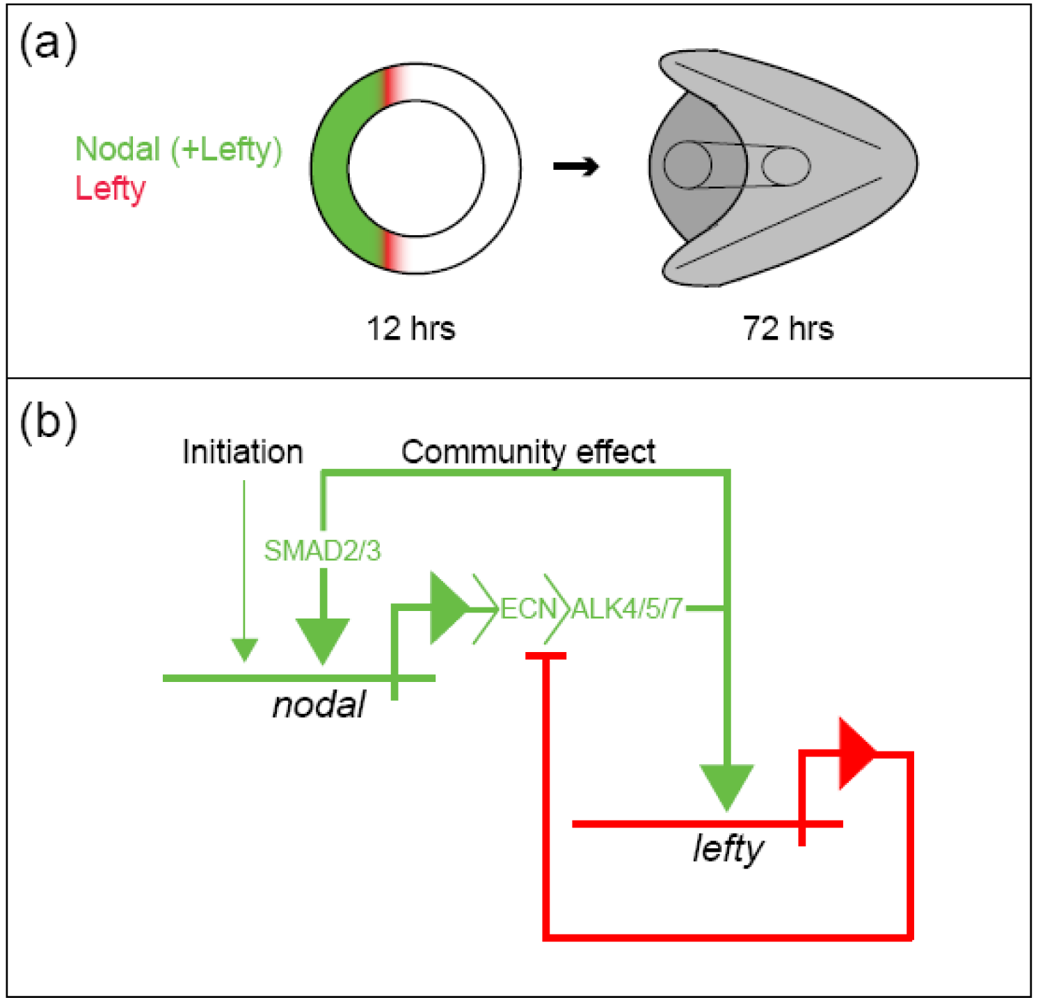
The sea urchin embryo produces a bilaterally symmetric larval form known as a pluteus (Figure 1). The primary axis defining the plane of bilateral symmetry is historical information that is preformed in the structurally polarized cytoplasm of the egg, whereas the secondary axis emerges epigenetically via the activities of two genes: nodal and lefty [9-12]. Both nodal and lefty encode diffusible extracellular proteins that are produced beginning at about the 60-120 cell morula stage. Specific interaction of Nodal protein with a cell surface receptor (‘Alk4/5/7’) leads to activation of an intracellular DNA-binding protein (‘Smad2/3’) that migrates to the cell nucleus and stimulates activation of the nodal gene, and hence production of more Nodal protein [12-16]. This occurs both within a cell that produces Nodal protein (‘autocrine’ signaling), and in its immediate neighbors (‘paracrine’ signaling) [12]. Thus activation of nodal establishes a positive feedback loop that both amplifies and maintains nodal activity. However, transduction of the Nodal signal also activates lefty, whose protein product competitively inhibits the interaction of Nodal with its receptor [11,17-20]. In this way nodal activity is controlled by negative feedback from lefty. Because Lefty protein is more diffusible than Nodal, this epigenetic circuit embodies a ‘reaction-diffusion’ pattern formation process involving local activation and lateral inhibition [21,22]. Such processes, which are ubiquitous at many different scales in nature (e.g., in ecosystems [23]), have been shown both theoretically and experimentally to produce emergent spatial information (i.e., to break symmetry), either out of initially homogeneous conditions, which are inherently unstable [24], or in response to subtle initial anisotropies [25].
In the sea urchin embryo a graded distribution of maternal mitochondria often prefigures the secondary axis of the embryo, and contributes to activation of nodal via hydrogen peroxide, a byproduct of mitochondrial respiration that functions as an intracellular signal in many contexts [26-28]. The mitochondrial gradient, historical information that emerged developmentally during oogenesis, influences the oral-aboral polariazation of the embryo by contributing to the anisotropic zygotic activation of nodal; it is thus somewhat preformative. However, development of the secondary axis is only stochastically influenced by mitochondrial distribution, and not strictly dependent on it (i.e., the mitochondrial gradient prefigures axial polarity in a significant majority of embryos, but not in all embryos) [27], which contrasts to the situation in other ontogenetic models (e.g., fruit flies and nematode worms, or the primary axis of the sea urchin embryo) wherein axis specification requires maternal mRNA or protein determinants that are spatially localized during oogenesis or early embryogenesis. The development of the latter systems is highly preformative in that it is more tightly constrained by the historical information developed maternally than is specification of the secondary axis of the sea urchin embryo.
The epigenetic circuit involving nodal and lefty makes use of the deeply historical genetic information recorded in DNA (the nucleotide sequences of nodal and lefty) to promote the de novo emergence of spatial information. Two types of genetically encoded historical information can be distinguished: structural and cis-regulatory. The structural information encoded in a gene represents the linear sequence of amino acids that constitute the protein product of that gene, in this case either Nodal or Lefty. The cis-regulatory information encoded in a gene consists of sequence-specific binding sites for molecules (typically proteins) whose presence or absence at such sites influences the production of the protein product (i.e., the ‘expression’) of a given gene. From the foregoing discussion of the regulatory circuit comprising nodal and lefty, it might be surmised that operation of this circuit requires a regulatory sequence in each gene that engages the Smad2/3 protein activated in response to Nodal signaling (Figure 2B). It is conceivable that this would be all that the circuit needs to break symmetry in the embryo, requiring for its initial condition only that the zygote provide a threshold amount of activated Smad2/3 protein, which given the instability of the homogeneous state [24] could even be distributed more or less homogeneously. In reality however a number of additional sequences that bind other regulatory proteins are present that contribute to the regulation of both nodal and lefty, for example, maternal proteins that are likely to respond to mitochondrial redox signaling [15,16,16].
The nodal-lefty gene regulatory circuit is a relatively simple example of how historical (genetic) information is used cybernetically to develop emergent (spatial) information within the context of a developing embryo (in this case the secondary axis of the embryo). It is obvious that normal embryogenesis depends on the functional integrity of both the genetic structural information encoding the Nodal and Lefty proteins, and the genetic cis-regulatory information that controls their expression. However, it should be noted that the two types of genetic information are qualitatively very different, and this difference has important ramifications. Cis-regulatory information generally takes the form of short (and usually ‘degenerate’) sequences whose positioning and orientation within the context of the gene is relatively unconstrained, compared to the much longer, more highly constrained sequences that encode proteins. Thus, whatever flexibility is available in the ‘genetic program of development’ largely involves regulatory information, and the evolution of form is likely to depend largely on changes in cis-regulatory sequences [29-32].
The heritable reproducibility of ontogeny is explained by the fact that the active developmental emergence of anatomical and physiological information is controlled by the passive historical information within the DNA structure, which responds algorithmically to the environmental context of the egg, zygote, and embryo. The logic of the algorithmic program is determined by the architecture of a gene regulatory network [31], which integrates multiple regulatory circuits (such as the nodal/lefty circuit described above). Gene regulatory networks are defined by the interactions between regulatory genes (sequence-specific DNA binding proteins such as Smad2/3 that control gene expression, and signaling proteins such as Nodal that control the activities of the DNA-binding proteins) via the regulatory sequences that control their expression. Regulatory sequences are often clustered within modules that execute Boolean logic functions (AND, OR, NOT, etc.) contingent on the presence or absence of specific proteins binding to the sequences, which control the production of the protein encoded by the gene to which the cis-regulatory modules are linked [31]. In the sea urchin, the nodal/lefty circuit is gateway into a larger gene regulatory network that encodes the ontogenetic program of ectodermal cell differentiation along the secondary axis of the embryo [33,34].
Developmental gene regulatory networks increase the determinacy of cell fate along the trajectory of ontogeny. The causality manifested by this system is cybernetic: Regulatory states specifying cell fate are maintained in mutually exclusive domains by a complex system of positive and negative feedback [35]. The oral ectoderm regulatory state is established by the positive feedback community effect of Nodal, which is localized by negative feedback from Lefty; and this regulatory state initiates aboral ectoderm specification by activating bmp2/4 within prospective oral ectoderm. BMP2/4 signaling is only active at a distance from its source however (via long-range diffusion) because Nodal also activates chordin, a short-range BMP2/4 antagonist, within oral ectoderm. The resulting long-range BMP2/4 signaling activates the aboral ectoderm regulatory state that is then maintained by activities of several transcription factors (Tbx2/3, IrxA, Dlx, and Ixh2.9) that are mutually-reinforcing by virtue of feedback loops encoded within the GRN (Figure 3). Thus, the formal logic of the gene regulatory network produces deterministic responses to the contingent regulatory state of a given nucleus (the set of regulatory proteins that are active therein at a given moment), and in this way progressively transforms the progeny of a relatively indeterminate stem cell (e.g., the zygote), with numerous potential fates, into many differentiated cells, whose determined fate is to play a highly specific role in the physiological and anatomical functions of a specialized tissue. In other words, the historical information encoded in the network (via the physical structure of DNA) is used to produce, de novo, the emergent information manifested in the anatomy and physiology of the mature organism.
The developmental emergence of information via historically encoded logic occurs hierarchically. For ontogenetic systems it is reasonable to choose the cell as the focal level of the organizational hierarchy, since the cell is the fundamental unit of life, and ontogeny typically begins with a single cell (the zygote). As described above the fate of the cell is controlled by the genome housed within the cell nucleus, which reflexively engenders and responds to a succession of transient nuclear regulatory states via the developmental gene regulatory network described above. However, the regulatory state of the nucleus also depends on the location of the cell within the developing embryo, and on the environmental cues received by the cell because of that location. Some of these cues are provided by intercellular signals (such that provided by Nodal describe above), receipt of which changes the nuclear regulatory state by activating a latent regulatory protein (e.g., Smad2/3).
As development proceeds differentiated cells form tissues with definitive structural attributes. For example, many tissues are composed of epithelia wherein the cells are polarized along an apical-basal axis defined by the basal location of a collagen-containing extracellular matrix, and linked together with intercellular junctions, thus forming an integrated unit. Mechanical forces (e.g., stretching) experienced by the tissue-and hence by the cells within the tissue-influence the regulatory state within nucleus of each cell by activating latent regulatory proteins [36-38]. Finally, the fully formed organism provides systemic hormonal cues that affect the regulatory states of nuclei throughout the body or within specific organs in response to environmental cues (e.g., diet, pheromones, light, temperature etc.), as well as structural ‘niches’ that provide signals required for the active maintenance of multi-potent stem cells required for regenerative tissue homeostasis. Thus, development spans multiple hierarchical levels of organization (cell, tissue, and organism), and continues throughout the lifespan of an organism. At every level and stage the emergent information produced developmentally depends on, and is constrained by, historical information: Both that inherited from previous stages of development (e.g., tissue structure), as well as that bequeathed by previous generations in the form of DNA (itself a product of evolutionary development).
3. Developmental Genomics and Information Theory
The thesis of this review is that, in general, information is a product of development, which locally reduces indeterminacy (breaks symmetry) via cybernetic causality (positive and negative feedback). The developmental production of information can be qualitatively inferred by considering the trajectory of ontogeny (Figure 1): The relative indeterminacy of early developmental stages is manifested by the fact that in general, it is more difficult to describe (or identify) a species by the appearance of its egg or blastula than by that of the mature organism. However, the developmental emergence of information can also be quantified using the mathematics of information theory [39,40]. For ontogenetic systems this is now possible using genome-wide measurements of gene expression.
Recall that the indeterminacy of a system is represented statistically by its entropy:
In ontogeny, the outcome of the process of cell fate specification is differentiation, which at the molecular level manifests as a defined set of gene products present in a given cell, each of which is associated with a metabolic process occurring within the cell. For a multipotent stem or progenitor cell of indeterminate fate (i.e., a cell that will produce progeny of various different fates) the set of genes that is expressed is not only qualitatively different than those expressed in its determined progeny, but also quantitatively different, having higher entropy—many more processes are active (at relatively low levels on average) in undetermined cells than in differentiated cells. This can be seen for example in hematopoietic stem cells [41], which promiscuously express many non-hematopoietic genes, and many more genes (in total) than are expressed in their differentiated progeny, which in turn express fewer genes, and at higher levels (in support of their specialized functions).
It is now possible to use deep RNA sequencing (RNAseq) to accurately measure the expression levels of all of a cell's genes, which allows us to express the entropy of gene expression as:
Where ai is the abundance of transcript i in sequence ‘reads per kilobase of gene model per million sequence reads’ (RPKM) [42], and N is the total number of RPKM. Although equation 4 has not yet been put to use in studies of ontogeny or stem-cell based regeneration, a prediction is that when it is, multi-potent progenitor cells will invariably be found to have a higher HGE value than their developed progeny, reflecting their higher level of indeterminacy. Preliminary data obtained in this laboratory thus far indicate that this is the case. Gene expression entropy may thus provide a predictive measure of the regenerative capacity of isolated cells and tissue samples.
A characteristic of pluripotency that has received increasing attention is the relatively ‘open’ state of chromatin in progenitor cells compared to their differentiated descendents [41]. In addition to well-known epigenetic modifications of DNA and histones, this openness involves chromosomal topology, which is developmentally dynamic [43,44]. Interestingly, chromosome topology is mutually linked to gene expression, as shown by a recent information theoretic analysis that used distance matrices to measure correlated changes in gene expression and intranuclear chromosome association networks [45].
Information theory has also been put to fruitful use in modeling the development of cancer (e.g., [46-48]), a process characterized by increasing informational and/or physical entropy. For example Levine and colleagues implemented an information theoretic ‘suprisal analysis’ of gene expression data from in vitro cultured cells to determine the extent to which physical entropy is submaximal owing to informational constraints within those cells [46]. For this analysis the Lagrange method of ‘undetermined multipliers’ was used to identify gene expression patterns (phenotypes) representing the relevant constraints in cells that at different stages of carcinogenesis, as well as the relative importance of each phenotype in the transformation process [46].
Finally, in a very interesting recent study Francois and Siggia used an information theoretic measure to quantify fitness of computationally simulated developmental gene regulatory networks that were allowed to evolve in silico [49]. Remarkably, this analysis showed that standard (incremental) Darwinian selection for increased fitness can produce gene regulatory networks that developmentally generate axial segmentation, much as occurs in arthropods and vertebrates with Hox genes [30], and involving similar rules and phenomenology [49].
4. Evolutionary Development
From a developmental perspective, the laws of physics can be viewed as historical information produced by the very early development of the universe following the big bang [6]. The rules of chemistry encapsulated in the periodic table of the elements are historical information that developed somewhat later within the context of stellar systems. Biological phenomena developed relatively recently by a still unknown process that managed to traverse the ‘cybernetic cut’, the divide between ‘chance contingency’ (which generates descriptive historical information) and ‘choice contingency’ (the goal-directed use of prescriptive or algorithmically anticipatory historical information) that distinguishes life from non-life [50]. Understanding how this occurred remains a fundamental challenge, a ‘holy grail’ of biological science. One approach that may eventually bear fruit begins with growth of the metabolic cycles and complex chemical networks that were likely to be energetically favored by the reducing conditions present during the pre-biotic development of earth [51,52]. However, it remains unclear whether this is sufficient to produce the historical information-containing polymers that potentiated Darwinian selection.
However the cybernetic cut was traversed, because it was, biology is essentially a science of semiotics: The prescriptive signification and teleonomic (goal-directed) use of historical information [53]. After the emergence of such information (now embodied by RNA and DNA), biological information flow became increasingly constrained within selected lineages by genetically-recorded history. As shown above, the development of emergent information in an organism (which includes physiological responses to environmental changes that maintain homeostasis) is logically controlled by historical information encoded in the nucleotide sequence of its genomic DNA. That history goes very deep: the genomic repertoire of structural information (protein coding sequences) is largely the same among all animals from sponges to humans, and entire gene regulatory circuits have been conserved among distantly related species that diverged over 500 million years ago [54].
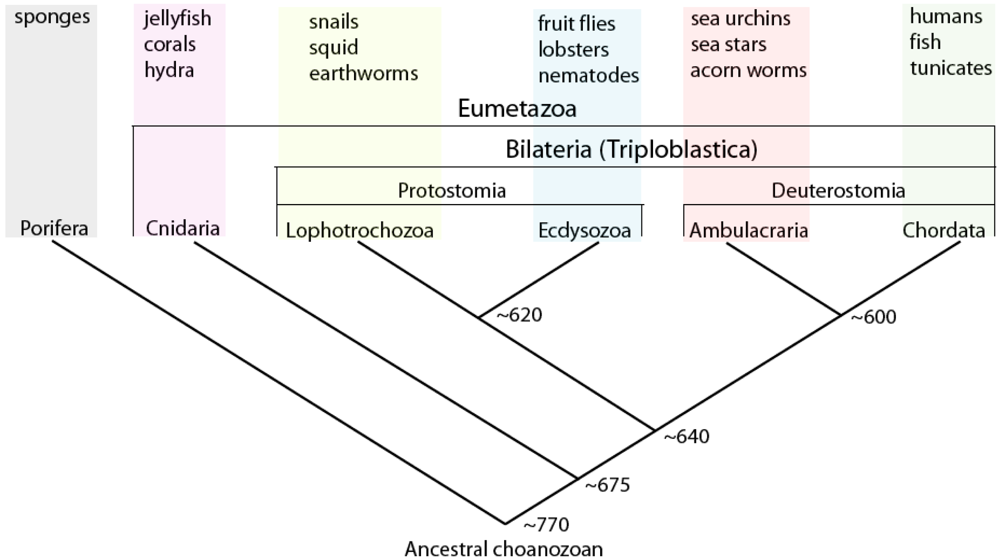
Indeed, the extent to which animals are evolvable might be understood by considering the structure of gene regulatory networks in terms of both modularity and pleiotropy [32]. Here it is interesting to note that the nodal-lefty gene regulatory circuit shown in Figure 2 is conserved at least throughout deuterostomes (chordates and ambulacraians; Figure 4), and used in multiple disparate ontogenetic contexts [even within the same organism; in sea urchin embryos (as in vertebrate embryos) it is also used to specify the left-right axis of the larva, which controls the internally nested development of the juvenile]; it therefore can be thought of a gene regulatory module or ‘plug-in’ [31] used for breaking symmetry in multicellular tissues. The ‘downstream’ processes dependent upon the operation of this module vary considerably, depending on organism-specific genetic cis-regulatory architecture. It should be obvious that for any given regulatory circuit, the fewer the processes that are dependent on its structure/function, the more freedom it will have to evolve [56].
It is thus reasonable to surmise that early during the development of life on earth, the accumulation of historical information—for example, as manifested by the ‘depth’ of gene regulatory networks— was shallower and less constraining—that is, life was less developmentally informed than it is now and had a higher level of indeterminacy [57,58]. As such it was more developmentally plastic, a notion that is consistent with the spectacular anatomical diversity generated by the ‘Cambrian explosion’ [59-61]. In this context it is interesting to consider the role of microRNAs (short processed gene transcripts that sequence-specifically interfere with the translation of specific mRNAs into protein) in evolutionary development [62-64]. It was recently proposed by Peterson and colleagues [64] that microRNAs play a key role in developmental canalization [65], that is, the buffering of ontogeny against epigenetic noise associated with transcriptional control of gene expression. It can be argued that the evolution of increased organismal complexity—such as that manifested by recently evolved animals (e.g., humans) compared to those that evolved much earlier (e.g., sponges)—required increasingly precise control of gene expression. MicroRNAs provide such control, serving to increase proteomic determinacy deriving from relatively noisy transcriptomes. For example in vertebrates Nodal expression is controlled by a microRNA (miR-430) that is activated in response to Nodal signaling (another feedback control on the latter) [66]. Such control increases the influence of genetically inherited information, augmenting developmental reproducibility which in turn augments the power of Darwinian selection [64]. Importantly, the phylogenetic distribution of microRNAs is consistent with this idea: unlike the genomic repertoire of protein-coding sequences, which is more or less the same among all animals, the genomic repertoire of microRNAs increased over the course of phylogenetic evolution with the additions of new sequences and little loss of old sequences [62,63]. Thus, microRNA diversity increased in concert with the evolution of anatomical complexity, a developmental emergence of information at the phylogenetic level.
5. Summary and Conclusions
Information refers to structural constraints that reduce uncertainty, and thus to that which motivates science: knowledge, predictability and causality. My thesis is that information manifests development, which is defined as a systems-level trajectory of change that increases determinacy. Development occurs when the events of history produce, and are recorded within, cyclic configurations of processes that engender feedback, which selectively reduces local degrees of freedom. This phenomenology is epitomized by the operation of gene regulatory networks underlying organismal ontogeny, but extends throughout ecosystems [40,67]. In conclusion, I suggest that scientific inquiries into origins—for example, the origin of life—need to obtain a historical record that is complete enough to allow elucidation of both the relevant feedback cycles, and the systemic context that favored their emergence.

Acknowledgments
I thank Kevin Peterson and Joel Graber for helpful comments and suggestions. The work in my laboratory reviewed here was funded by the NIH (R01 ES-016722).
References and Notes
- Salthe, S.N. Development and Evolution: Complexity and Change in Biology; MIT Press: Cambridge, MA, USA, 1993. [Google Scholar]
- Valentine, J.W.; May, C.L. Hierarchies in biology and paleontology. Paleobiology 1996, 22, 23–33. [Google Scholar]
- Coffman, J.A. Developmental ascendency: from bottom-up to top-down control. Biol. Theor. 2006, 1, 165–178. [Google Scholar]
- Coffman, J.A. On causality in nonlinear complex systems: the developmentalist perspective. In Handbook of the Philosophy of Science: Philosophy of Complex Systems; Hooker, C., Ed.; Elsevier: Amsterdam, The Netherlands, 2011; Volume 10, pp. 287–309. [Google Scholar]
- Joyce, G.F. The antiquity of RNA-based evolution. Nature 2002, 418, 214–221. [Google Scholar]
- Carroll, S.M. Is our Universe natural? Nature 2006, 440, 1132–1136. [Google Scholar]
- Driesch, H. Entwicklungsmechanisme Studien. I. Der Werth der beiden ersten Furchungszellen in der Echinodermentwicklung. Experimentelle Erzeugen von Theil und Doppelbildung. Zeitschrift für wissenschaftliche Zoologie 1892, 53. [Google Scholar]
- Boveri, T. Uber mehrpolige Mitosen als Mittel zur Analzyse des Zellkerns. Verhandlungen der physicalish-medizinischen Gesselschaft zu Wurzburg. Neu. Folge. 1902, 35, 67–90. [Google Scholar]
- Duboc, V.; Rottinger, E.; Besnardeau, L.; Lepage, T. Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev. Cell 2004, 6, 397–410. [Google Scholar]
- Flowers, V.L.; Courteau, G.R.; Poustka, A.J.; Weng, W.; Venuti, J.M. Nodal/activin signaling establishes oral-aboral polarity in the early sea urchin embryo. Dev. Dyn. 2004, 231, 727–740. [Google Scholar]
- Duboc, V.; Lapraz, F.; Besnardeau, L.; Lepage, T. Lefty acts as an essential modulator of Nodal activity during sea urchin oral-aboral axis formation. Dev. Biol. 2008, 320, 49–59. [Google Scholar]
- Bolouri, H.; Davidson, E.H. The gene regulatory network basis of the “community effect,” and analysis of a sea urchin embryo example. Dev. Biol. 2010, 340, 170–178. [Google Scholar]
- Lapraz, F.; Rottinger, E.; Duboc, V.; Range, R.; Duloquin, L.; Walton, K.; Wu, S.Y.; Bradham, C.; Loza, M.A.; Hibino, T.; Wilson, K.; Poustka, A.; McClay, D.; Angerer, L.; Gache, C.; Lepage, T. RTK and TGF-beta signaling pathways genes in the sea urchin genome. Dev. Biol. 2006, 300, 132–152. [Google Scholar]
- Yaguchi, S.; Yaguchi, J.; Burke, R.D. Sp-Smad2/3 mediates patterning of neurogenic ectoderm by nodal in the sea urchin embryo. Dev. Biol. 2007, 302, 494–503. [Google Scholar]
- Nam, J.; Su, Y.-H.; Lee, P.Y.; Robertson, A.J.; Coffman, J.A.; Davidson, E.H. Cis-regulatory control of the nodal gene, initiator of the sea urchin oral ectoderm gene network. Dev. Biol. 2007, 306, 860–869. [Google Scholar]
- Range, R.; Lapraz, F.; Quirin, M.; Marro, S.; Besnardeau, L.; Lepage, T. Cis-regulatory analysis of nodal and maternal control of dorsal-ventral axis formation by Univin, a TGF-{beta} related to Vg1. Development 2007, 134, 3649–3664. [Google Scholar]
- Chen, Y.; Schier, A.F. Lefty proteins are long-range inhibitors of squint-mediated nodal signaling. Curr. Biol. 2002, 12, 2124–2128. [Google Scholar]
- Sakuma, R.; Ohnishi, Y.; Meno, C.; Fujii, H.; Juan, H.; Takeuchi, J.; Ogura, T.; Li, E.; Miyazono, K.; Hamada, H. Inhibition of Nodal signalling by Lefty mediated through interaction with common receptors and efficient diffusion. Genes Cells 2002, 7, 401–412. [Google Scholar]
- Chen, C.; Shen, M.M. Two modes by which Lefty proteins inhibit nodal signaling. Curr. Biol. 2004, 14, 618–624. [Google Scholar]
- Cheng, S.K.; Olale, F.; Brivanlou, A.H.; Schier, A.F. Lefty blocks a subset of TGFbeta signals by antagonizing EGF-CFC coreceptors. PLoS Biol. 2004, 2, E30. [Google Scholar]
- Juan, H.; Hamada, H. Roles of nodal-lefty regulatory loops in embryonic patterning of vertebrates. Genes Cells 2001, 6, 923–930. [Google Scholar]
- Solnica-Krezel, L. Vertebrate development: taming the nodal waves. Curr. Biol. 2003, 13, R7–R9. [Google Scholar]
- van de Koppel, J.; Gascoigne, J.C.; Theraulaz, G.; Rietkerk, M.; Mooij, W.M.; Herman, P.M. Experimental evidence for spatial self-organization and its emergent effects in mussel bed ecosystems. Science 2008, 322, 739–742. [Google Scholar]
- Turing, A.M. The chemical basis of morphogenesis. Bull. Math. Biol. 1952, 52, 153–97. [Google Scholar]
- Gierer, A.; Meinhardt, H. A theory of biological pattern formation. Kybernetik 1972, 12, 30–39. [Google Scholar]
- Coffman, J.A.; McCarthy, J.J.; Dickey-Sims, C.; Robertson, A.J. Oral-aboral axis specification in the sea urchin embryo II. Mitochondrial distribution and redox state contribute to establishing polarity in Strongylocentrotus purpuratus. Dev. Biol. 2004, 273, 160–171. [Google Scholar]
- Coffman, J.A.; Coluccio, A.; Planchart, A.; Robertson, A.J. Oral-aboral axis specification in the sea urchin embryo III. Role of mitochondrial redox signaling via H(2)O(2). Dev. Biol. 2009, 330, 123–130. [Google Scholar]
- Coffman, J.A. Mitochondria and metazoan epigenesis. Semin. Cell Dev. Biol. 2009, 20, 321–329. [Google Scholar]
- Davidson, E.H. Genomic Regulatory Systems: Development and Evolution; Academic Press: San Diego, CA, USA, 2001. [Google Scholar]
- Carroll, S.B.; Grenier, J.K.; Weatherbee, S.D. From DNA to Diversity; Blackwell: Malden, MA, USA, 2001. [Google Scholar]
- Davidson, E.H. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution; Academic Press/Elsevier: San Diego, CA, USA, 2006. [Google Scholar]
- Davidson, E.H.; Erwin, D.H. Gene regulatory networks and the evolution of animal body plans. Science 2006, 311, 796–800. [Google Scholar]
- Su, Y.H.; Li, E.; Geiss, G.K.; Longabaugh, W.J.; Kramer, A.; Davidson, E.H. A perturbation model of the gene regulatory network for oral and aboral ectoderm specification in the sea urchin embryo. Dev. Biol. 2009, 329, 410–421. [Google Scholar]
- Lapraz, F.; Besnardeau, L.; Lepage, T. Patterning of the dorsal-ventral axis in echinoderms: Insights into the evolution of the BMP-chordin signaling network. PLoS Biol. 2009, 7, e1000248. [Google Scholar]
- Davidson, E.H.; McClay, D.R.; Hood, L. Regulatory gene networks and the properties of the developmental process. Proc. Natl. Acad Sci. USA 2003, 100, 1475–1480. [Google Scholar]
- Iqbal, J.; Zaidi, M. Molecular regulation of mechanotransduction. Biochem. Biophys. Res. Commun. 2005, 328, 751–755. [Google Scholar]
- Knobloch, T.J.; Madhavan, S.; Nam, J.; Agarwal, S., Jr.; Agarwal, S. Regulation of chondrocytic gene expression by biomechanical signals. Crit. Rev. Eukaryot. Gene Expr. 2008, 18, 139–150. [Google Scholar]
- Ziros, P.G.; Basdra, E.K.; Papavassiliou, A.G. Runx2: of bone and stretch. Int. J. Biochem. Cell Biol. 2008, 40, 1659–1663. [Google Scholar]
- Shannon, C.E. A mathematical theory of communication. Bell Sys. Tech. J. 1948, 27. [Google Scholar]
- Ulanowicz, R.E. Growth and Development: Ecosystems Phenomenology; Springer-Verlag: New York, NY, USA, 1986. [Google Scholar]
- Akashi, K.; He, X.; Chen, J.; Iwasaki, H.; Niu, C.; Steenhard, B.; Zhang, J.; Haug, J.; Li, L. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood 2003, 101, 383–389. [Google Scholar]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar]
- Alvarez, M.; Rhodes, S.J.; Bidwell, J.P. Context-dependent transcription: all politics is local. Gene 2003, 313, 43–57. [Google Scholar]
- Kim, S.H.; McQueen, P.G.; Lichtman, M.K.; Shevach, E.M.; Parada, L.A.; Misteli, T. Spatial genome organization during T-cell differentiation. Cytogenet. Genome. Res. 2004, 105, 292–301. [Google Scholar]
- Rajapakse, I.; Perlman, M.D.; Scalzo, D.; Kooperberg, C.; Groudine, M.; Kosak, S.T. The emergence of lineage-specific chromosomal topologies from coordinate gene regulation. Proc. Natl. Acad Sci. USA 2009, 106, 6679–6684. [Google Scholar]
- Remacle, F.; Kravchenko-Balasha, N.; Levitzki, A.; Levine, R.D. Information-theoretic analysis of phenotype changes in early stages of carcinogenesis. Proc. Natl. Acad Sci. USA 2010, 107, 10324–10329. [Google Scholar]
- Berretta, R.; Moscato, P. Cancer biomarker discovery: The entropic hallmark. PLoS One 2010, 5, e12262. [Google Scholar]
- Teschendorff, A.E.; Severini, S. Increased entropy of signal transduction in the cancer metastasis phenotype. BMC Syst. Biol. 2010, 4, 104. [Google Scholar]
- Francois, P.; Siggia, E.D. Predicting embryonic patterning using mutual entropy fitness and in silico evolution. Development 2010, 137, 2385–2395. [Google Scholar]
- Abel, D.L. The ‘cybernetic cut’: Progressing from description to prescription in systems theory. Open Cybern. Syst. J. 2008, 2, 252–262. [Google Scholar]
- Smith, E.; Morowitz, H.J. Universality in intermediary metabolism. Proc. Natl. Acad Sci. USA 2004, 101, 13168–13173. [Google Scholar]
- Copley, S.D.; Smith, E.; Morowitz, H.J. The origin of the RNA world: Co-evolution of genes and metabolism. Bioorg. Chem. 2007, 35, 430–443. [Google Scholar]
- Hoffmeyer, J. Biosemiotics: Towards a new synthesis in biology. Eur. J. Semiotic Studies 1997, 9, 355–376. [Google Scholar]
- Hinman, V.F.; Nguyen, A.T.; Cameron, R.A.; Davidson, E.H. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc. Natl. Acad Sci. USA 2003, 100, 13356–13361. [Google Scholar]
- Peterson, K.J.; Cotton, J.A.; Gehling, J.G.; Pisani, D. The Ediacaran emergence of bilaterians: Congruence between the genetic and the geological fossil records. Philos. Trans. R Soc. Lond. B Biol. Sci. 2008, 363, 1435–1443. [Google Scholar]
- Schank, J.C.; Wimsatt, W.C. Generative entrenchment and evolution. PSA: Proc. Biennial. Mtg. Phil. Sci. Ass. 1986, 2, 33–60. [Google Scholar]
- Newman, S.A.; Muller, G.B. Epigenetic mechanisms of character origination. J. Exp. Zool. 2000, 288, 304–317. [Google Scholar]
- Newman, S.A. The pre-Mendelian, pre-Darwinian world: Shifting relations between genetic and epigenetic mechanisms in early multicellular evolution. J. Biosci. 2005, 30, 75–85. [Google Scholar]
- Gould, S.J. Wonderful Life: The Burgess Shale and the Nature of History; Norton: New York, NY, USA, 1989. [Google Scholar]
- Peterson, K.J.; McPeek, M.A.; Evans, D.A.D. Tempo and mode of early animal evolution: Inferences from rocks, Hox, and molecular clocks. Paleobiology 2005, 31, 36–55. [Google Scholar]
- Briggs, D.E.G.; Fortey, R.A. Wonderful strife: Systematics, stem groups, and the phylogenetic signal of the Cambrian radiation. Paleobiology 2005, 31, 94–112. [Google Scholar]
- Sempere, L.F.; Cole, C.N.; McPeek, M.A.; Peterson, K.J. The phylogenetic distribution of metazoan microRNAs: Insights into evolutionary complexity and constraint. J. Exp. Zool. B Mol. Dev. Evol. 2006, 306, 575–588. [Google Scholar]
- Wheeler, B.M.; Heimberg, A.M.; Moy, V.N.; Sperling, E.A.; Holstein, T.W.; Heber, S.; Peterson, K.J. The deep evolution of metazoan microRNAs. Evol. Dev. 2009, 11, 50–68. [Google Scholar]
- Peterson, K.J.; Dietrich, M.R.; McPeek, M.A. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. Bioessays 2009, 31, 736–747. [Google Scholar]
- Waddington, C.H. Canalization of development and genetic assimilation of acquired characters. Nature 1959, 183, 1654–1655. [Google Scholar]
- Choi, W.Y.; Giraldez, A.J.; Schier, A.F. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 2007, 318, 271–274. [Google Scholar]
- Ulanowicz, R.E. Ecology, the Ascendent Perspective; Columbia University Press: New York, NY, USA, 1997. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Coffman, J.A. Information as a Manifestation of Development. Information 2011, 2, 102-116. https://doi.org/10.3390/info2010102
Coffman JA. Information as a Manifestation of Development. Information. 2011; 2(1):102-116. https://doi.org/10.3390/info2010102
Chicago/Turabian StyleCoffman, James A. 2011. "Information as a Manifestation of Development" Information 2, no. 1: 102-116. https://doi.org/10.3390/info2010102
APA StyleCoffman, J. A. (2011). Information as a Manifestation of Development. Information, 2(1), 102-116. https://doi.org/10.3390/info2010102