Towards Initial Indications for a Thiol-Based Redox Control of Arabidopsis 5-Aminolevulinic Acid Dehydratase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth and Mutant Lines
2.2. Cloning, Expression and Purification of Recombinant ALAD and TRX f1
2.3. Protein Extraction
2.4. Gel-Shift Assays
2.5. ALAD Activity Assay with Recombinant Protein and Plant Extracts
2.6. Bimolecular Fluorescence Complementation Assay
2.7. Pull-Down Experiments
3. Results
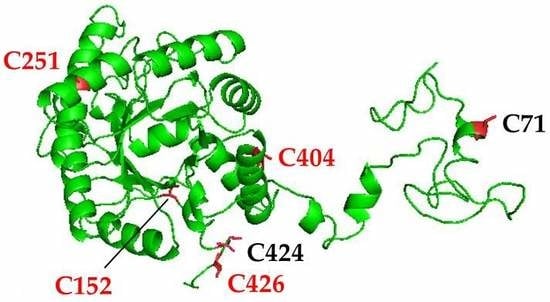
3.1. Structural Analysis and Protein Sequence Alignment Reveal Four Highly Conserved Cysteine Residues in Arabidopsis ALAD
3.2. Posttranslational Stability of ALAD in TRX and NTRC-Deficient Arabidopsis Seedlings
3.3. ALAD Interacts with TRX and NTRC
3.4. Redox-Dependent Structural Modifications of Recombinant ALAD
4. Discussion
4.1. Redox-Dependent Modification of Alad Stability and Activity in Arabidopsis
4.2. Redox-Dependent Modification of Recombinant ALAD
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Aa | amino acid residues |
| ALA | 5-aminolevulinic acid |
| ALAD | 5-aminolevulinic acid dehydratase |
| BiFC | bimolecular fluorescence complementation |
| BLAST | Basic Local Alignment Search Tool |
| CHLG | chlorophyll synthase |
| CHLI | subunit of the Mg chelatase |
| CHLM | Mg protoporphyrin methyltransferase |
| DTT | dithiothreitol |
| GluTR | glutamyl-tRNA reductase |
| GSAAT | glutamate-1-semialdehyde aminotransferase |
| mPEG-MAL | methoxypolyethylene glycol maleimide 5000 |
| NEM | N-ethylmaleimide |
| NTRC | NADPH-dependent thioredoxin reductase C |
| PBG | porphobilinogen |
| POR | protochlorophyllide oxidoreductase |
| PPOX1/2 | protoporphyrinogen IX oxidase |
| ROS | reactive oxygen species |
| RuBisCo1,5 | ribulose-bisphosphate carboxylase large subunit |
| TBS | tetrapyrrole biosynthesis |
| TRX | thioredoxin |
| YFP | yellow-fluorescence protein |
References
- Richter, A.S.; Grimm, B. Thiol-based redox control of enzymes involved in the tetrapyrrole biosynthesis pathway in plants. Front. Plant Sci. 2013, 4, 371. [Google Scholar] [CrossRef] [PubMed]
- Stenbaek, A.; Jensen, P.E. Redox regulation of chlorophyll biosynthesis. Phytochemistry 2010, 71, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Busch, A.W.; Montgomery, B.L. Interdependence of tetrapyrrole metabolism, the generation of oxidative stress and the mitigative oxidative stress response. Redox Biol. 2015, 4, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, B.B. The Path to Thioredoxin and Redox Regulation Beyond Chloroplasts. Plant Cell Physiol. 2017, 58, 1826–1832. [Google Scholar] [CrossRef] [PubMed]
- Michelet, L.; Zaffagnini, M.; Morisse, S.; Sparla, F.; Perez-Perez, M.E.; Francia, F.; Danon, A.; Marchand, C.H.; Fermani, S.; Trost, P.; et al. Redox regulation of the Calvin-Benson cycle: Something old, something new. Front. Plant Sci. 2013, 4, 470. [Google Scholar] [CrossRef] [PubMed]
- Serrato, A.J.; Fernandez-Trijueque, J.; Barajas-Lopez, J.D.; Chueca, A.; Sahrawy, M. Plastid thioredoxins: A “one-for-all” redox-signaling system in plants. Front. Plant Sci. 2013, 4, 463. [Google Scholar] [CrossRef] [PubMed]
- Serrato, A.J.; Perez-Ruiz, J.M.; Spinola, M.C.; Cejudo, F.J. A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 43821–43827. [Google Scholar] [CrossRef] [PubMed]
- Perez-Ruiz, J.M.; Naranjo, B.; Ojeda, V.; Guinea, M.; Cejudo, F.J. NTRC-dependent redox balance of 2-Cys peroxiredoxins is needed for optimal function of the photosynthetic apparatus. Proc. Natl. Acad. Sci. USA 2017, 114, 12069–12074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spinola, M.C.; Perez-Ruiz, J.M.; Pulido, P.; Kirchsteiger, K.; Guinea, M.; Gonzalez, M.; Cejudo, F.J. NTRC new ways of using NADPH in the chloroplast. Physiol. Plant. 2008, 133, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.S.; Peter, E.; Rothbart, M.; Schlicke, H.; Toivola, J.; Rintamaki, E.; Grimm, B. Posttranslational influence of NADPH-dependent thioredoxin reductase C on enzymes in tetrapyrrole synthesis. Plant. Physiol. 2013, 162, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Fan, T.; Liu, Y.; Rothbart, M.; Yu, J.; Zhou, S.; Grimm, B.; Luo, M. Thioredoxin redox regulates ATPase activity of magnesium chelatase CHLI subunit and modulates redox-mediated signaling in tetrapyrrole biosynthesis and homeostasis of reactive oxygen species in pea plants. Plant. Physiol. 2012, 159, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.S.; Perez-Ruiz, J.M.; Cejudo, F.J.; Grimm, B. Redox-control of chlorophyll biosynthesis mainly depends on thioredoxins. FEBS Lett. 2018, 592, 3111–3115. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.S.; Wang, P.; Grimm, B. Arabidopsis Mg-Protoporphyrin IX Methyltransferase Activity and Redox Regulation Depend on Conserved Cysteines. Plant Cell Physiol. 2016, 57, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Da, Q.; Wang, P.; Wang, M.; Sun, T.; Jin, H.; Liu, B.; Wang, J.; Grimm, B.; Wang, H.B. Thioredoxin and NADPH-Dependent Thioredoxin Reductase C Regulation of Tetrapyrrole Biosynthesis. Plant Physiol. 2017, 175, 652–666. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.E.; Reid, J.D.; Hunter, C.N. Modification of cysteine residues in the ChlI and ChlH subunits of magnesium chelatase results in enzyme inactivation. Biochem. J. 2000, 352 Pt 2, 435–441. [Google Scholar] [CrossRef]
- Stenbaek, A.; Hansson, A.; Wulff, R.P.; Hansson, M.; Dietz, K.J.; Jensen, P.E. NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Lett. 2008, 582, 2773–2778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balmer, Y.; Koller, A.; del Val, G.; Manieri, W.; Schurmann, P.; Buchanan, B.B. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. USA 2003, 100, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, M.; Florencio, F.J. Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc. Natl. Acad. Sci. USA 2003, 100, 16107–16112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spencer, P.; Jordan, P.M. Characterization of the two 5-aminolaevulinic acid binding sites, the A- and P-sites, of 5-aminolaevulinic acid dehydratase from Escherichia coli. Biochem. J. 1995, 305 Pt 1, 151–158. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Wang, W.; Chen, D.; Ji, Q.; Jing, Y.; Wang, H.; Lin, R. Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 2012, 24, 1984–2000. [Google Scholar] [CrossRef] [PubMed]
- Boese, Q.F.; Spano, A.J.; Li, J.M.; Timko, M.P. Aminolevulinic acid dehydratase in pea (Pisum sativum L.). Identification of an unusual metal-binding domain in the plant enzyme. J. Biol. Chem. 1991, 266, 17060–17066. [Google Scholar] [PubMed]
- Kervinen, J.; Dunbrack, R.L., Jr.; Litwin, S.; Martins, J.; Scarrow, R.C.; Volin, M.; Yeung, A.T.; Yoon, E.; Jaffe, E.K. Porphobilinogen synthase from pea: Expression from an artificial gene, kinetic characterization, and novel implications for subunit interactions. Biochemistry 2000, 39, 9018–9029. [Google Scholar] [CrossRef] [PubMed]
- Kokona, B.; Rigotti, D.J.; Wasson, A.S.; Lawrence, S.H.; Jaffe, E.K.; Fairman, R. Probing the oligomeric assemblies of pea porphobilinogen synthase by analytical ultracentrifugation. Biochemistry 2008, 47, 10649–10656. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, E.K. The Remarkable Character of Porphobilinogen Synthase. ACC Chem. Res. 2016, 49, 2509–2517. [Google Scholar] [CrossRef] [PubMed]
- Balange, A.P.; Lambert, C. In vitro activation of δ-aminolevulinate dehydratase from far-red irradiated radish (Raphanus sativus L.) seedlings by thioredoxin f. Plant Sci. Lett. 1983, 32, 253–259. [Google Scholar] [CrossRef]
- Thormahlen, I.; Ruber, J.; von Roepenack-Lahaye, E.; Ehrlich, S.M.; Massot, V.; Hummer, C.; Tezycka, J.; Issakidis-Bourguet, E.; Geigenberger, P. Inactivation of thioredoxin f1 leads to decreased light activation of ADP-glucose pyrophosphorylase and altered diurnal starch turnover in leaves of Arabidopsis plants. Plant Cell Environ. 2013, 36, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Thormahlen, I.; Meitzel, T.; Groysman, J.; Ochsner, A.B.; von Roepenack-Lahaye, E.; Naranjo, B.; Cejudo, F.J.; Geigenberger, P. Thioredoxin f1 and NADPH-Dependent Thioredoxin Reductase C Have Overlapping Functions in Regulating Photosynthetic Metabolism and Plant Growth in Response to Varying Light Conditions. Plant Physiol. 2015, 169, 1766–1786. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; von Heijne, G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999, 8, 978–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Muthuramalingam, M.; Dietz, K.J.; Stroher, E. Thiol-disulfide redox proteomics in plant research. Methods Mol. Biol. 2010, 639, 219–238. [Google Scholar] [CrossRef] [PubMed]
- Mauzerall, D.; Granick, S. The occurrence and determination of delta-amino-levulinic acid and porphobilinogen in urine. J. Biol. Chem. 1956, 219, 435–446. [Google Scholar] [PubMed]
- Frankenberg, N.; Erskine, P.T.; Cooper, J.B.; Shoolingin-Jordan, P.M.; Jahn, D.; Heinz, D.W. High resolution crystal structure of a Mg2+-dependent porphobilinogen synthase. J. Mol. Biol. 1999, 289, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Frere, F.; Schubert, W.D.; Stauffer, F.; Frankenberg, N.; Neier, R.; Jahn, D.; Heinz, D.W. Structure of porphobilinogen synthase from Pseudomonas aeruginosa in complex with 5-fluorolevulinic acid suggests a double Schiff base mechanism. J. Mol. Biol. 2002, 320, 237–247. [Google Scholar] [CrossRef]
- Coates, L.; Beaven, G.; Erskine, P.T.; Beale, S.I.; Avissar, Y.J.; Gill, R.; Mohammed, F.; Wood, S.P.; Shoolingin-Jordan, P.; Cooper, J.B. The X-ray structure of the plant like 5-aminolaevulinic acid dehydratase from Chlorobium vibrioforme complexed with the inhibitor laevulinic acid at 2.6 A resolution. J. Mol. Biol. 2004, 342, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Coates, L.; Beaven, G.; Erskine, P.T.; Beale, S.I.; Wood, S.P.; Shoolingin-Jordan, P.M.; Cooper, J.B. Structure of Chlorobium vibrioforme 5-aminolaevulinic acid dehydratase complexed with a diacid inhibitor. Acta Crystallogr. D Biol. Crystallogr. 2005, 61, 1594–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Z.Y.; Chang, C.C.; Lu, X.; Chen, J.; Li, B.L.; Chang, T.Y. The disulfide linkage and the free sulfhydryl accessibility of acyl-coenzyme A:cholesterol acyltransferase 1 as studied by using mPEG5000-maleimide. Biochemistry 2005, 44, 6537–6546. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wittmann, D.; Kløve, S.; Wang, P.; Grimm, B. Towards Initial Indications for a Thiol-Based Redox Control of Arabidopsis 5-Aminolevulinic Acid Dehydratase. Antioxidants 2018, 7, 152. https://doi.org/10.3390/antiox7110152
Wittmann D, Kløve S, Wang P, Grimm B. Towards Initial Indications for a Thiol-Based Redox Control of Arabidopsis 5-Aminolevulinic Acid Dehydratase. Antioxidants. 2018; 7(11):152. https://doi.org/10.3390/antiox7110152
Chicago/Turabian StyleWittmann, Daniel, Sigri Kløve, Peng Wang, and Bernhard Grimm. 2018. "Towards Initial Indications for a Thiol-Based Redox Control of Arabidopsis 5-Aminolevulinic Acid Dehydratase" Antioxidants 7, no. 11: 152. https://doi.org/10.3390/antiox7110152
APA StyleWittmann, D., Kløve, S., Wang, P., & Grimm, B. (2018). Towards Initial Indications for a Thiol-Based Redox Control of Arabidopsis 5-Aminolevulinic Acid Dehydratase. Antioxidants, 7(11), 152. https://doi.org/10.3390/antiox7110152