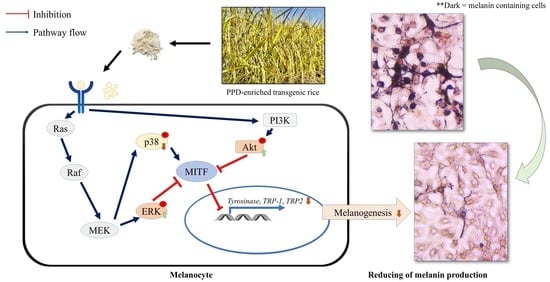
Protopanaxadiol-Enriched Rice Extracts Suppressed Oxidative and Melanogenic Activities in Melan-a Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. ABTS Radical Scavenging Ability Assay
2.3. Melan-a Cell Culture
2.4. Melan-a Cell Viability Assay
2.5. Melanin Excretion and Melanin Content Assays
2.6. L-DOPA Staining Assay
2.7. Cellular Tyrosinase Activity Assay
2.8. Morphological Appearance Using the Fontana–Masson Assay
2.9. Quantification of Gene Expression by Real-Time Polymerase Chain Reaction (PCR)
2.10. Western Blot Assay
2.11. Statistical Analysis
3. Results
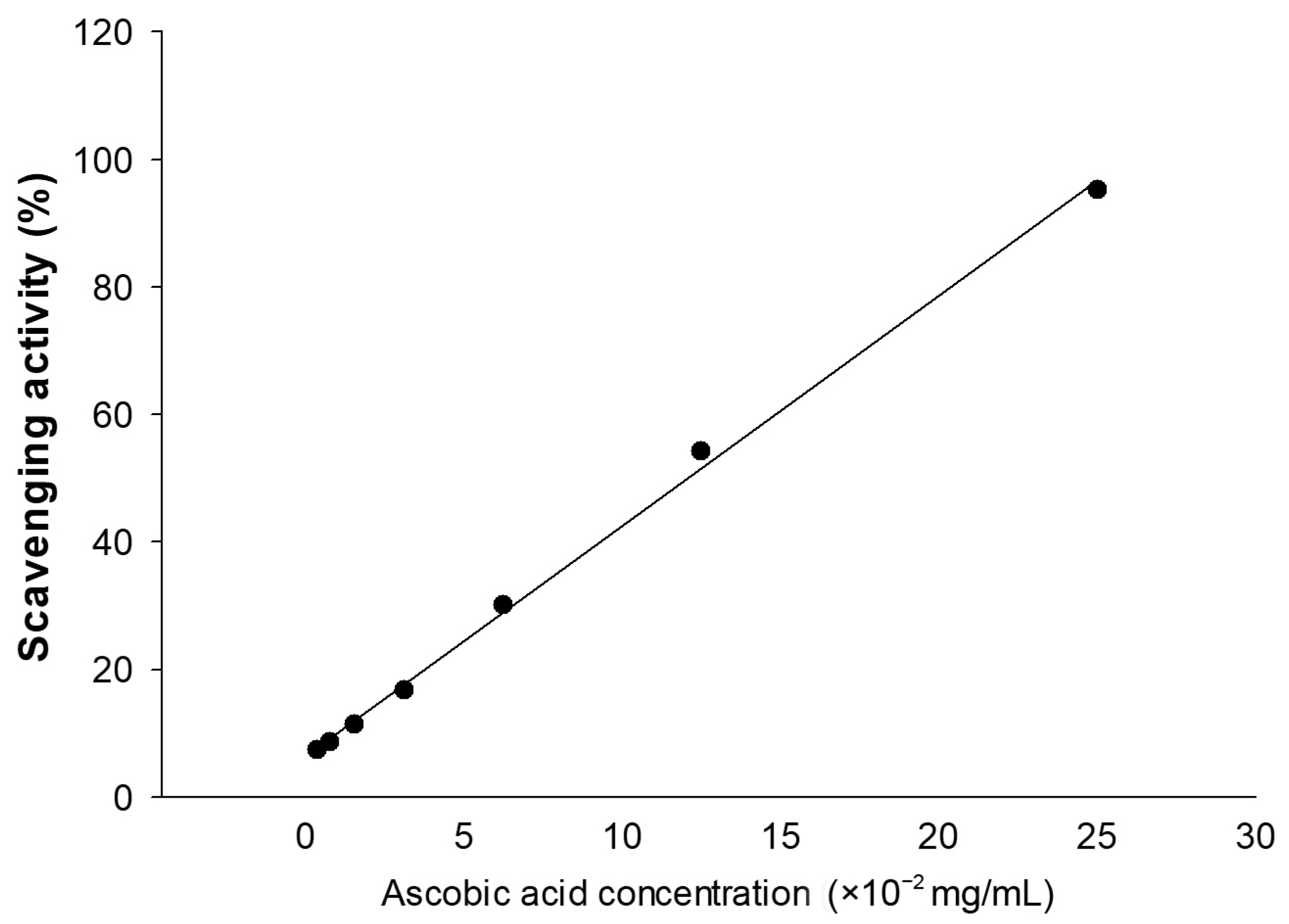
3.1. Effects of Transgenic Rice Seed Extracts on Antioxidant Activity
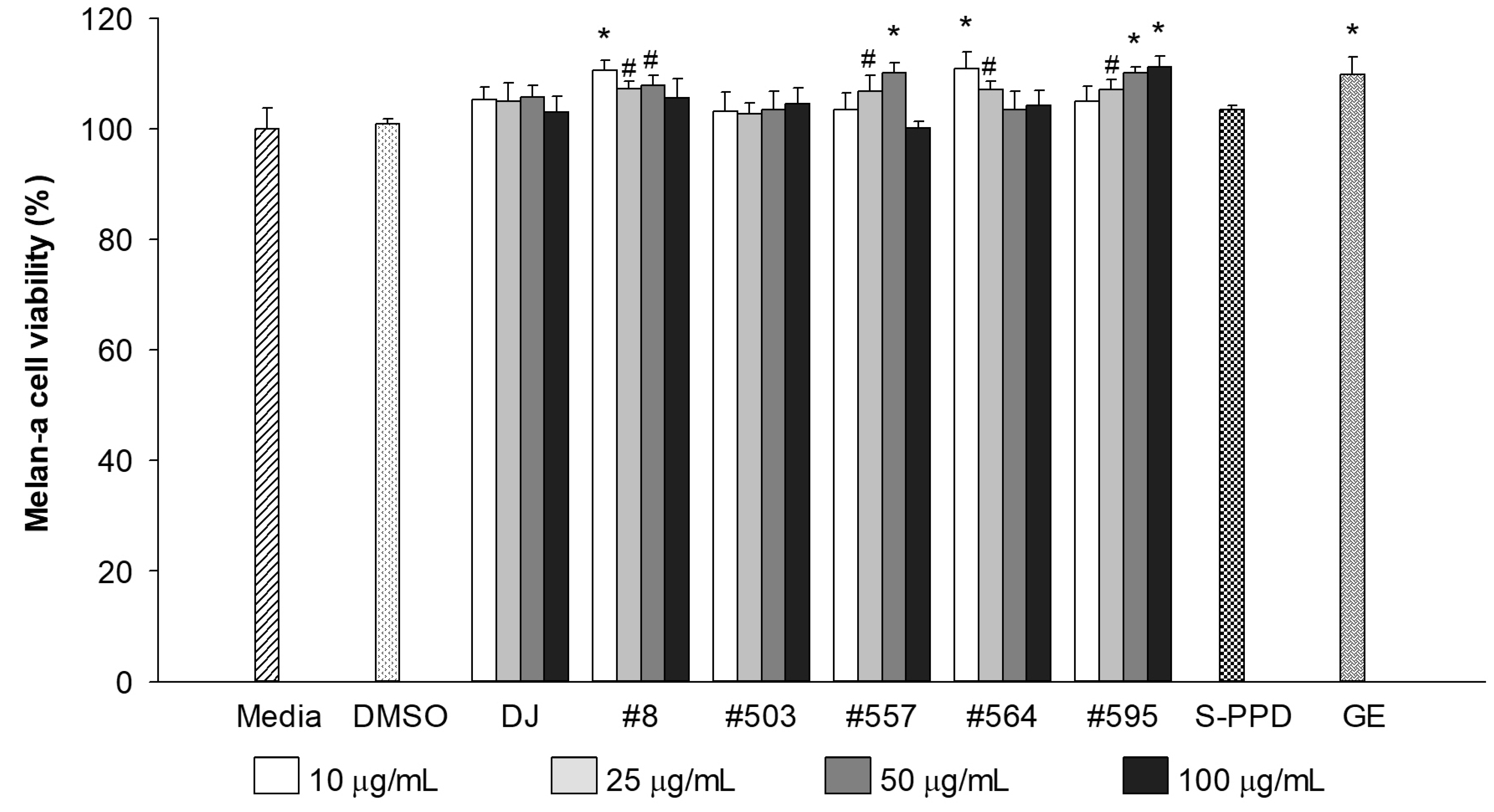
3.2. Effects of Transgenic Rice Seed Extracts on Melan-a Cell Viability
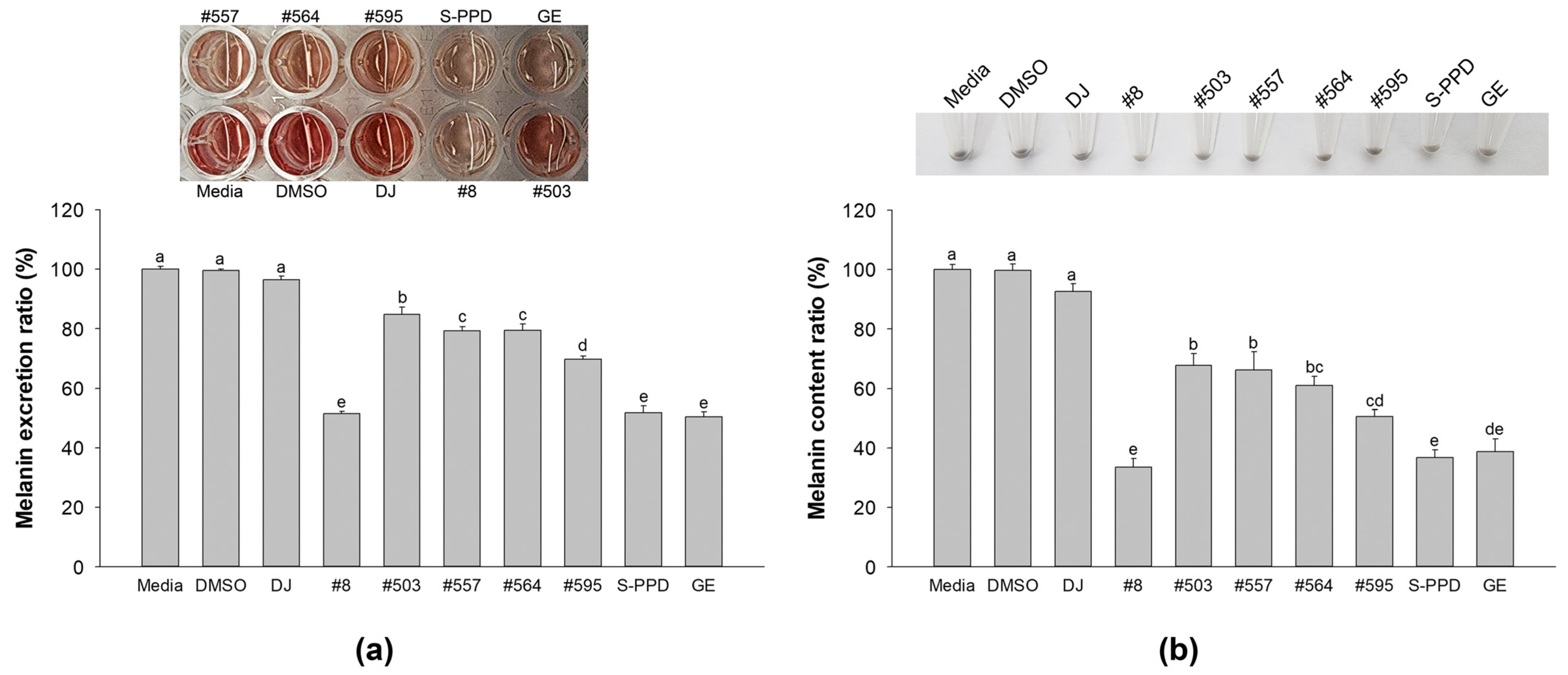
3.3. Effects of Transgenic Rice Seed Extracts on Melanin Content and Melanin Excretion
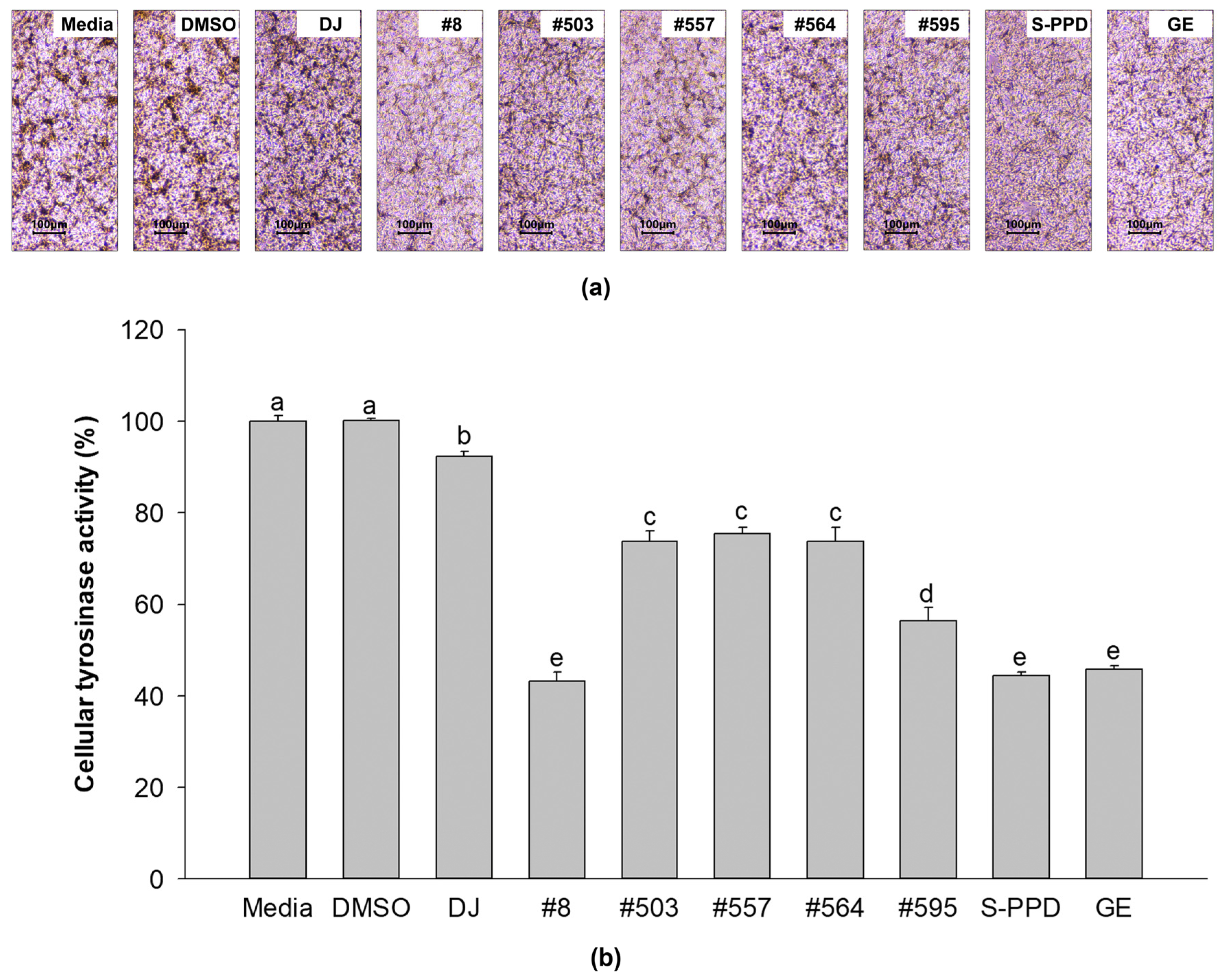
3.4. Effects of Transgenic Rice Seed Extracts on the Cellular Tyrosinase Activity
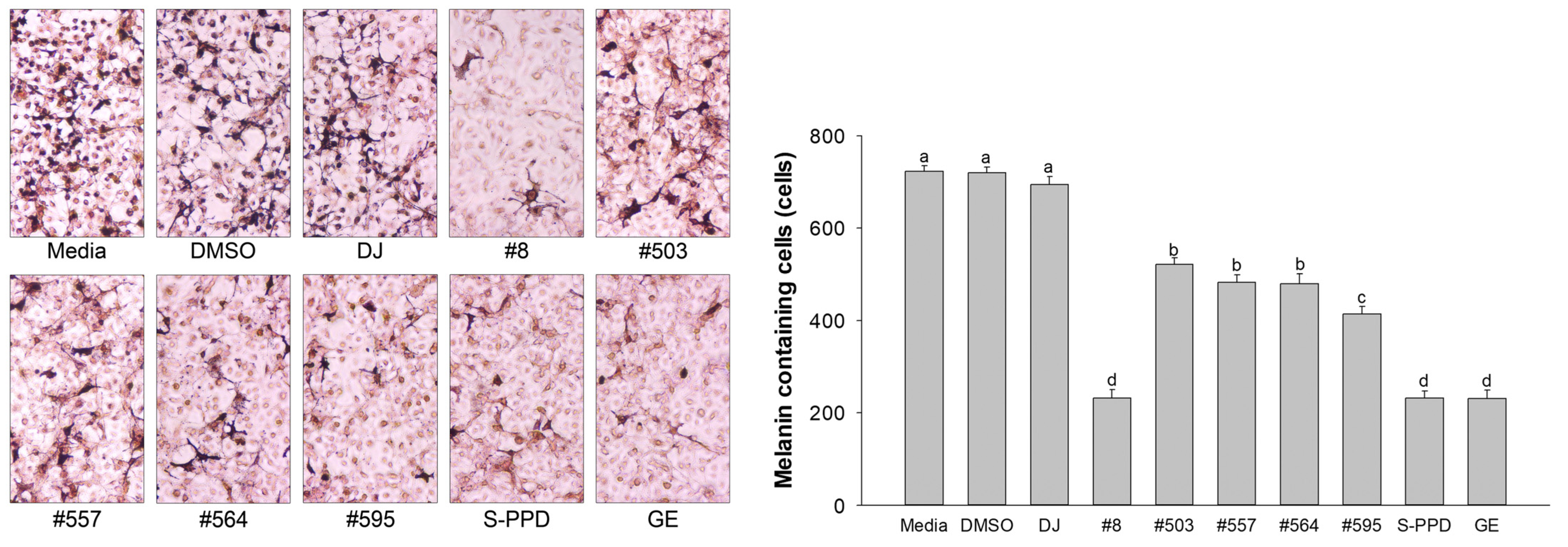
3.5. Effects of Transgenic Rice Seed Extracts on Melanin-Containing Melan-a Cells
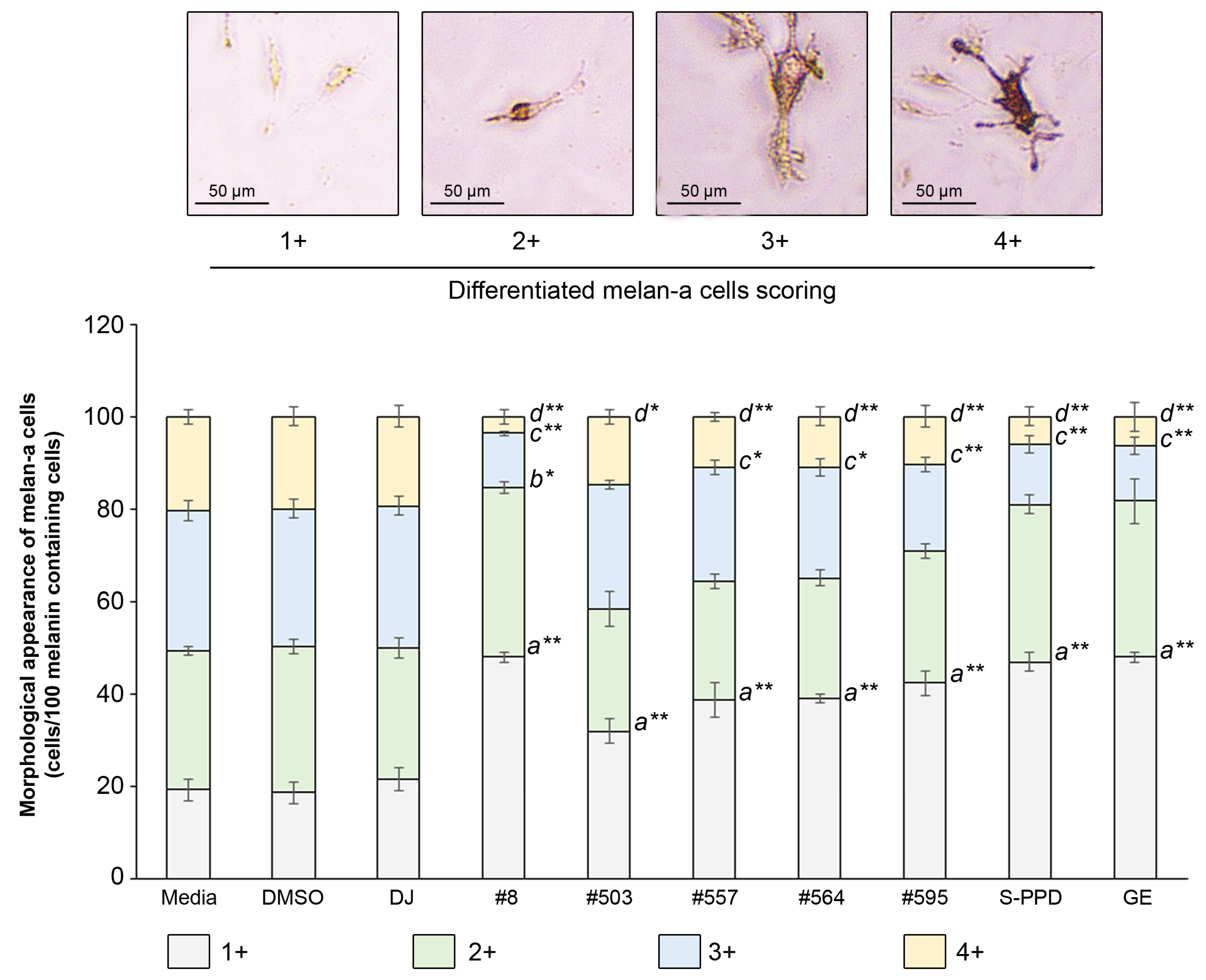
3.6. Effects of Transgenic Rice Seed Extracts on the Morphological Appearance of Melan-a Cells
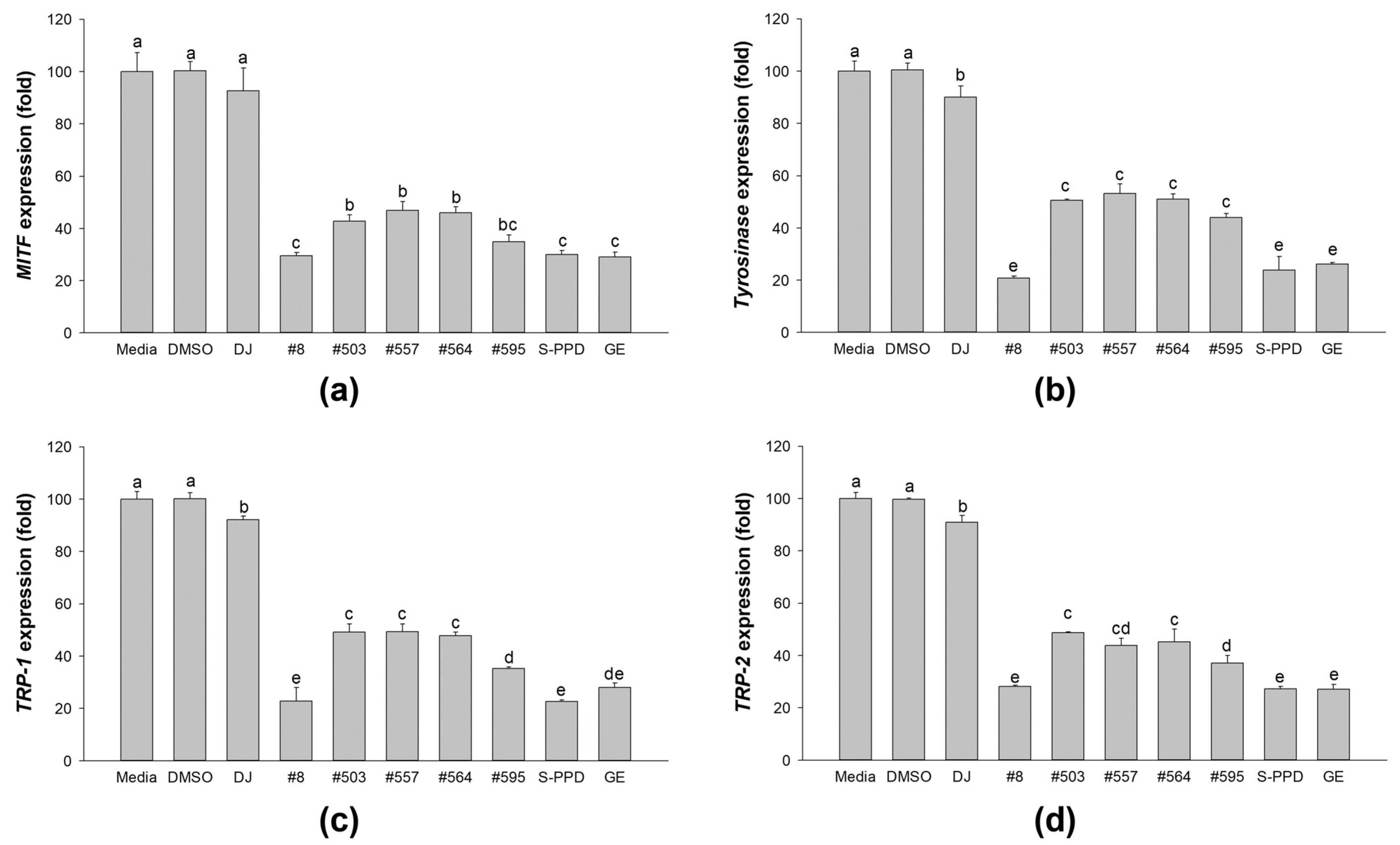
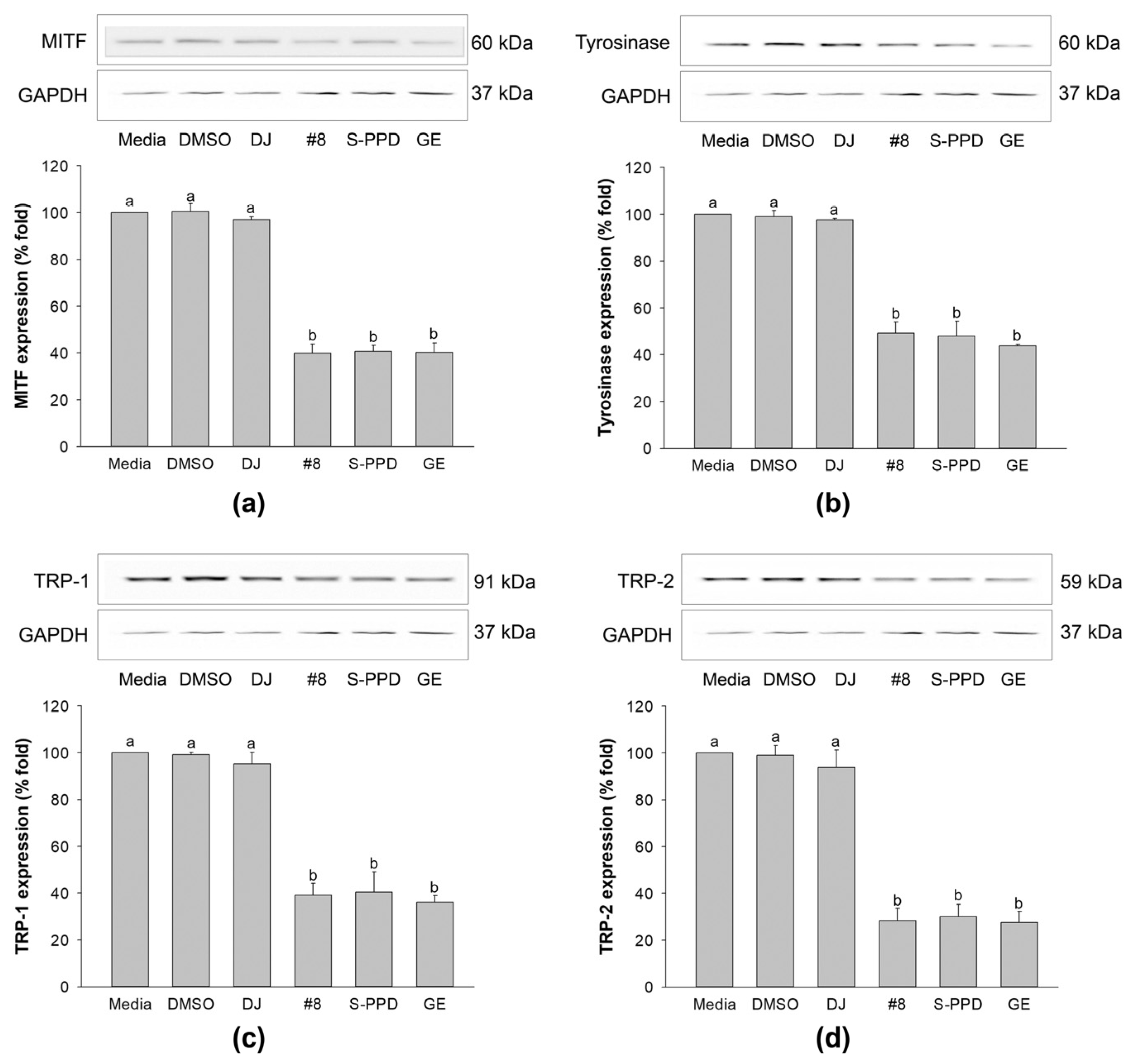
3.7. Effects of Transgenic Rice Seed Extracts on Melanogenic-Related Gene Expression
3.8. Effects of Transgenic Rice Seed Extracts on Melanogenesis-Related Proteins
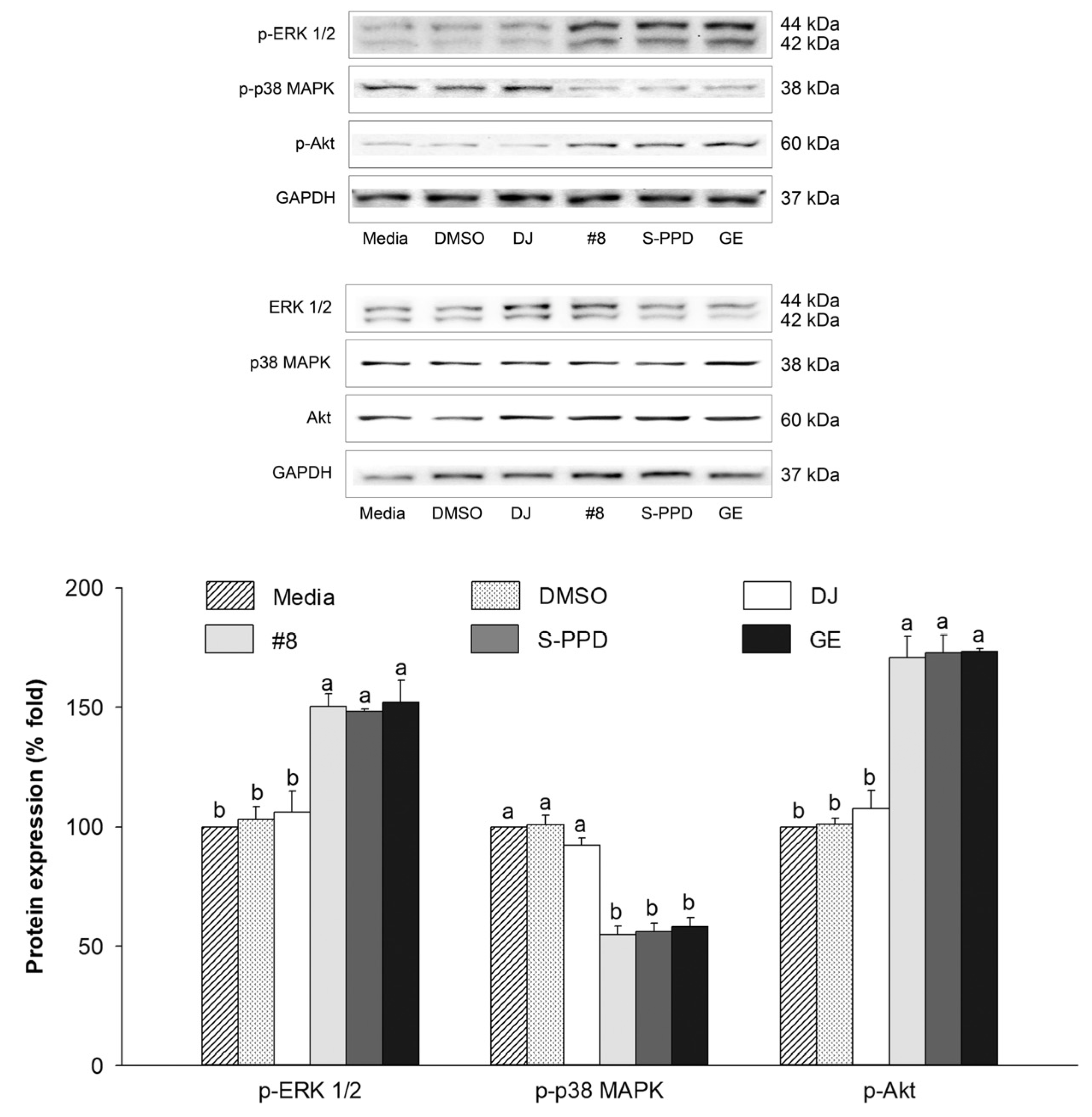
3.9. Effects of Transgenic Rice Seed Extracts on the MAPKs (ERK 1/2 and p38) and PI3K/Akt Signaling Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Swann, G. The skin is the body’s largest organ. J. Vis. Commun. Med. 2010, 33, 148–149. [Google Scholar] [CrossRef]
- You, L.; Cho, J.Y. The regulatory role of Korean ginseng in skin cells. J. Ginseng. Res. 2021, 45, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Gould, J. Superpowered skin. Nature 2018, 563, S84. [Google Scholar] [CrossRef] [Green Version]
- Ortonne, J.P. Normal and abnormal skin color. Ann. Dermatol. Venereol. 2012, 139, S125–S129. [Google Scholar] [CrossRef] [PubMed]
- D’Mello, S.A.N.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling pathways in melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathee, P.; Kumar, S.; Kumar, D.; Kumari, B.; Yadav, S.S. Skin hyperpigmentation and its treatment with herbs: An alternative method. Futur. J. Pharm. Sci. 2021, 7, 132. [Google Scholar] [CrossRef]
- Sotiropoulou, G.; Zingkou, E.; Pampalakis, G. Redirecting drug repositioning to discover innovative cosmeceuticals. Exp. Dermatol. 2021, 30, 628–644. [Google Scholar] [CrossRef]
- Gillbro, J.M.; Olsson, M.J. The melanogenesis and mechanisms of skin-lightening agents—Existing and new approaches. Int. J. Cosmet. Sci. 2011, 33, 210–221. [Google Scholar] [CrossRef]
- Gonzalez, N.; Perez, M. Natural cosmeceutical ingredients for hyperpigmentation. J. Drugs Dermatol. 2016, 15, 26–34. [Google Scholar]
- Kim, K. Effect of ginseng and ginsenosides on melanogenesis and their mechanism of action. J. Ginseng. Res. 2015, 39, 1–6. [Google Scholar] [CrossRef]
- Kang, K.S.; Yokozawa, T.; Kim, H.Y.; Park, J.H. Study on the nitric oxide scavenging effects of ginseng and its compounds. J. Agric. Food Chem. 2006, 54, 2558–2562. [Google Scholar] [CrossRef]
- Shin, J.Y.; Song, J.Y.; Yun, Y.S.; Yang, H.O.; Rhee, D.K.; Pyo, S. Immunostimulating effects of acidic polysaccharides extract of Panax ginseng on macrophage function. Immunopharmacol. Immunotoxicol. 2002, 24, 469–482. [Google Scholar] [CrossRef]
- Du, X.F.; Jiang, C.Z.; Wu, C.F.; Won, E.K.; Choung, S.Y. Synergistic immunostimulating activity of pidotimod and red ginseng acidic polysaccharide against cyclophosphamide-induced immunosuppression. Arch. Pharm. Res. 2008, 31, 1153–1159. [Google Scholar] [CrossRef]
- Xu, L.L.; Han, T.; Wu, J.Z.; Zhang, Q.Y.; Zhang, H.; Huang, B.K.; Rahman, K.; Qin, L.P. Comparative research of chemical constituents, antifungal and antitumor properties of ether extracts of Panax ginseng and its endophytic fungus. Phytomedicine 2009, 16, 609–616. [Google Scholar] [CrossRef]
- Radad, K.; Gille, G.; Liu, L.; Rausch, W.D. Use of ginseng in medicine with emphasis on neurodegenerative disorders. J. Pharmacol. Sci. 2006, 100, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Naval, M.V.; Gómez-Serranillos, M.P.; Carretero, M.E.; Villar, A.M. Neuroprotective effect of a ginseng (Panax ginseng) root extract on astrocytes primary culture. J. Ethnopharmacol. 2007, 112, 262–270. [Google Scholar] [CrossRef]
- Atri, C.; Guerfali, F.Z.; Laouini, D. Role of human macrophage polarization in inflammation during infectious diseases. Int. J. Mol. Sci. 2018, 19, 1801. [Google Scholar] [CrossRef] [Green Version]
- Kang, K.B.; Jayakodi, M.; Lee, Y.S.; Nguyen, V.B.; Park, H.-S.; Koo, H.J.; Choi, I.Y.; Kim, D.H.; Chung, Y.J.; Ryu, B.; et al. Identification of candidate UDP-glycosyltransferases involved in protopanaxadiol-type ginsenoside biosynthesis in Panax ginseng. Sci. Rep. 2018, 8, 11744. [Google Scholar] [CrossRef] [Green Version]
- Siddiqi, M.Z.; Ximenes, H.A.; Song, B.-K.; Park, H.Y.; Lee, W.H.; Han, H.; Im, W.-T. Enhanced production of ginsenoside Rh2(S) from PPD-type major ginsenosides using BglSk cloned from Saccharibacillus kuerlensis together with two glycosidase in series. Saudi J. Biol. Sci. 2021, 28, 4668–4676. [Google Scholar] [CrossRef]
- Wang, C.Z.; Zhang, Z.; Wan, J.Y.; Zhang, C.F.; Anderson, S.; He, X.; Yu, C.; He, T.C.; Qi, L.W.; Yuan, C.S. Protopanaxadiol, an active ginseng metabolite, significantly enhances the effects of fluorouracil on colon cancer. Nutrients 2015, 7, 799–814. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, H.; Sung, J.H.; Matsumiya, S.; Uchiyama, M. Main ginseng saponin metabolites formed by intestinal bacteria. Planta Med. 1996, 62, 453–457. [Google Scholar] [CrossRef]
- Bae, E.A.; Park, S.Y.; Kim, D.H. Constitutive beta-glucosidases hydrolyzing ginsenoside Rb1 and Rb2 from human intestinal bacteria. Biol. Pharm. Bull. 2000, 23, 1481–1485. [Google Scholar] [CrossRef]
- Oh, S.J.; Oh, Y.; Ryu, I.W.; Kim, K.; Lim, C.J. Protective properties of ginsenoside Rb3 against UV-B radiation-induced oxidative stress in HaCaT keratinocytes. Biosci. Biotechnol. Biochem. 2016, 80, 95–103. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.Y.; Park, K.G.; Namgoong, S.; Han, S.K.; Jeong, S.H.; Dhong, E.S.; Kim, W.K. Effects of Panax ginseng extract on human dermal fibroblast proliferation and collagen synthesis. Int. Wound J. 2016, 13 (Suppl. S1), 42–46. [Google Scholar] [CrossRef]
- Hwang, Y.P.; Choi, J.H.; Kim, H.G.; Choi, J.M.; Hwang, S.K.; Chung, Y.C.; Jeong, H.G. Cultivated ginseng suppresses ultraviolet B-induced collagenase activation via mitogen-activated protein kinases and nuclear factor κB/activator protein-1-dependent signaling in human dermal fibroblasts. Nutr. Res. 2012, 32, 428–438. [Google Scholar] [CrossRef]
- Wang, L.; Lu, A.P.; Yu, Z.L.; Wong, R.N.; Bian, Z.X.; Kwok, H.H.; Yue, P.Y.; Zhou, L.M.; Chen, H.; Xu, M.; et al. The melanogenesis-inhibitory effect and the percutaneous formulation of ginsenoside Rb1. AAPS PharmSciTech 2014, 15, 1252–1262. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.Y.; Jeong, Y.T.; Jeong, S.C.; Lee, M.K.; Min, J.W.; Lee, J.W.; Kim, G.S.; Lee, S.E.; Ahn, Y.S.; Kang, H.C.; et al. Melanin biosynthesis inhibition effects of ginsenoside Rb2 isolated from Panax ginseng berry. J. Microbiol. Biotechnol. 2015, 25, 2011–2015. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, W.J.; Chang, S.E.; Lee, G.Y. Antimelanogenic effect of ginsenoside Rg3 through extracellular signal-regulated kinase-mediated inhibition of microphthalmia-associated transcription factor. J. Ginseng. Res. 2015, 39, 238–242. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.E.; Park, J.I.; Myung, C.H.; Hwang, J.S. Inhibitory effects of ginsenosides on basic fibroblast growth factor-induced melanocyte proliferation. J. Ginseng. Res. 2017, 41, 268–276. [Google Scholar] [CrossRef] [Green Version]
- Han, J.Y.; Baek, S.H.; Jo, H.J.; Yun, D.W.; Choi, Y.E. Genetically modified rice produces ginsenoside aglycone (protopanaxadiol). Planta 2019, 250, 1103–1110. [Google Scholar] [CrossRef]
- Monmai, C.; Kim, J.-S.; Baek, S.-H. Transgenic rice seed extracts exert immunomodulatory effects by modulating immune-related biomarkers in RAW264.7 macrophage cells. Nutrients 2022, 14, 4143. [Google Scholar] [CrossRef]
- Lee, Y.; Kantayos, V.; Kim, J.S.; Rha, E.S.; Son, Y.J.; Baek, S.H. Inhibitory effects of protopanaxadiol-producing transgenic rice seed extracts on RANKL-induced osteoclast differentiation. Life 2022, 12, 1886. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Park, J.-Y.; Song, M.W.; Kim, K.-T.; Paik, H.-D. Improved antioxidative, anti-inflammatory, and antimelanogenic effects of fermented hydroponic ginseng with Bacillus strains. Antioxidants 2022, 11, 1848. [Google Scholar] [CrossRef]
- Lee, M.-H.; Lin, Y.-P.; Hsu, F.-L.; Zhan, G.-R.; Yen, K.-Y. Bioactive constituents of Spatholobus suberectus in regulating tyrosinase-related proteins and mRNA in HEMn cells. Phytochemistry 2006, 67, 1262–1270. [Google Scholar] [CrossRef]
- Rodboon, T.; Okada, S.; Suwannalert, P. Germinated riceberry rice enhanced protocatechuic acid and vanillic acid to suppress melanogenesis through cellular oxidant-related tyrosinase activity in B16 cells. Antioxidants 2020, 9, 247. [Google Scholar] [CrossRef] [Green Version]
- Al-Okbi, S.Y. Nutraceuticals of anti-inflammatory activity as complementary therapy for rheumatoid arthritis. Toxicol. Ind. Health 2014, 30, 738–749. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Lyu, J.-L.; Kuo, Y.-H.; Chiu, C.-Y.; Wen, K.-C.; Chiang, H.-M. The anti-melanogenesis effect of 3,4-dihydroxybenzalacetone through downregulation of melanosome maturation and transportation in B16F10 and human epidermal melanocytes. Int. J. Mol. Sci. 2021, 22, 2823. [Google Scholar] [CrossRef]
- Deng, J.; Liu, Y.; Duan, Z.; Zhu, C.; Hui, J.; Mi, Y.; Ma, P.; Ma, X.; Fan, D.; Yang, H. Protopanaxadiol and protopanaxatriol-type saponins ameliorate glucose and lipid metabolism in type 2 diabetes mellitus in high-fat diet/streptozocin-induced mice. Front. Pharmacol. 2017, 8, 506–516. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.; Sorn, S.; Baek, S.; Jang, S.; Kim, S. Antioxidant and apoptotic effects of Korean white ginseng extracted with the same ratio of protopanaxadiol and protopanaxatriol saponins in human hepatoma HepG2 cells. Ann. N Y Acad. Sci. 2009, 1171, 217–227. [Google Scholar] [CrossRef]
- Simon, J.D.; Peles, D.; Wakamatsu, K.; Ito, S. Current challenges in understanding melanogenesis: Bridging chemistry, biological control, morphology, and function. Pigment. Cell Melanoma Res. 2009, 22, 563–579. [Google Scholar] [CrossRef] [PubMed]
- Cichorek, M.; Wachulska, M.; Stasiewicz, A.; Tymińska, A. Skin melanocytes: Biology and development. Postepy Dermatol. Alergol. 2013, 30, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Hirobe, T. How are proliferation and differentiation of melanocytes regulated? Pigment. Cell Melanoma Res. 2011, 24, 462–478. [Google Scholar] [CrossRef]
- Chung, Y.C.; Kim, Y.B.; Kim, B.S.; Hyun, C.-G. Anti-melanogenic effects of bergamottin via mitogen-activated protein kinases and protein kinase B signaling pathways. Nat. Prod. Commun. 2019, 14. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Chu, J.H.; Wang, H.; Xu, H.; Chou, G.X.; Leung, A.K.; Fong, W.F.; Yu, Z.L. Involvement of p38 MAPK signaling pathway in the anti-melanogenic effect of San-bai-tang, a Chinese herbal formula, in B16 cells. J. Ethnopharmacol. 2010, 132, 533–535. [Google Scholar] [CrossRef]
- Jang, J.Y.; Lee, J.H.; Kang, B.W.; Chung, K.T.; Choi, Y.H.; Choi, B.T. Dichloromethane fraction of Cimicifuga heracleifolia decreases the level of melanin synthesis by activating the ERK or AKT signaling pathway in B16F10 cells. Exp. Dermatol. 2009, 18, 232–237. [Google Scholar] [CrossRef]
- Jin, M.L.; Park, S.Y.; Kim, Y.H.; Park, G.; Son, H.J.; Lee, S.J. Suppression of α-MSH and IBMX-induced melanogenesis by cordycepin via inhibition of CREB and MITF, and activation of PI3K/Akt and ERK-dependent mechanisms. Int. J. Mol. Med. 2012, 29, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Akaberi, M.; Emami, S.A.; Vatani, M.; Tayarani-Najaran, Z. Evaluation of antioxidant and anti-melanogenic activity of different extracts of Aerial Parts of N. Sintenisii in murine melanoma B16F10 cells. Iran. J. Pharm. Res. 2018, 17, 225–235. [Google Scholar] [PubMed]
- Baek, S.H.; Lee, S.H. Sesamol decreases melanin biosynthesis in melanocyte cells and zebrafish: Possible involvement of MITF via the intracellular cAMP and p38/JNK signalling pathways. Exp. Dermatol. 2015, 24, 761–766. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.K.; Sarkar, C.; Mallick, S.; Saha, B.; Bera, R.; Bhadra, R. Human placental lipid induces melanogenesis through p38 MAPK in B16F10 mouse melanoma. Pigment. Cell Res. 2005, 18, 113–121. [Google Scholar] [CrossRef]
- Alam, M.B.; Bajpai, V.K.; Lee, J.; Zhao, P.; Byeon, J.H.; Ra, J.S.; Majumder, R.; Lee, J.S.; Yoon, J.I.; Rather, I.A.; et al. Inhibition of melanogenesis by jineol from Scolopendra subspinipes mutilans via MAP-Kinase mediated MITF downregulation and the proteasomal degradation of tyrosinase. Sci. Rep. 2017, 7, 45858. [Google Scholar] [CrossRef] [PubMed]

| Gene | Accession Number | Sequence (5′-3′) | Target Size (bp) |
|---|---|---|---|
| MITF | NM_001113198.2 | Forward primer: AGCGTGTATTTTCCCCACAG | 239 |
| Reverse primer: CCTTAGCTCGTTGCTGTTCC | |||
| TRP-1 | NM_031202.3 | Forward primer: TCTGGCCTCCAGTTACCAAC | 223 |
| Reverse primer: TCAGTGAGGAGAGGCTGGTT | |||
| TRP-2 | X63349.1 | Forward primer: ACCCTGTGTTTGTGGTCCTC | 186 |
| Reverse primer: GTTGCTCTGCGGTTAGGAAG | |||
| Tyrosinase | D00131.1 | Forward primer: CCAGAAGCCAATGCACCTAT | 193 |
| Reverse primer: CCAGATACGACTGGCCTTGT | |||
| GAPDH | NM_001289726.2 | Forward primer: AACTTTGGCATTGTGGAAGG | 223 |
| Reverse primer: ACACATTGGGGGTAGGAACA |
| Treatment | Concentration (mg/mL) | Antioxidant Activity | IC50 (mg/mL) | |
|---|---|---|---|---|
| ABTS Radical Scavenging (%) | Vitamin C Equivalent Antioxidant Capacity (VCEAC: mg/g Dry Weight) | |||
| DMSO | 0.1% | 0.08 ± 0.11 | − | − |
| DJ | 10 | 8.09 ± 1.20 d | 0.005 ± 0.003 d | Not in the range (<50%) |
| 25 | 11.86 ± 0.65 d | 0.015 ± 0.002 d | ||
| 50 | 23.81 ± 0.98 d | 0.048 ± 0.003 d | ||
| 100 | 39.83 ± 1.63 d | 0.093 ± 0.005 d | ||
| #8 | 10 | 25.81 ± 0.98 a | 0.054 ± 0.003 a | 37.90 ± 0.30 |
| 25 | 42.53 ± 1.09 a | 0.100 ± 0.003 a | ||
| 50 | 60.94 ± 0.76 a | 0.151 ± 0.002 a | ||
| 100 | 96.46 ± 1.31 a | 0.249 ± 0.004 a | ||
| #503 | 10 | 15.02 ± 3.16 c | 0.024 ± 0.009 c | 64.27 ± 0.15 |
| 25 | 25.35 ± 0.98 c | 0.052 ± 0.003 c | ||
| 50 | 42.91 ± 1.20 c | 0.101 ± 0.003 c | ||
| 100 | 71.49 ± 1.31 c | 0.180 ± 0.004 c | ||
| #557 | 10 | 16.10 ± 1.42 bc | 0.027 ± 0.004 bc | 60.41 ± 2.17 |
| 25 | 26.96 ± 1.09 c | 0.057 ± 0.003 c | ||
| 50 | 44.99 ± 1.31 c | 0.107 ± 0.004 c | ||
| 100 | 74.96 ± 1.63 c | 0.190 ± 0.005 c | ||
| #564 | 10 | 16.02 ± 0.87 bc | 0.027 ± 0.002 bc | 60.50 ± 1.22 |
| 25 | 27.27 ± 1.31 c | 0.058 ± 0.004 c | ||
| 50 | 44.14 ± 0.33 c | 0.104 ± 0.001 c | ||
| 100 | 75.27 ± 2.51 bc | 0.191 ± 0.007 bc | ||
| #595 | 10 | 21.57 ± 1.74 ab | 0.042 ± 0.005 ab | 50.73 ± 0.72 |
| 25 | 33.05 ± 2.07 b | 0.074 ± 0.006 b | ||
| 50 | 51.23 ± 0.98 b | 0.124 ± 0.003 b | ||
| 100 | 82.13 ± 2.83 b | 0.210 ± 0.008 b | ||
| S-PPD | 70 ng/mL | 21.73 ± 0.87 ab | 0.042 ± 0.002 ab | 275.82 ± 5.35 ng/mL |
| 175 ng/mL | 42.60 ± 2.07 a | 0.100 ± 0.006 a | ||
| 350 ng/mL | 62.87 ± 0.87 a | 0.156 ± 0.002 a | ||
| 700 ng/mL | 94.61 ± 0.65 a | 0.244 ± 0.002 a | ||
| GE | 10 mg/mL | 26.43 ± 0.33 a | 0.055 ± 0.001 a | 38.26 ± 0.63 |
| 25 mg/mL | 42.06 ± 0.44 a | 0.099 ± 0.001 a | ||
| 50 mg/mL | 59.40 ± 1.20 a | 0.147 ± 0.003 a | ||
| 100 mg/mL | 94.07 ± 1.42 a | 0.243 ± 0.004 a | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monmai, C.; Kim, J.-S.; Promyot, K.; Baek, S.-H. Protopanaxadiol-Enriched Rice Extracts Suppressed Oxidative and Melanogenic Activities in Melan-a Cells. Antioxidants 2023, 12, 166. https://doi.org/10.3390/antiox12010166
Monmai C, Kim J-S, Promyot K, Baek S-H. Protopanaxadiol-Enriched Rice Extracts Suppressed Oxidative and Melanogenic Activities in Melan-a Cells. Antioxidants. 2023; 12(1):166. https://doi.org/10.3390/antiox12010166
Chicago/Turabian StyleMonmai, Chaiwat, Jin-Suk Kim, Karantharat Promyot, and So-Hyeon Baek. 2023. "Protopanaxadiol-Enriched Rice Extracts Suppressed Oxidative and Melanogenic Activities in Melan-a Cells" Antioxidants 12, no. 1: 166. https://doi.org/10.3390/antiox12010166